Deep Learning-Based Multi-Lead ECG Reconstruction from Lead I with Metadata Integration and Uncertainty Estimation
Highlights
- A dual-branch deep learning model integrating Lead I signals and patient metadata improved 12-lead ECG reconstruction performance.
- Predictive uncertainty estimation using Monte Carlo dropout reflects waveform reliability.
- Metadata integration enhances the model performance.
- Uncertainty heatmaps provide interpretable reliability information for clinical use.
Abstract
1. Introduction
- Propose a dual-branch deep learning framework that reconstructs standard 12-lead ECGs from a strict single-lead (Lead I) input, demonstrating the feasibility of single-lead adaptation for wearable ECG applications.
- Demonstrate that integrating clinically interpretable metadata significantly improves reconstruction performance, particularly in diagnostically important waveform components such as the QRS complex and T wave.
- Introduce predictive uncertainty estimation into ECG reconstruction using Monte Carlo dropout, enabling reliability-aware interpretation that complements conventional accuracy metrics and enhances clinical trustworthiness.
2. Materials and Methods
2.1. Dataset and Preprocessing
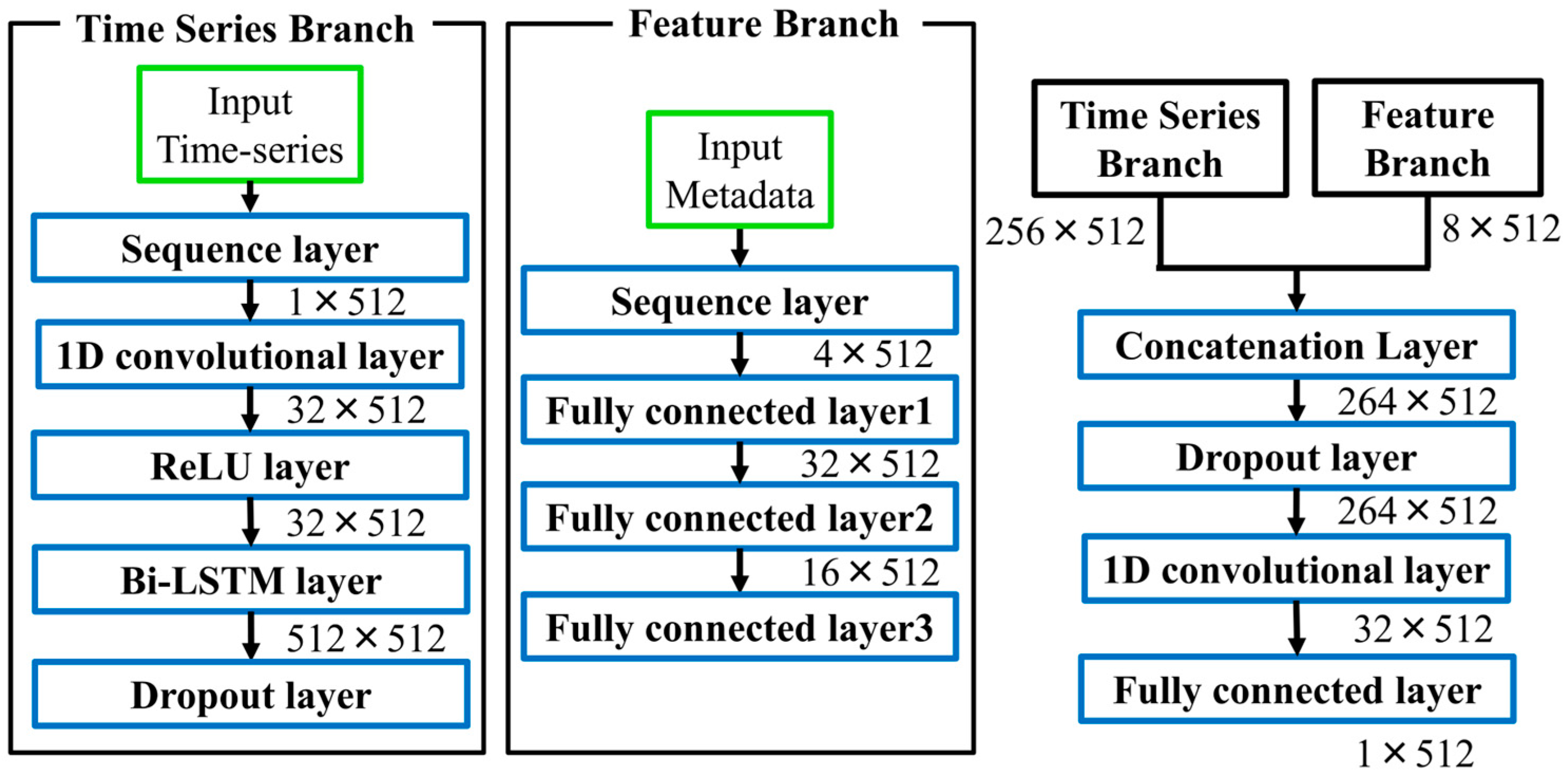
2.2. Model Architecture and Training
2.3. Experimental Conditions and Evaluation
- Single-feature integration: Each metadata feature was individually added to Lead I.
- Pairwise integration: Two features that showed superior performance in the single-feature experiments were combined.
- Full integration: All the metadata features were incorporated.
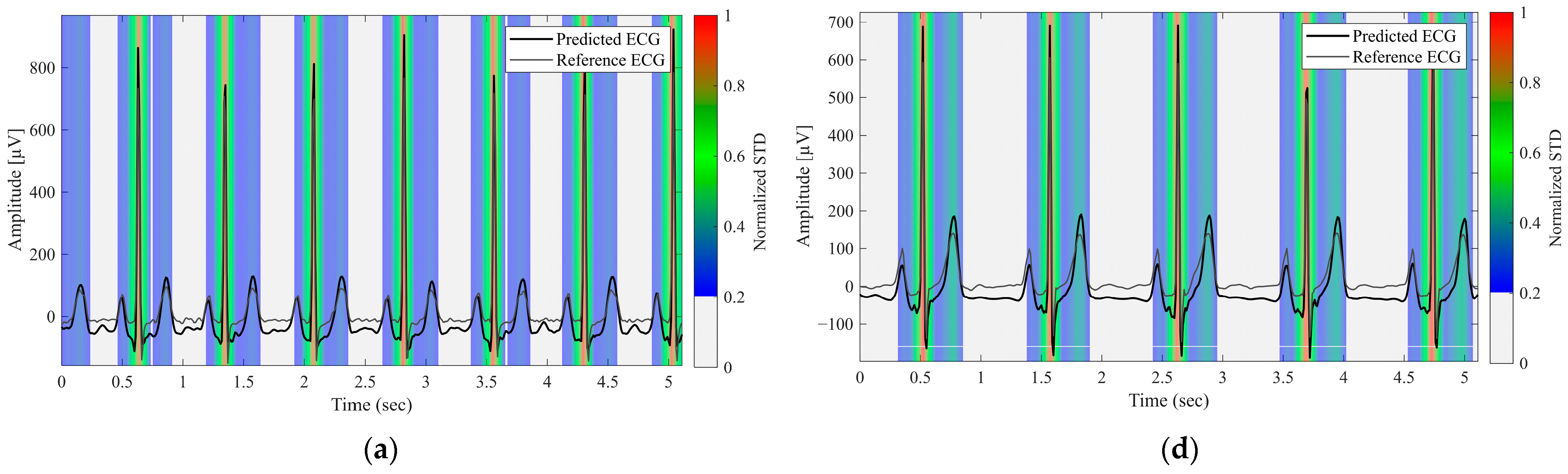
2.4. Uncertainty Visualization and Reliability Assessment
3. Results
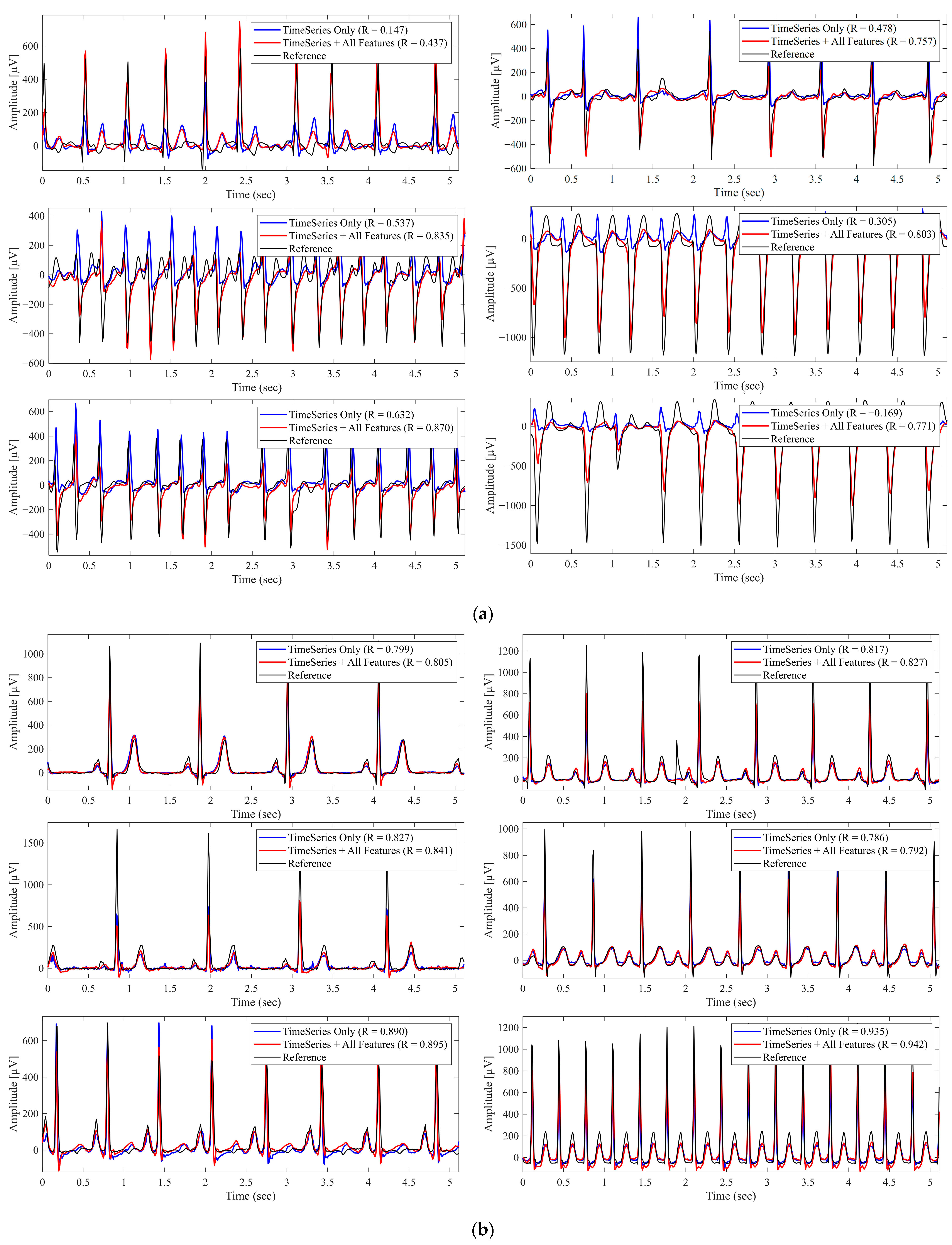
3.1. Impact of Metadata on Reconstruction Performance
3.2. Segment-Wise Analysis of Metadata Effects
3.3. Comparison with Existing Models
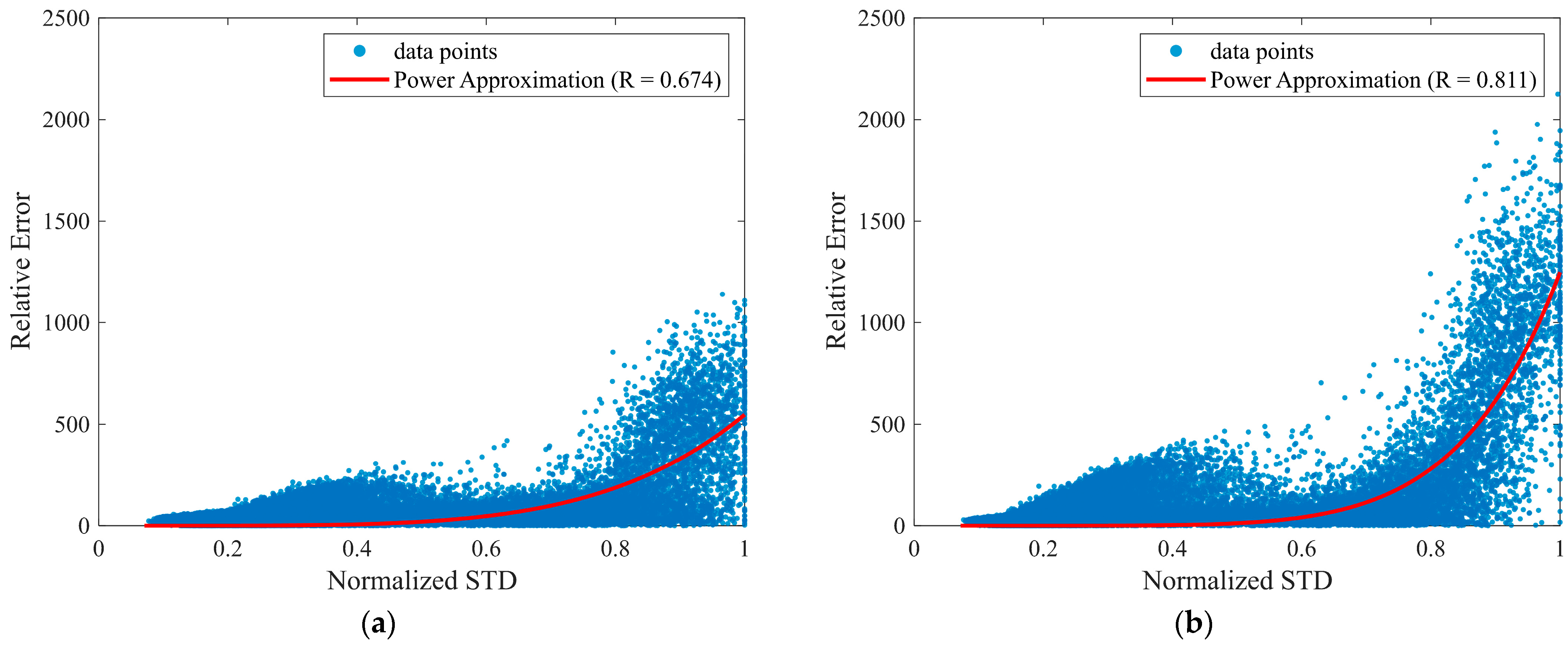
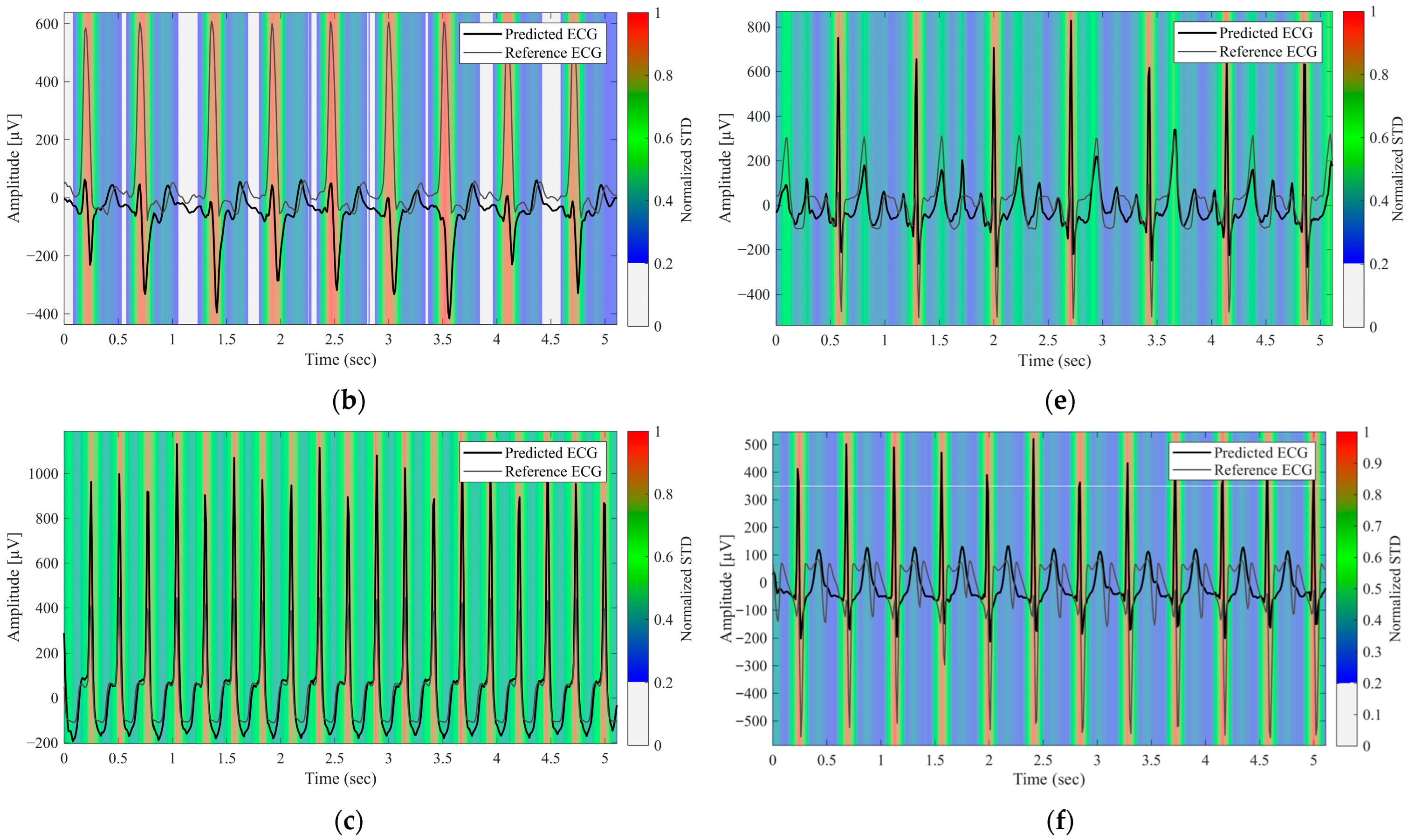
3.4. Evaluation of Predictive Uncertainty
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ECG | Electrocardiogram |
| AI | Artificial Intelligence |
| CNN | Convolutional Neural Network |
| RNN | Recurrent Neural Network |
| LSTM | Long Short-Term Memory |
| Bi-LSTM | Bidirectional Long Short-Term Memory |
| 1D-CNN | One-Dimensional Convolutional Neural Network |
| MC | Monte Carlo |
| RMSE | Root Mean Squared Error |
| R | Pearson’s Correlation Coefficient |
| SSIM | Structural Similarity Index |
| ReLU | Rectified Linear Unit |
| BPM | Beats Per Minute |
| RAxis | Right Axis (ventricular depolarization axis) |
| TAxis | T-wave Axis (ventricular repolarization axis) |
| Hz | Hertz |
| s | Seconds |
References
- Surawicz, B.; Childers, R.; Deal, B.J.; Gettes, L.S. AHA/ACCF/HRS Recommendations for the Standardization and Interpretation of the Electrocardiogram. J. Am. Coll. Cardiol. 2009, 53, 976–981. [Google Scholar] [CrossRef]
- Chou’s Electrocardiography in Clinical Practice; Elsevier: Amsterdam, The Netherlands, 2008. [CrossRef]
- Rizzo, C.; Monitillo, F.; Iacoviello, M. 12-Lead Electrocardiogram Features of Arrhythmic Risk: A Focus on Early Repolarization. World J. Cardiol. 2016, 8, 447. [Google Scholar] [CrossRef]
- Rafie, N.; Kashou, A.H.; Noseworthy, P.A. ECG Interpretation: Clinical Relevance, Challenges, and Advances. Hearts 2021, 2, 505–513. [Google Scholar] [CrossRef]
- Comprehensive Electrocardiology; Macfarlane, P.W., van Oosterom, A., Pahlm, O., Kligfield, P., Janse, M., Camm, J., Eds.; Springer: London, UK, 2010; ISBN 978-1-84882-045-6. [Google Scholar]
- Serhani, M.A.; El Kassabi, H.T.; Ismail, H.; Nujum Navaz, A. ECG Monitoring Systems: Review, Architecture, Processes, and Key Challenges. Sensors 2020, 20, 1796. [Google Scholar] [CrossRef] [PubMed]
- Ifenze, O.M.; Okanumee, C.J.; Nkuma-Udah, K.I.; Mbonu, E.S.; Ndubuka, G.I.N. Wearable Technology in Cardiovascular Health Monitoring: Current Trends and Future Directions. Eur. J. Biol. Med. Sci. Res. 2025, 13, 37–60. [Google Scholar] [CrossRef]
- Qtaish, A.; Al-Shrouf, A. A Portable IoT-Cloud ECG Monitoring System for Healthcare. IJCSNS Int. J. Comput. Sci. Netw. Secur. 2022, 22, 269–275. [Google Scholar] [CrossRef]
- Steinhubl, S.R.; Waalen, J.; Edwards, A.M.; Ariniello, L.M.; Mehta, R.R.; Ebner, G.S.; Carter, C.; Baca-Motes, K.; Felicione, E.; Sarich, T.; et al. Effect of a Home-Based Wearable Continuous ECG Monitoring Patch on Detection of Undiagnosed Atrial Fibrillation. JAMA 2018, 320, 146. [Google Scholar] [CrossRef]
- Perez, M.V.; Mahaffey, K.W.; Hedlin, H.; Rumsfeld, J.S.; Garcia, A.; Ferris, T.; Balasubramanian, V.; Russo, A.M.; Rajmane, A.; Cheung, L.; et al. Large-Scale Assessment of a Smartwatch to Identify Atrial Fibrillation. N. Engl. J. Med. 2019, 381, 1909–1917. [Google Scholar] [CrossRef]
- Guo, S.-L.; Han, L.-N.; Liu, H.-W.; Si, Q.-J.; Kong, D.-F.; Guo, F.-S. The Future of Remote ECG Monitoring Systems. J. Geriatr. Cardiol. 2016, 13, 528–530. [Google Scholar] [CrossRef]
- Haveman, M.E.; van Rossum, M.C.; Vaseur, R.M.E.; van der Riet, C.; Schuurmann, R.C.L.; Hermens, H.J.; de Vries, J.-P.P.M.; Tabak, M. Continuous Monitoring of Vital Signs with Wearable Sensors During Daily Life Activities: Validation Study. JMIR Form. Res. 2022, 6, e30863. [Google Scholar] [CrossRef]
- Edenbrandt, L.; Pahlm, O. Vectorcardiogram Synthesized from a 12-Lead ECG: Superiority of the Inverse Dower Matrix. J. Electrocardiol. 1988, 21, 361–367. [Google Scholar] [CrossRef]
- Kors, J.A.; Van Herpen, G.; Sittig, A.C.; Van Bemmel, J.H. Reconstruction of the Frank Vectorcardiogram from Standard Electrocardiographic Leads: Diagnostic Comparison of Different Methods. Eur. Heart J. 1990, 11, 1083–1092. [Google Scholar] [CrossRef]
- Somani, S.; Russak, A.J.; Richter, F.; Zhao, S.; Vaid, A.; Chaudhry, F.; De Freitas, J.K.; Naik, N.; Miotto, R.; Nadkarni, G.N.; et al. Deep Learning and the Electrocardiogram: Review of the Current State-of-the-Art. Europace 2021, 23, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Houssein, E.H.; Kilany, M.; Hassanien, A.E. ECG Signals Classification: A Review. Int. J. Intell. Eng. Inform. 2017, 5, 376. [Google Scholar] [CrossRef]
- Sridhar, C.; Lih, O.S.; Jahmunah, V.; Koh, J.E.W.; Ciaccio, E.J.; San, T.R.; Arunkumar, N.; Kadry, S.; Rajendra Acharya, U. Accurate Detection of Myocardial Infarction Using Non Linear Features with ECG Signals. J. Ambient. Intell. Humaniz. Comput. 2021, 12, 3227–3244. [Google Scholar] [CrossRef]
- Hebiguchi, K.; Togo, H.; Hirata, A. Reducing Lead Requirements for Wearable ECG: Chest Lead Reconstruction with 1D-CNN and Bi-LSTM. Inform. Med. Unlocked 2025, 53, 101624. [Google Scholar] [CrossRef]
- Sanjo, K.; Hebiguchi, K.; Tang, C.; Rashed, E.A.; Kodera, S.; Togo, H.; Hirata, A. Sensitivity of Electrocardiogram on Electrode-Pair Locations for Wearable Devices: Computational Analysis of Amplitude and Waveform Distortion. Biosensors 2024, 14, 153. [Google Scholar] [CrossRef] [PubMed]
- Savostin, A.; Koshekov, K.; Ritter, Y.; Savostina, G.; Ritter, D. 12-Lead ECG Reconstruction Based on Data From the First Limb Lead. Cardiovasc. Eng. Technol. 2024, 15, 346–358. [Google Scholar] [CrossRef]
- Seo, H.C.; Yoon, G.W.; Joo, S.; Nam, G.B. Multiple Electrocardiogram Generator with Single-Lead Electrocardiogram. Comput. Methods Programs Biomed. 2022, 221, 106858. [Google Scholar] [CrossRef]
- Rajotte, K.J.; Islam, B.; Huang, X.; McManus, D.D.; Clancy, E.A. ECG Statement Classification and Lead Reconstruction Using CNN-Based Models. IEEE J. Biomed. Health Inform. 2025, 1–11. [Google Scholar] [CrossRef]
- Lence, A.; Granese, F.; Fall, A.; Hanczar, B.; Salem, J.E.; Zucker, J.D.; Prifti, E. ECGrecover: A Deep Learning Approach for Electrocardiogram Signal Completion. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Toronto, ON, Canada, 3–7 August 2025; Association for Computing Machinery: New York, NY, USA, 2025; Volume 1, pp. 2359–2370. [Google Scholar]
- Manimaran, G.; Puthusserypady, S.; Domínguez, H.; Atienza, A.; Bardram, J.E. NERULA: A Dual-Pathway Self-Supervised Learning Framework for Electrocardiogram Signal Analysis. In International Conference on Innovative Techniques and Applications of Artificial Intelligence; Springer: Cham, Switzerland, 2024. [Google Scholar]
- Petmezas, G.; Stefanopoulos, L.; Kilintzis, V.; Tzavelis, A.; Rogers, J.A.; Katsaggelos, A.K.; Maglaveras, N. State-of-the-Art Deep Learning Methods on Electrocardiogram Data: Systematic Review. JMIR Med. Inform. 2022, 10, e38454. [Google Scholar] [CrossRef] [PubMed]
- Gal, Y.; Hron, J.; Kendall, A. Concrete Dropout. arXiv 2017. [Google Scholar] [CrossRef]
- Gal, Y.; Uk, Z.A. Dropout as a Bayesian Approximation: Representing Model Uncertainty in Deep Learning Zoubin Ghahramani. arXiv 2016. [Google Scholar] [CrossRef]
- Zahari, R.; Cox, J.; Obara, B. Uncertainty-Aware Image Classification on 3D CT Lung. Comput. Biol. Med. 2024, 172, 108324. [Google Scholar] [CrossRef]
- Abdar, M.; Samami, M.; Dehghani Mahmoodabad, S.; Doan, T.; Mazoure, B.; Hashemifesharaki, R.; Liu, L.; Khosravi, A.; Acharya, U.R.; Makarenkov, V.; et al. Uncertainty Quantification in Skin Cancer Classification Using Three-Way Decision-Based Bayesian Deep Learning. Comput. Biol. Med. 2021, 135, 104418. [Google Scholar] [CrossRef] [PubMed]
- Lemay, A.; Hoebel, K.; Bridge, C.P.; Befano, B.; De Sanjosé, S.; Egemen, D.; Rodriguez, A.C.; Schiffman, M.; Campbell, J.P.; Kalpathy-Cramer, J. Improving the Repeatability of Deep Learning Models with Monte Carlo Dropout. NPJ Digit. Med. 2022, 5, 174. [Google Scholar] [CrossRef]
- Leibig, C.; Allken, V.; Ayhan, M.S.; Berens, P.; Wahl, S. Leveraging Uncertainty Information from Deep Neural Networks for Disease Detection. Sci. Rep. 2017, 7, 17816. [Google Scholar] [CrossRef] [PubMed]
- Kendall, A.; Gal, Y. What Uncertainties Do We Need in Bayesian Deep Learning for Computer Vision? arXiv 2017. [Google Scholar] [CrossRef]
- Zheng, J.; Zhang, J.; Danioko, S.; Yao, H.; Guo, H.; Rakovski, C. A 12-Lead Electrocardiogram Database for Arrhythmia Research Covering More than 10,000 Patients. Sci. Data 2020, 7, 48. [Google Scholar] [CrossRef]
- Guo, Y.; Tang, Q.; Chen, Z.; Li, S. UNet-BiLSTM: A Deep Learning Method for Reconstructing Electrocardiography from Photoplethysmography. Electronics 2024, 13, 1869. [Google Scholar] [CrossRef]
- He, R.; Liu, Y.; Wang, K.; Zhao, N.; Yuan, Y.; Li, Q.; Zhang, H. Automatic Cardiac Arrhythmia Classification Using Combination of Deep Residual Network and Bidirectional LSTM. IEEE Access 2019, 7, 102119–102135. [Google Scholar] [CrossRef]
- Stahlschmidt, S.R.; Ulfenborg, B.; Synnergren, J. Multimodal Deep Learning for Biomedical Data Fusion: A Review. Brief Bioinform 2022, 23, bbab569. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J.L. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Chen, J.; Wu, W.; Liu, T.; Hong, S. Multi-Channel Masked Autoencoder and Comprehensive Evaluations for Reconstructing 12-Lead ECG from Arbitrary Single-Lead ECG. npj Cardiovasc. Health 2024, 1, 34. [Google Scholar] [CrossRef]
- Zhan, Z.; Chen, J.; Li, K.; Huang, L.; Xu, L.; Bian, G.B.; Millham, R.; de Albuquerque, V.H.C.; Wu, W. Conditional Generative Adversarial Network Driven Variable-Duration Single-Lead to 12-Lead Electrocardiogram Reconstruction. Biomed. Signal Process Control 2024, 95, 106377. [Google Scholar] [CrossRef]
- Thygesen, K.; Alpert, J.S.; Jaffe, A.S.; Chaitman, B.R.; Bax, J.J.; Morrow, D.A.; White, H.D. Fourth Universal Definition of Myocardial Infarction (2018). Circulation 2018, 138, e618–e651. [Google Scholar] [CrossRef]
- Srivastava, A.; Sheet, D.; Patra, A. L2G-ECG: Learning to Generate Missing Leads in ECG Signals Using Adversarial Autoencoder. IEEE J. Biomed. Health Inform. 2025, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lan, E. Performer: A Novel PPG-to-ECG Reconstruction Transformer for a Digital Biomarker of Cardiovascular Disease Detection. In Proceedings of the 2023 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 3–7 January 2023. [Google Scholar]
- Kan, C.; Ye, Z.; Zhou, H.; Cheruku, S.R. DG-ECG: Multi-Stream Deep Graph Learning for the Recognition of Disease-Altered Patterns in Electrocardiogram. Biomed. Signal Process Control 2023, 80, 104388. [Google Scholar] [CrossRef]
- Kline, A.; Wang, H.; Li, Y.; Dennis, S.; Hutch, M.; Xu, Z.; Wang, F.; Cheng, F.; Luo, Y. Multimodal Machine Learning in Precision Health: A Scoping Review. NPJ Digit. Med. 2022, 5, 171. [Google Scholar] [CrossRef]
- Attia, Z.I.; Noseworthy, P.A.; Lopez-Jimenez, F.; Asirvatham, S.J.; Deshmukh, A.J.; Gersh, B.J.; Carter, R.E.; Yao, X.; Rabinstein, A.A.; Erickson, B.J.; et al. An Artificial Intelligence-Enabled ECG Algorithm for the Identification of Patients with Atrial Fibrillation during Sinus Rhythm: A Retrospective Analysis of Outcome Prediction. Lancet 2019, 394, 861–867. [Google Scholar] [CrossRef]
- Raghunath, S.; Pfeifer, J.M.; Ulloa-Cerna, A.E.; Nemani, A.; Carbonati, T.; Jing, L.; vanMaanen, D.P.; Hartzel, D.N.; Ruhl, J.A.; Lagerman, B.F.; et al. Deep Neural Networks Can Predict New-Onset Atrial Fibrillation From the 12-Lead ECG and Help Identify Those at Risk of Atrial Fibrillation–Related Stroke. Circulation 2021, 143, 1287–1298. [Google Scholar] [CrossRef]
- Ribeiro, A.H.; Ribeiro, M.H.; Paixão, G.M.M.; Oliveira, D.M.; Gomes, P.R.; Canazart, J.A.; Ferreira, M.P.S.; Andersson, C.R.; Macfarlane, P.W.; Meira, W.; et al. Automatic Diagnosis of the 12-Lead ECG Using a Deep Neural Network. Nat. Commun. 2020, 11, 1760. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhang, F. A Novel Real-Time Detection and Classification Method for ECG Signal Images Based on Deep Learning. Sensors 2024, 24, 5087. [Google Scholar] [CrossRef] [PubMed]
- Bi, S.; Lu, R.; Xu, Q.; Zhang, P. Accurate Arrhythmia Classification with Multi-Branch, Multi-Head Attention Temporal Convolutional Networks. Sensors 2024, 24, 8124. [Google Scholar] [CrossRef] [PubMed]
| Feature | Category | Range/Criteria |
|---|---|---|
| RAxis/TAxis | Normal | |
| Left Deviation | ||
| Right Deviation | ||
| Extreme Deviation | ||
| QRS Duration | Normal | <120 ms |
| Prolonged | ≥120 ms | |
| Ventricular Rate | Bradycardia | <60 bpm |
| Normal | 60–100 bpm | |
| Tachycardia | <100 ms | |
| QRS Count | Low (Bradycardia) | <10 counts |
| Normal | 10–16 counts | |
| High (Tachycardia) | ≥17 counts |
| Input Data | Correlation Coefficients (R) | RMSE [mV] |
|---|---|---|
| Time Series Only | 0.763 ± 0.134 | 0.119 ± 0.084 |
| Time Series + RAxis | * 0.787 ± 0.111 | * 0.113 ± 0.078 |
| Time Series + TAxis | * 0.788 ± 0.110 | * 0.112 ± 0.078 |
| Time Series + VentricularRate | * 0.785 ± 0.113 | * 0.114 ± 0.079 |
| Time Series + QRSCount | * 0.788 ± 0.111 | * 0.113 ± 0.077 |
| Time Series + QRSDuration | * 0.788 ± 0.110 | * 0.112 ± 0.077 |
| Input Data | Correlation Coefficients (R) | RMSE [mV] |
|---|---|---|
| Time Series Only | 0.763 ± 0.134 | 0.119 ± 0.084 |
| Time Series + RAxis + QRSDuration | * 0.788 ± 0.111 | * 0.112 ± 0.077 |
| Time Series + RAxis + QRSCount | * 0.791 ± 0.109 | * 0.111 ± 0.077 |
| Time Series + RAxis + TAxis | * 0.789 ± 0.110 | * 0.115 ± 0.078 |
| Time Series + TAxis+ QRSDuration | * 0.788 ± 0.111 | * 0.114 ± 0.076 |
| Time Series + TAxis+ QRSCount | * 0.789 ± 0.111 | * 0.114 ± 0.078 |
| Time Series + QRSDuration+ QRSCount | * 0.789 ± 0.110 | * 0.114 ± 0.078 |
| Time Series + All Features | * 0.792 ± 0.109 | * 0.111 ± 0.076 |
| Input Data | P Wave | QRS Complex | ST Segments | T Wave | Ave |
|---|---|---|---|---|---|
| Time Series only | 0.571 | 0.761 | 0.590 | 0.668 | 0.648 |
| Time Series + RAxis | 0.575 | 0.801 | 0.591 | 0.672 | 0.660 |
| Time Series + TAxis | 0.574 | 0.764 | 0.597 | 0.721 | 0.664 |
| Time Series + QRSDuration | 0.573 | 0.806 | 0.597 | 0.675 | 0.663 |
| Time Series + All Features | 0.578 | 0.811 | 0.593 | 0.725 | 0.677 |
| Reference | Dataset | Method | ECG Duration (Frequency) | Correlation Coefficients (R) |
|---|---|---|---|---|
| Hebiguchi et al. [18] | PTB-XL | Bi-LSTM + CNN | 5.12 s (100 Hz) | 0.78 |
| Chen, Jiarong et al. [38] | MCMA | 10 s (500 Hz) | 0.77 | |
| Savostin, Alexey et al. [20] | U-Net | 1.3 s (100 Hz) | 0.80 | |
| Seo et al. [21] | PTB-XL, SPH&CU DB | U-Net | 10 s (500 Hz) | 0.76 |
| Zhan et al. [39] | SPH&CU DB, CPSC2018 | cGAN | 2.05 s (500 Hz) | 0.74 |
| Proposed method | SPH&CU DB | Dual Branch (Bi-LSTM + CNN) | 5.12 s (100 Hz) | 0.79 |
| Model | Time Series | RAxis | Other Features | Correlation Coefficients (R) | RMSE [mV] | SSIM |
|---|---|---|---|---|---|---|
| Proposed Method (Dual Branch: BiLSTM + CNN) | ✓ | ** 0.763 ± 0.134 | ** 0.119 ± 0.084 | ** 0.740 ± 0.153 | ||
| ✓ | ✓ | *,** 0.787 ± 0.111 | * 0.113 ± 0.078 | * 0.796 ± 0.141 | ||
| ✓ | ✓ | ✓ | *,** 0.792 ± 0.109 | *,** 0.111 ± 0.076 | *,** 0.804 ± 0.129 | |
| Baseline Method (BiLSTM + CNN) | ✓ | ✓ | 0.778 ± 0.122 | 0.116 ± 0.052 | 0.782 ± 0.149 | |
| ✓ | ✓ | ✓ | 0.782 ± 0.121 | 0.115 ± 0.052 | 0.785 ± 0.146 | |
| Baseline Method (U-Net) | ✓ | 0.771 ± 0.121 | 0.116 ± 0.079 | 0.758 ± 0.148 | ||
| ✓ | ✓ | 0.781 ± 0.114 | 0.114 ± 0.078 | 0.792 ± 0.146 | ||
| ✓ | ✓ | ✓ | 0.784 ± 0.113 | 0.114 ± 0.077 | 0.795 ± 0.139 |
| Normalized STD Quantile | Relative Error (RMSE) |
|---|---|
| Q1 | 50.8 ± 39.5 |
| Q2 | 90.7 ± 70.9 |
| Q3 | 112.5 ± 95.2 |
| Q4 | 403.0 ± 302.1 |
| Lead | Correlation Coefficients (R) |
|---|---|
| II | 0.674 |
| III | 0.552 |
| aVr | 0.738 |
| aVl | 0.642 |
| aVf | 0.591 |
| V1 | 0.776 |
| V2 | 0.684 |
| V3 | 0.675 |
| V4 | 0.750 |
| V5 | 0.807 |
| V6 | 0.811 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Nakanishi, R.; Hirata, A.; Kubota, Y. Deep Learning-Based Multi-Lead ECG Reconstruction from Lead I with Metadata Integration and Uncertainty Estimation. Sensors 2026, 26, 212. https://doi.org/10.3390/s26010212
Nakanishi R, Hirata A, Kubota Y. Deep Learning-Based Multi-Lead ECG Reconstruction from Lead I with Metadata Integration and Uncertainty Estimation. Sensors. 2026; 26(1):212. https://doi.org/10.3390/s26010212
Chicago/Turabian StyleNakanishi, Ryuichi, Akimasa Hirata, and Yoshiki Kubota. 2026. "Deep Learning-Based Multi-Lead ECG Reconstruction from Lead I with Metadata Integration and Uncertainty Estimation" Sensors 26, no. 1: 212. https://doi.org/10.3390/s26010212
APA StyleNakanishi, R., Hirata, A., & Kubota, Y. (2026). Deep Learning-Based Multi-Lead ECG Reconstruction from Lead I with Metadata Integration and Uncertainty Estimation. Sensors, 26(1), 212. https://doi.org/10.3390/s26010212







