Improvement of Bioactive Compound Classification through Integration of Orthogonal Cell-Based Biosensing Methods
Abstract
:1. Introduction
2. Results and Discussion
2.1 Algal responses to model compounds
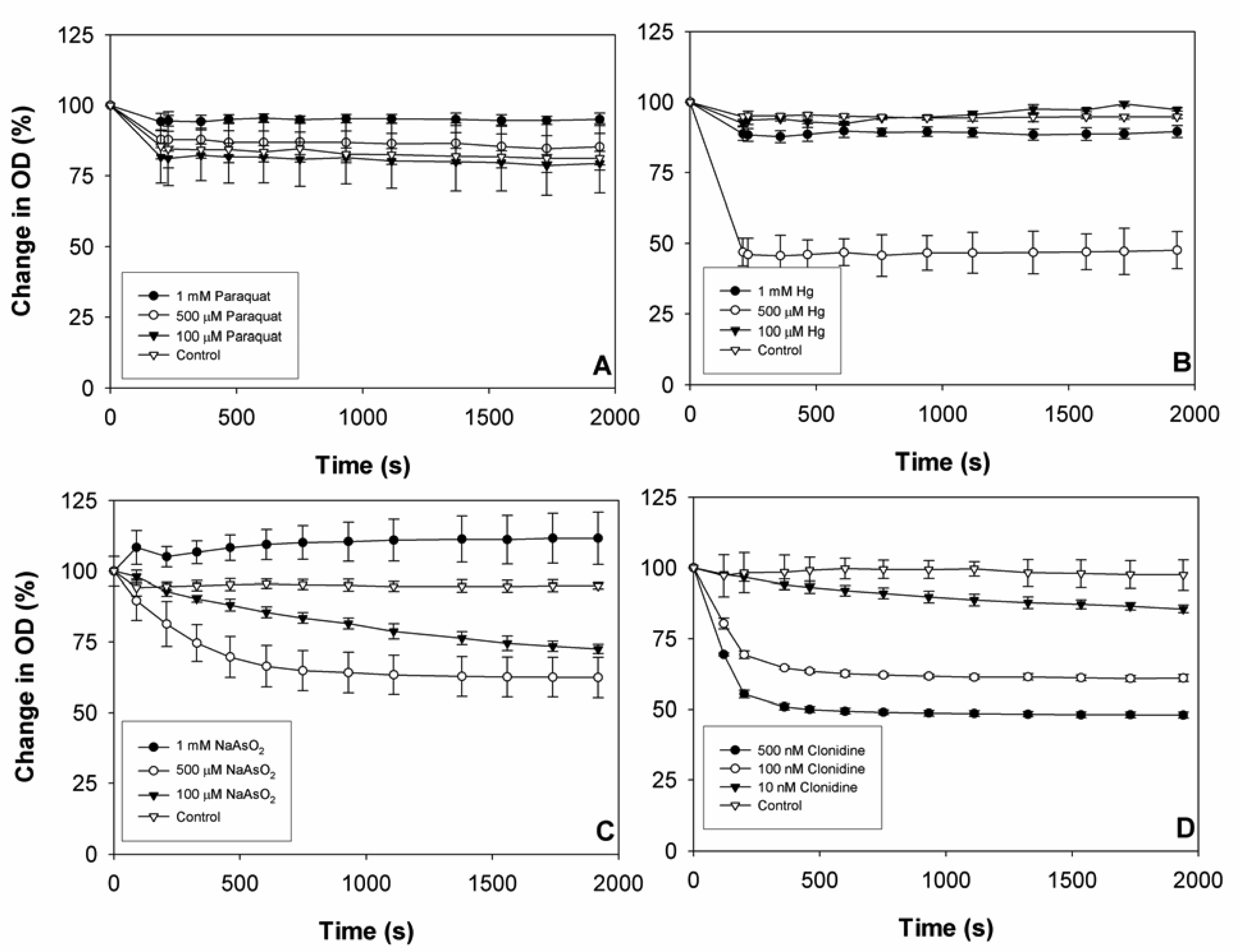
2.2 Chromatophore response to model compounds
2.3 Classification of algal and chromatophore system responses
3. Conclusions
4. Experimental Section
4.1 Chemicals and Solutions
4.2 Cell Culture
4.3 Bioactive Compound Preparation
4.4 Algal Cell Fluorescence Measurements
4.5 Chromatophore Optical Density Measurements
4.6 Data Analysis
Acknowledgments
References and Notes
- Karlsson, J.; Andersson, R.G.; Askelof, P.; Elwing, H.; Granstrom, M.; Grundtrom, N.; Lunstrom, I.; Ohman, L. The melanophore aggregating response of isolated fish scales: a very rapid and sensitive diagnosis of whooping cough. FEMS Microbiology Letters 1991, 82, 169–176. [Google Scholar]
- Elwing, H.; Karlsson, J.O.; Grundstrom, N.; Gustafsson, A.L.; von Schenck, H.; Sundgren, H.; Odman, S; Andersson, R.G.; Lunstrom, I. Fish scales as biosensors for catecholamines. Biosensors & Bioelectronics 1990, 5, 449–459. [Google Scholar]
- Chaplen, F.W.R.; Upson, R.J.; McFadden, P.N.; Kolodziej, W.J. Fish chromatophores as cytosensors in a microscale device: Detection of environmental toxins and bacterial pathogens. Pigment Cell Res. 2002, 15, 19–26. [Google Scholar]
- Altamirano, A.; Garcia-Villada, L.; Agrelo, M.; Sanchez-Martin, L.; Martin-Otero, L.; Flores-Moya, A.; Rico, M.; Lopez-Rodas, V.; Costas, E. A novel approach to improve specificity of algal biosensors using wild-type and resistant mutants: an application to detect TNT. Biosensors & Bioelectronics 2004, 19, 1319–1323. [Google Scholar]
- Rizzuto, M.; Polcaro, C.; Desiderio, C.; Koblizek, M.; Pilloton, R.; Giardi, M.T. Herbicide monitoring in surface water samples with a photosystem-II based biosensor, Proceedings of the 2nd workshop on Chemical Sensors and Biosensors; Mazzei, F., Pilloton, R., Eds.; E.N.E.A.: Rome, 2000; pp. 346–357.
- Naessens, M.; Leclerc, J.C.; Tran-Minh, C. Fiber optic biosensor using Chlorella vulgaris for determination of toxic components. Ecotoxicology and Environmental Safety 2000, 46, 181–185. [Google Scholar]
- Rodriguez, M., Jr.; Sanders, C.A.; Greenbaum, E. Biosensors for rapid monitoring of primary-source drinking water using naturally occurring photosynthesis. Biosensors & Bioelectronics 2002, 17. [Google Scholar]
- Bernier, M; Popovic, R; Carpentier, R. Mercury inhibition at the donor side of photosystem II is reversed by chloride. FEBS Lett. 1993, 321, 19–23. [Google Scholar]
- Chen, G.; Way, J.; Armour, S.; Watson, C.; Queen, K.; Jayawickreme, C.K.; Chen, C.J.; Kenakin, T. Use of constitutive G-protein coupled receptor activity for drug discovery. Molecular Pharmacology 1999, 57, 125–134. [Google Scholar]
- Tseng, C.H. The potential biological mechanisms of arsenic-induced diabetes mellitus. Toxicol. Appl. Pharmacol. 2004, 197(2), 67–83. [Google Scholar]
- Shobha Rani, A; Sudharsan, R.; Reddy, T.N.; Reddy, P.U.M.; Raju, T.N. Effect of arsenite on certain aspects of protein metabolism in fresh water teleost, Tilapia mossambica. J. Environ. Biol. 2001, 22(2), 101–104. [Google Scholar]
- Wong, P.K. Effects of 2,4-D, glyphosate and paraquat on growth, photosynthesis and chlorophyll-a synthesis of Scenedesmus quadricauda Berb 614. Chemosphere 2000, 41(1-2), 177–82. [Google Scholar]
- Elstner, E.F.; Osswald, W. Chlorophyll photobleaching and Ethane production in Dichlorophenyldimethylurea-(DCMU) or Paraquat- treated Euglena gracilis cells. Z. Naturforsch. 1980, 35c, 129–135. [Google Scholar]
- Narayanan, A. Use of Elicitor Sets to Characterize Cellular Signal Transduction Networks. M.S. Thesis, Oregon State University, 2003. [Google Scholar]
- Martinez, A.M.; Kak, A.C. PCA versus LDA. IEEE Transactions on Pattern Analysis and Machine Intelligence 2001, 23(2), 228–233. [Google Scholar]
- Vissvesvaran, G. Classification of Toxins using Orthogonal Sensing Techniques. M.S. Thesis, Oregon State University, 2003. [Google Scholar]



| Case# | Toxin | [X] | a | B | c | a1 | b1 | c1 |
|---|---|---|---|---|---|---|---|---|
| 1 | Paraquat | 100 μM | 43.75 | 0.005 | 57.03 | 19.1 | 0.0061 | 80.44 |
| 2 | Paraquat | 100 μM | 53.92 | 0.0028 | 47.08 | 16.1 | 0.0043 | 82.76 |
| 3 | Paraquat | 100 μM | 59.56 | 0.0027 | 42.56 | 19.2 | 0.004 | 79.49 |
| 4 | NaAsO2 | 1 mM | 50.28 | 0.003 | 47.37 | -8.2 | 0.0008 | 109.54 |
| 5 | NaAsO2 | 1 mM | 57.43 | 0.0024 | 41.5 | -25.15 | 0.0013 | 123.65 |
| 6 | NaAsO2 | 500 μM | 100.68 | 0.0002 | 1.27 | -7.6 | 0.0042 | 106.32 |
| 7 | NaAsO2 | 500 μM | 43.97 | 0.0007 | 57.75 | 46.47 | 0.0023 | 66.17 |
| 8 | NaAsO2 | 100 μM | 15.68 | 0.0008 | 85.6 | 49.28 | 0.0021 | 63.04 |
| 9 | NaAsO2 | 100 μM | 16.35 | 0.0007 | 84.14 | 38.98 | 0.0008 | 65.87 |
| 10 | HgCl2 | 1 mM | 53.53 | 0.011 | 47.74 | 8.9 | 0.008 | 91.14 |
| 11 | HgCl2 | 1 mM | 49.92 | 0.013 | 48.85 | 12.8 | 0.0064 | 97.4 |
| 12 | HgCl2 | 1 mM | 44.54 | 0.0029 | 42.04 | 0.6 | -0.0006 | 86.87 |
| 13 | HgCl2 | 500 μM | 41.43 | 0.0092 | 60.31 | 55.56 | 0.01 | 44.41 |
| 14 | HgCl2 | 500 μM | 34.34 | 0.0014 | 61.1 | 0.066 | -0.0018 | 52.78 |
| 15 | HgCl2 | 500 μM | 55 | 0.0024 | 45.24 | 58.61 | 0.0088 | 41.3 |
| 16 | HgCl2 | 100 μM | 46.4 | 0.0012 | 43.15 | 0.44 | -0.0013 | 92.52 |
| 17 | HgCl2 | 100 μM | 46.87 | 0.0005 | 54.26 | 0.42 | -0.0016 | 92.03 |
| 18 | Clonidine | 500 nM | -14.65 | 0.0006 | 100.06 | 53.26 | 0.0061 | 48.05 |
| 19 | Clonidine | 500 nM | 12.8 | 0.0013 | 88.42 | 52.02 | 0.0055 | 49.18 |
| 20 | Clonidine | 500 nM | 13.07 | 0.0048 | 88.5 | 54.21 | 0.0055 | 47.25 |
| 21 | Clonidine | 100 nM | 13.05 | 0.0016 | 99.71 | 41.81 | 0.0042 | 60.1 |
| 22 | Clonidine | 100 nM | 16.05 | 0.0014 | 87.23 | 41.27 | 0.0047 | 60.46 |
| Actual Type | Group Size | Predicted Type | |||
|---|---|---|---|---|---|
| Paraquat | Sodium Arsenite | Mercury | Clonidine | ||
| Paraquat | 3 | 3 (100.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| Sodium Arsenite | 6 | 0 (0.00%) | 6 (100.00%) | 0 (0.00%) | 0 (0.00%) |
| Mercury | 8 | 1 (12.50%) | 0 (0.00%) | 7 (87.5%) | 0 (0.00%) |
| Clonidine | 5 | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 5 (100.00%) |
| Actual Type | Group Size | Predicted Type | |||
|---|---|---|---|---|---|
| Paraquat | Sodium Arsenite | Mercury | Clonidine | ||
| Paraquat | 3 | 3 (100.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| Sodium Arsenite | 6 | 1 (16.67%) | 5 (83.33%) | 0 (0.00%) | 0 (0.00%) |
| Mercury | 8 | 1 (12.50%) | 1 (12.50%) | 6 (75.00%) | 0 (0.00%) |
| Clonidine | 5 | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 5 (100.00%) |
© 2007 by MDPI ( http://www.mdpi.org). Reproduction is permitted for noncommercial purposes.
Share and Cite
Chaplen, F.W.R.; Vissvesvaran, G.; Henry, E.C.; Jovanovic, G.N. Improvement of Bioactive Compound Classification through Integration of Orthogonal Cell-Based Biosensing Methods. Sensors 2007, 7, 38-51. https://doi.org/10.3390/s7010038
Chaplen FWR, Vissvesvaran G, Henry EC, Jovanovic GN. Improvement of Bioactive Compound Classification through Integration of Orthogonal Cell-Based Biosensing Methods. Sensors. 2007; 7(1):38-51. https://doi.org/10.3390/s7010038
Chicago/Turabian StyleChaplen, Frank W. R., Ganesh Vissvesvaran, Eric C. Henry, and Goran N. Jovanovic. 2007. "Improvement of Bioactive Compound Classification through Integration of Orthogonal Cell-Based Biosensing Methods" Sensors 7, no. 1: 38-51. https://doi.org/10.3390/s7010038
APA StyleChaplen, F. W. R., Vissvesvaran, G., Henry, E. C., & Jovanovic, G. N. (2007). Improvement of Bioactive Compound Classification through Integration of Orthogonal Cell-Based Biosensing Methods. Sensors, 7(1), 38-51. https://doi.org/10.3390/s7010038




