Ultra-Fast Low Concentration Detection of Candida Pathogens Utilizing High Resolution Micropore Chips
Abstract
:1. Introduction
2. Methods and Materials
2.1. Micropore Fabrication and Flow Cell
2.2. Cell Preparation
2.3. Micropore Characterization
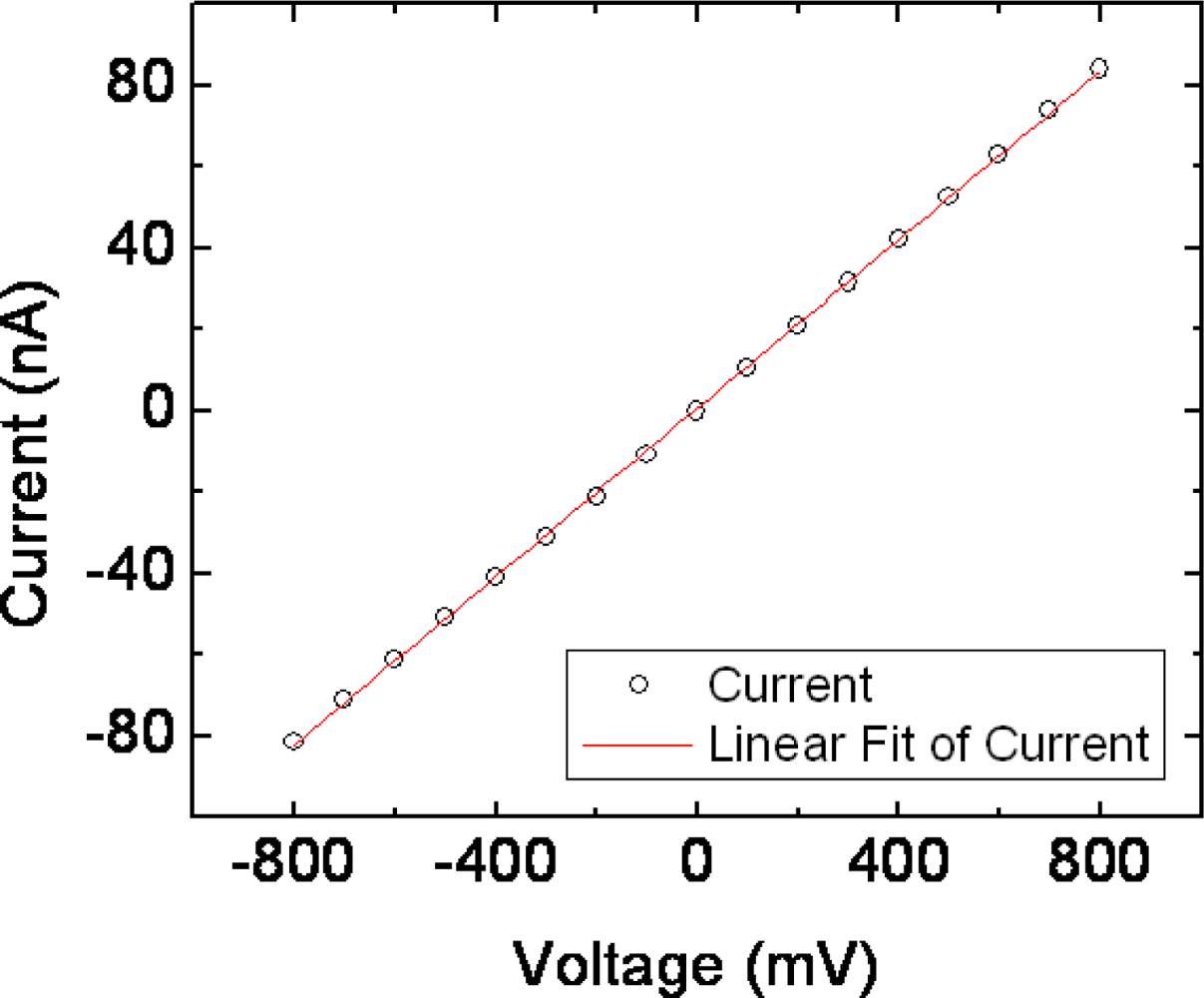
3. Results and Discussion
4. Conclusions
Acknowledgments
References and Notes
- Wisplinghoff, H.; Bischoff, T.; Tallent, S.M.; Seifert, H.; Wenzel, R.P.; Edmond, M.B. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis 2004, 39, 309–317. [Google Scholar]
- Pappas, P.G.; Rex, J.H.; Sobel, J.D.; Filler, S.G.; Dismukes, W.E.; Walsh, T.J.; Edwards, J.E. Infectious Diseases Society of America.Guidelines for treatment of candidiasis. Clin. Infect. Dis 2004, 38, 161–189. [Google Scholar]
- George, B.J.; Horvath, L.L.; Hospenthal, D.R. Effect of inoculum size on detection of Candida growth by the BACTEC 9240 automated blood culture system using aerobic and anaerobic media. J. Clin. Microbiol 2005, 43, 433–435. [Google Scholar]
- Horvath, L.L.; George, B.J.; Hospenthal, D.R. Detection of fifteen species of Candida in an automated blood culture system. J. Clin. Microbiol 2007, 45, 3062–3064. [Google Scholar]
- Horvath, L.L.; George, B.J.; Murray, C.K.; Harrison, L.S.; Hospenthal, D.R. Direct comparison of the BACTEC 9240 and BacT/ALERT 3D automated blood culture systems for candida growth detection. J. Clin. Microbiol 2005, 42, 115–118. [Google Scholar]
- McMullan, R.; Metwally, L.; Coyle, P.V.; Hedderwick, S.; McCloskey, B.; O'Neill, H.J.; Patterson, C.C.; Thompson, G.; Webb, C.H.; Hay, R.J. A prospective clinical trial of a real-time polymerase chain reaction assay for the diagnosis of candidemia in nonneutropenic, critically ill adults. Clin. Infect. Dis 2008, 46, 890–896. [Google Scholar]
- Coulter, W. High speed automatic blood cell counter and cell size analyzer. Proc. Natl. Elect. Conf 1956, 12, 1034–1040. [Google Scholar]
- DeBloise, R.; Bean, C.P. Counting and sizing of submicron particles by the resistive pulse technique. Rev. Sci. Instr 1970, 41, 909–916. [Google Scholar]
- Carbonaro, A.; Sohn, L.L. A resistive-pulse sensor chip for multianalyte immunoassays. Lab Chip 2005, 5, 1155–1160. [Google Scholar]
- Zhang, Z.; Zhe, J.; Chandra, S.; Hu, J. An electronic pollen detection method using Coulter counting principle. Atmos. Environ 2005, 39, 5446–5453. [Google Scholar]
- Maissel, L.; Glang, R. Handbook of Thin Film Technology; McGraw-Hill: New York, U.S.A., 1970. [Google Scholar]
- Smith, D.L. Thin-Film Deposition: Principles and Practice; McGraw-Hill Professional: New York, U.S.A., 1994. [Google Scholar]
- Kim, M.J.; McNally, B.; Murata, K.; Meller, A. Characterisitics of solid-state nanometer pores fabricated using transmission electron microscope (TEM). Nanotechnology 2007, 18, 205302–205306. [Google Scholar]
- Campbell, S.A. The Science and Engineering of Microelectronic Fabrication; Oxford University Press: Oxford, U.K., 1996. [Google Scholar]
- Madou, M. Fundamentals of Microfabrication, 2nd Edition ed; CRC Press: New York, U.S.A., 1997. [Google Scholar]
- Wonderlin, W.F. Optimizing planar lipid bilayer single-channel recordings for high resolution with rapid voltage steps. Biophys. J 1990, 58, 289–297. [Google Scholar]
- Wanunu, M.; Meller, A. Chemically modified solid-state nanopores. Nano Lett 2007, 7, 1580–1585. [Google Scholar]






© 2009 by the authors; licensee MDPI, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mulero, R.; Lee, D.H.; Kutzler, M.A.; Jacobson, J.M.; Kim, M.J. Ultra-Fast Low Concentration Detection of Candida Pathogens Utilizing High Resolution Micropore Chips. Sensors 2009, 9, 1590-1598. https://doi.org/10.3390/s90301590
Mulero R, Lee DH, Kutzler MA, Jacobson JM, Kim MJ. Ultra-Fast Low Concentration Detection of Candida Pathogens Utilizing High Resolution Micropore Chips. Sensors. 2009; 9(3):1590-1598. https://doi.org/10.3390/s90301590
Chicago/Turabian StyleMulero, Rafael, Dong Heun Lee, Michele A. Kutzler, Jeffrey M. Jacobson, and Min Jun Kim. 2009. "Ultra-Fast Low Concentration Detection of Candida Pathogens Utilizing High Resolution Micropore Chips" Sensors 9, no. 3: 1590-1598. https://doi.org/10.3390/s90301590
APA StyleMulero, R., Lee, D. H., Kutzler, M. A., Jacobson, J. M., & Kim, M. J. (2009). Ultra-Fast Low Concentration Detection of Candida Pathogens Utilizing High Resolution Micropore Chips. Sensors, 9(3), 1590-1598. https://doi.org/10.3390/s90301590



