MDEU-Net: Medical Image Segmentation Network Based on Multi-Head Multi-Scale Cross-Axis
Abstract
1. Introduction
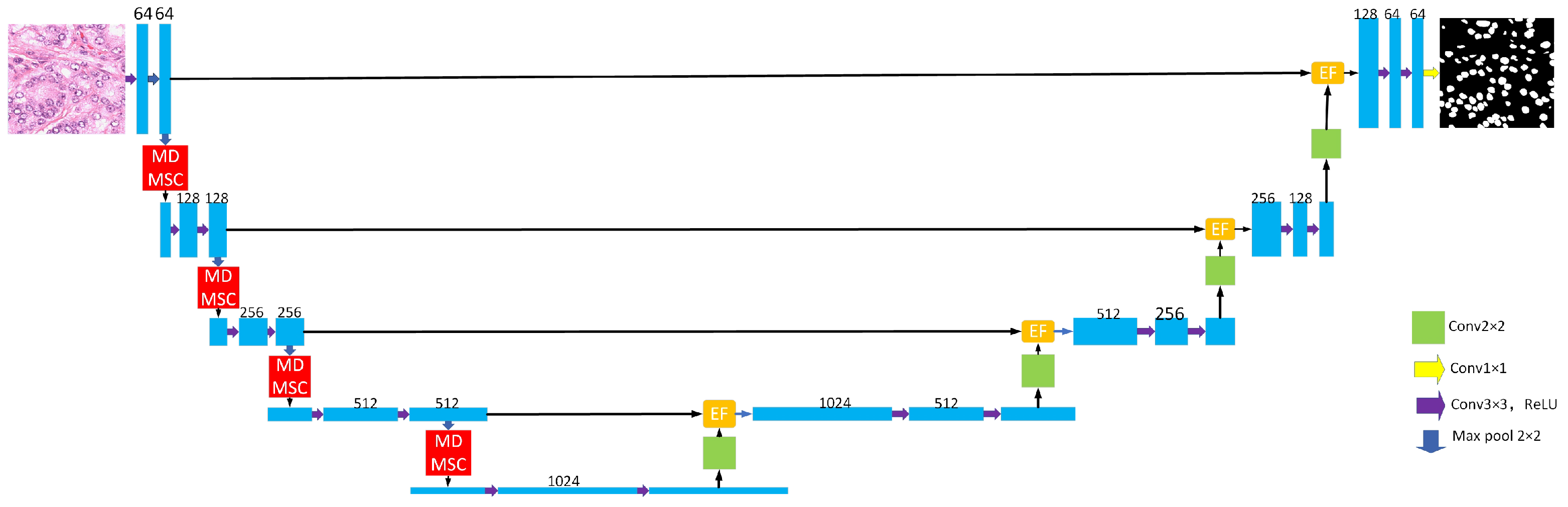
2. Network Architecture
2.1. General Organization
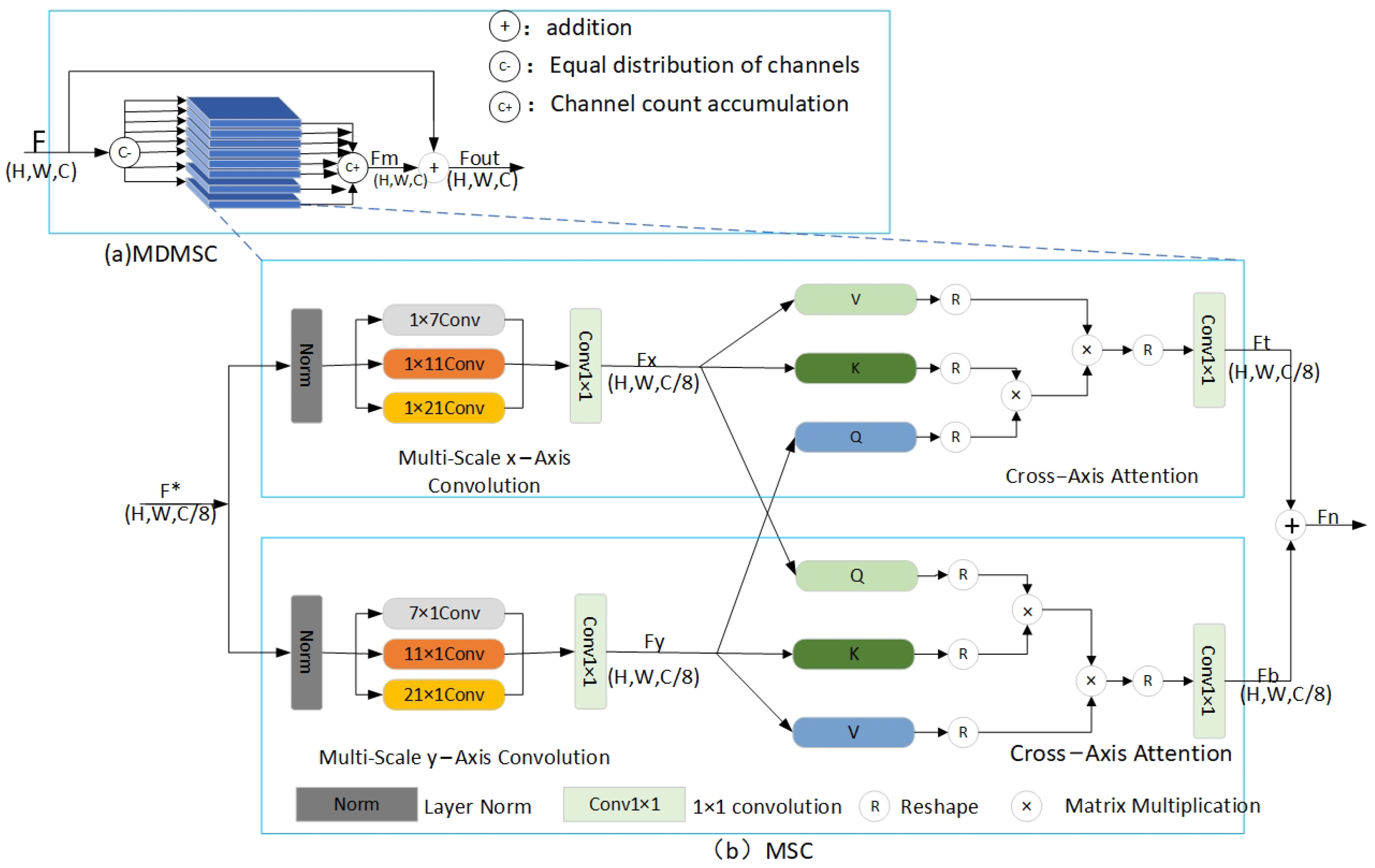
2.2. Multi-Head Multi-Scale Cross-Axis Attention Module
2.3. Efficient Feature Fusion Module
3. Experiments and Discussion of Results
3.1. Experimental Preparation
3.1.1. Experimental Data
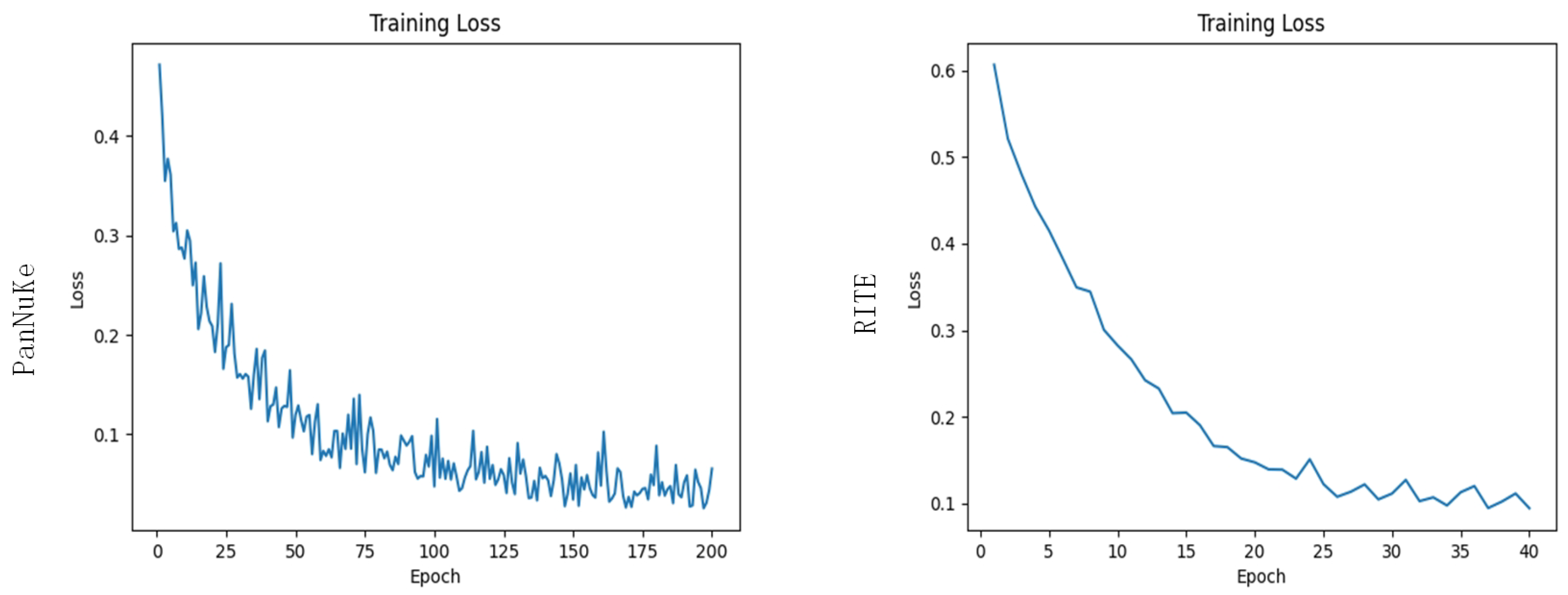
3.1.2. Experimental Methods
3.2. Experimental Evaluation Indicators and Results
3.2.1. Evaluation Indicators
| The Real Situation | Projected Results | |
|---|---|---|
| Standard Practice | Counter-Example | |
| standard practice | TP (True Positive) | FN (False Negative) |
| counter-example | FP (False Positive) | TN (True Negative) |
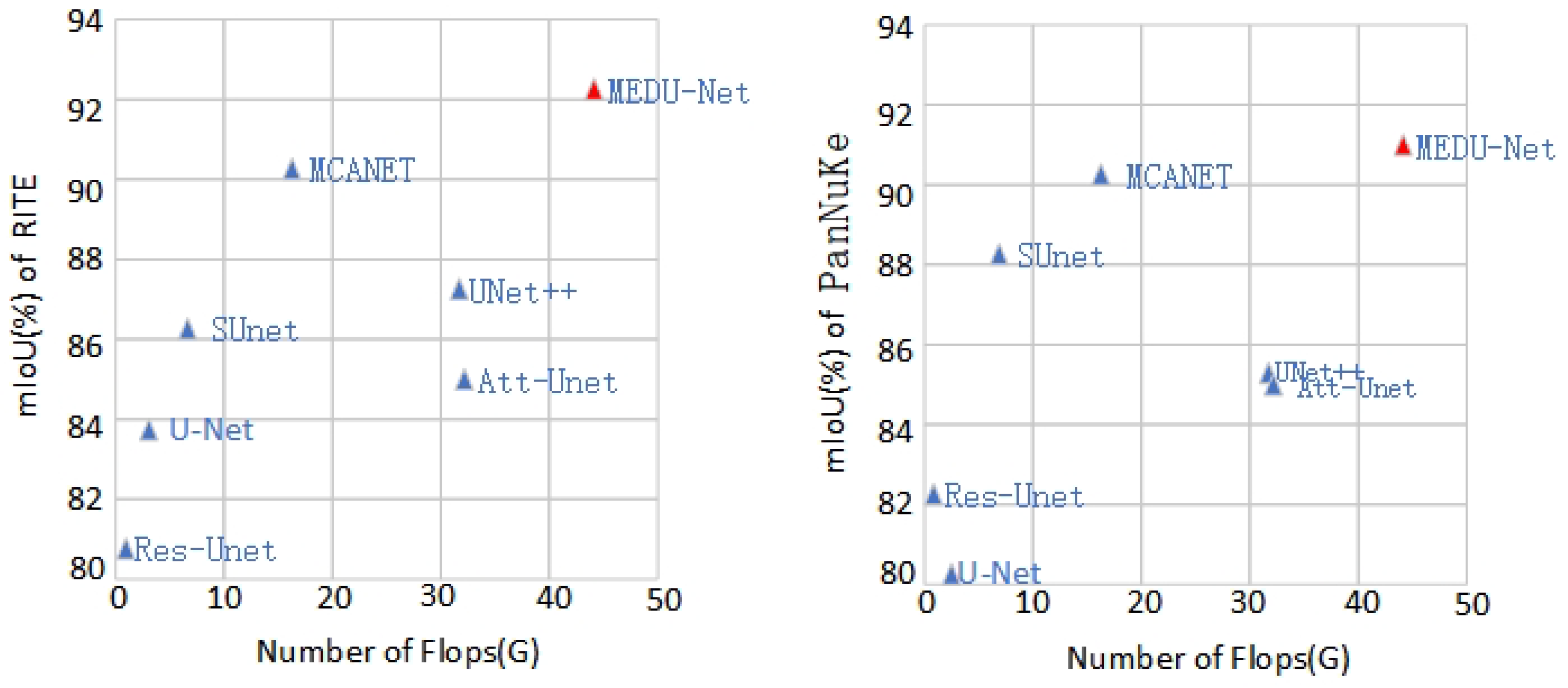
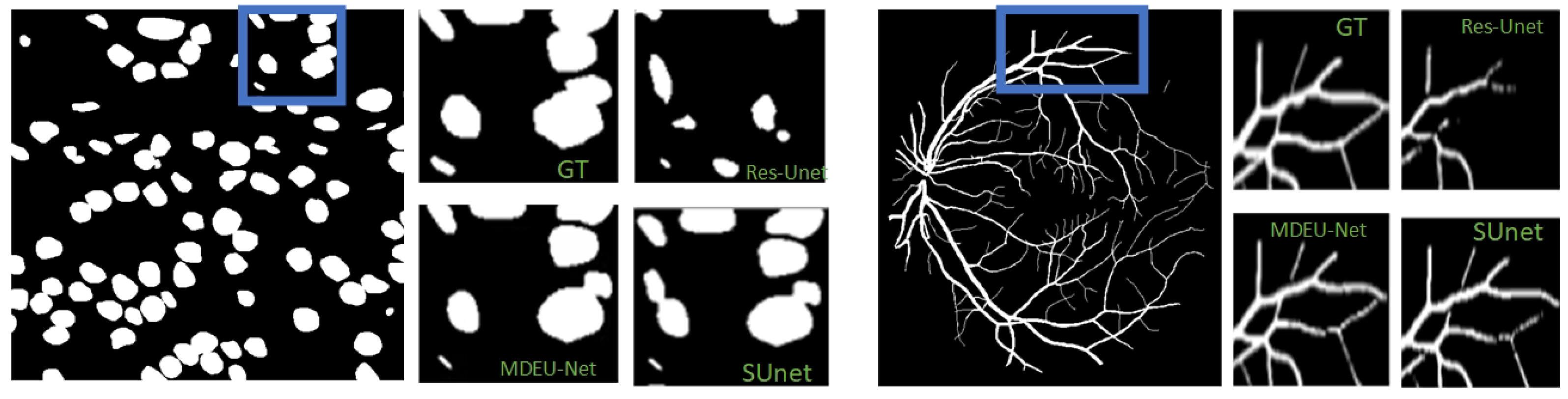
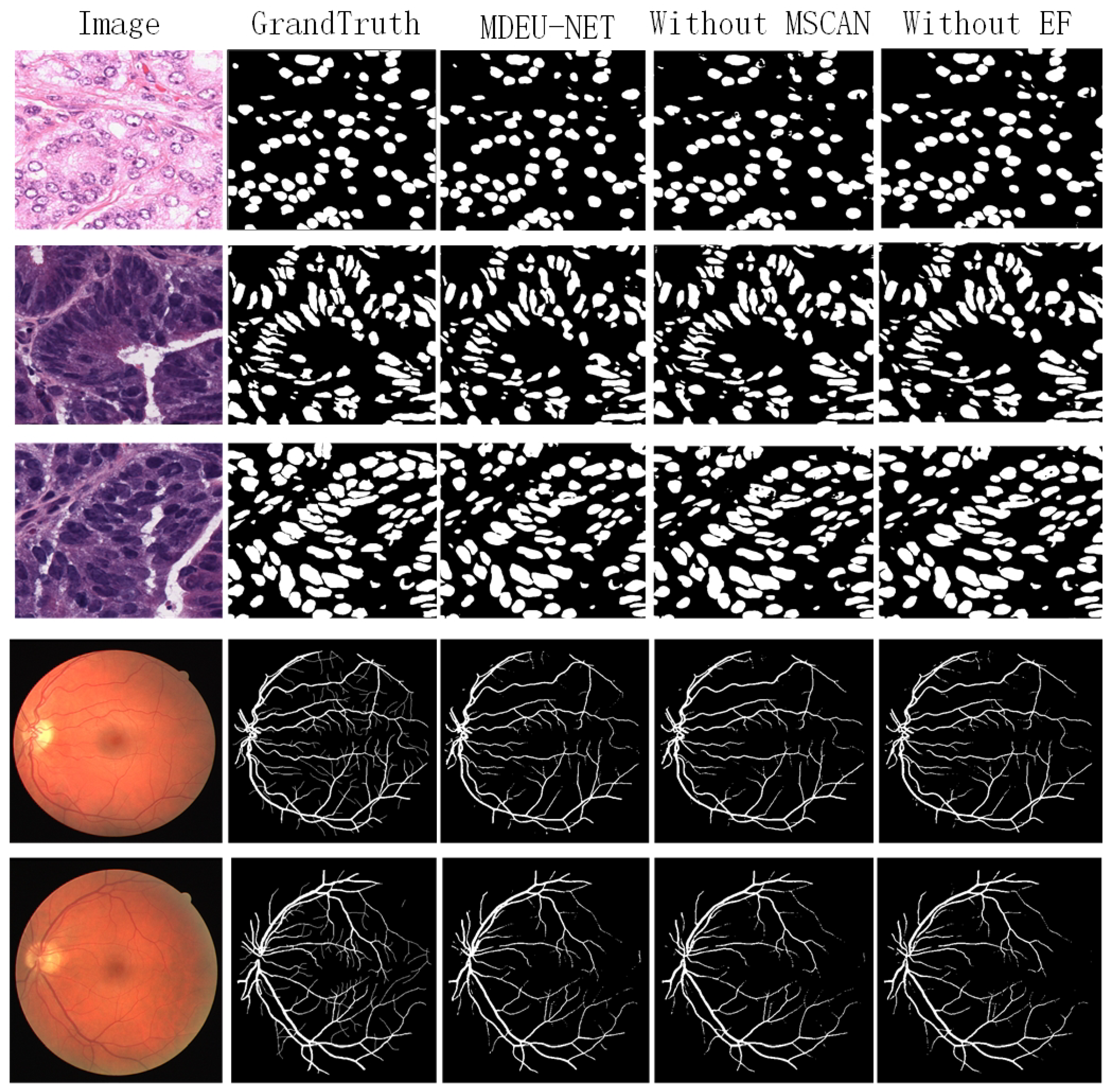
3.2.2. Experimental Results and Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kaloumenou, M.; Skotadis, E.; Lagopati, N.; Efstathopoulos, E.; Tsoukalas, D. Breath analysis: A promising tool for disease diagnosis—The role of sensors. Sensors 2022, 22, 1238. [Google Scholar] [CrossRef]
- Wu, J.; Fang, H.; Shang, F.; Yang, D.; Wang, Z.; Gao, J.; Yang, Y.; Xu, Y. SeATrans: Learning segmentation-assisted diagnosis model via transformer. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 7–11 October 2024; Springer: Berlin/Heidelberg, Germany, 2022; pp. 677–687. [Google Scholar]
- Jiang, H.; Diao, Z.; Shi, T.; Zhou, Y.; Wang, F.; Hu, W.; Zhu, X.; Luo, S.; Tong, G.; Yao, Y.D. A review of deep learning-based multiple-lesion recognition from medical images: Classification, detection and segmentation. Comput. Biol. Med. 2023, 157, 106726. [Google Scholar] [CrossRef]
- Gu, Y.; Wu, Q.; Tang, H.; Mai, X.; Shu, H.; Li, B.; Chen, Y. Lesam: Adapt segment anything model for medical lesion segmentation. IEEE J. Biomed. Health Inform. 2024, 28, 6031–6041. [Google Scholar] [CrossRef]
- Bagley, J.C.; Phillips, A.K.; Buchanon, S.; O’Neil, P.E.; Huff, E.S. Incidence and effects of anomalies and hybridization on Alabama freshwater fish index of biotic integrity results. Environ. Monit. Assess. 2025, 197, 50. [Google Scholar] [CrossRef]
- Cohen, A.B.; Diamant, I.; Klang, E.; Amitai, M.; Greenspan, H. Automatic detection and segmentation of liver metastatic lesions on serial CT examinations. In Medical Imaging 2014: Computer-Aided Diagnosis, Proceedings of the SPIE Medical Imaging, San Diego, CA, USA, 15–20 February 2014; SPIE: San Diego, CA, USA, 2014; Volume 9035, pp. 327–334. [Google Scholar]
- Kattenborn, T.; Leitloff, J.; Schiefer, F.; Hinz, S. Review on Convolutional Neural Networks (CNN) in vegetation remote sensing. ISPRS J. Photogramm. Remote Sens. 2021, 173, 24–49. [Google Scholar] [CrossRef]
- Rajaraman, S.; Antani, S.K.; Poostchi, M.; Silamut, K.; Hossain, M.A.; Maude, R.J.; Jaeger, S.; Thoma, G.R. Pre-trained convolutional neural networks as feature extractors toward improved malaria parasite detection in thin blood smear images. PeerJ 2018, 6, e4568. [Google Scholar] [CrossRef]
- Antonelli, M.; Reinke, A.; Bakas, S.; Farahani, K.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B.; Ronneberger, O.; Summers, R.M.; et al. The medical segmentation decathlon. Nat. Commun. 2022, 13, 4128. [Google Scholar] [CrossRef]
- Mazurowski, M.A.; Dong, H.; Gu, H.; Yang, J.; Konz, N.; Zhang, Y. Segment anything model for medical image analysis: An experimental study. Med. Image Anal. 2023, 89, 102918. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention, Proceedings of the MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Proceedings of the 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; Proceedings 4; Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–11. [Google Scholar]
- Liu, F.; Ren, X.; Zhang, Z.; Sun, X.; Zou, Y. Rethinking skip connection with layer normalization in transformers and resnets. arXiv 2021, arXiv:2105.07205. [Google Scholar]
- Oyedotun, O.K.; Shabayek, A.E.R.; Aouada, D.; Ottersten, B. Going deeper with neural networks without skip connections. In Proceedings of the 2020 IEEE International Conference on Image Processing (ICIP), Online, 25–28 October 2020; pp. 1756–1760. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Ding, Z.; Jiang, S.; Zhao, J. Take a close look at mode collapse and vanishing gradient in GAN. In Proceedings of the 2022 IEEE 2nd International Conference on Electronic Technology, Communication and Information (ICETCI), Online, 27–29 May 2022; pp. 597–602. [Google Scholar]
- Iqbal, S.; Khan, T.M.; Naqvi, S.S.; Naveed, A.; Meijering, E. TBConvL-Net: A hybrid deep learning architecture for robust medical image segmentation. Pattern Recognit. 2025, 158, 111028. [Google Scholar] [CrossRef]
- Song, W.; Wang, X.; Guo, Y.; Li, S.; Xia, B.; Hao, A. Centerformer: A novel cluster center enhanced transformer for unconstrained dental plaque segmentation. IEEE Trans. Multimed. 2024, 26, 10965–10978. [Google Scholar] [CrossRef]
- Wang, G.; Ma, Q.; Li, Y.; Mao, K.; Xu, L.; Zhao, Y. A skin lesion segmentation network with edge and body fusion. Appl. Soft Comput. 2025, 170, 112683. [Google Scholar] [CrossRef]
- Xia, J.; Cai, Z.; Heidari, A.A.; Ye, Y.; Chen, H.; Pan, Z. Enhanced moth-flame optimizer with quasi-reflection and refraction learning with application to image segmentation and medical diagnosis. Curr. Bioinform. 2023, 18, 109–142. [Google Scholar]
- Shao, H.; Zeng, Q.; Hou, Q.; Yang, J. Mcanet: Medical image segmentation with multi-scale cross-axis attention. arXiv 2023, arXiv:2312.08866. [Google Scholar]
- Sinha, A.; Dolz, J. Multi-scale self-guided attention for medical image segmentation. IEEE J. Biomed. Health Inform. 2020, 25, 121–130. [Google Scholar] [CrossRef]
- Feng, Y.; Cong, Y.; Xing, S.; Wang, H.; Ren, Z.; Zhang, X. GCFormer: Multi-scale feature plays a crucial role in medical images segmentation. Knowl. Based Syst. 2024, 300, 112170. [Google Scholar] [CrossRef]
- Lu, S.; Liu, M.; Yin, L.; Yin, Z.; Liu, X.; Zheng, W. The multi-modal fusion in visual question answering: A review of attention mechanisms. PeerJ Comput. Sci. 2023, 9, e1400. [Google Scholar] [CrossRef]
- Biswas, S.; Gogoi, A.; Biswas, M. Aspect Ratio Approximation for Simultaneous Minimization of Cross Axis Sensitivity Along Off-Axes for High-Performance Non-invasive Inertial MEMS. In Proceedings of the International Conference on Micro/Nanoelectronics Devices, Circuits and Systems, Silchar, India, 29–31 January 2023; Springer: Berlin/Heidelberg, Germany, 2023; pp. 463–469. [Google Scholar]
- Ding, Y.; Zhang, Z.; Zhao, X.; Hong, D.; Cai, W.; Yu, C.; Yang, N.; Cai, W. Multi-feature fusion: Graph neural network and CNN combining for hyperspectral image classification. Neurocomputing 2022, 501, 246–257. [Google Scholar] [CrossRef]
- Zhao, H.h.; Liu, H. Multiple classifiers fusion and CNN feature extraction for handwritten digits recognition. Granul. Comput. 2020, 5, 411–418. [Google Scholar] [CrossRef]
- Dai, Y.; Gieseke, F.; Oehmcke, S.; Wu, Y.; Barnard, K. Attentional feature fusion. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Online, 5–9 January 2021; pp. 3560–3569. [Google Scholar]
- Cordonnier, J.B.; Loukas, A.; Jaggi, M. Multi-head attention: Collaborate instead of concatenate. arXiv 2020, arXiv:2006.16362. [Google Scholar]
- Ilina, O.; Ziyadinov, V.; Klenov, N.; Tereshonok, M. A survey on symmetrical neural network architectures and applications. Symmetry 2022, 14, 1391. [Google Scholar] [CrossRef]
- Shen, K.; Guo, J.; Tan, X.; Tang, S.; Wang, R.; Bian, J. A study on relu and softmax in transformer. arXiv 2023, arXiv:2302.06461. [Google Scholar]
- Syed, A.S.; Sierra-Sosa, D.; Kumar, A.; Elmaghraby, A. A hierarchical approach to activity recognition and fall detection using wavelets and adaptive pooling. Sensors 2021, 21, 6653. [Google Scholar] [CrossRef]
- Guo, M.H.; Lu, C.Z.; Hou, Q.; Liu, Z.; Cheng, M.M.; Hu, S.M. Segnext: Rethinking convolutional attention design for semantic segmentation. Adv. Neural Inf. Process. Syst. 2022, 35, 1140–1156. [Google Scholar]
- Rehman, M.U.; Nizami, I.F.; Ullah, F.; Hussain, I. IQA Vision Transformed: A Survey of Transformer Architectures in Perceptual Image Quality Assessment. IEEE Access 2024, 12, 183369–183393. [Google Scholar] [CrossRef]
- Gluth, S.; Kern, N.; Kortmann, M.; Vitali, C.L. Value-based attention but not divisive normalization influences decisions with multiple alternatives. Nat. Hum. Behav. 2020, 4, 634–645. [Google Scholar] [CrossRef]
- Huang, H.; Zhou, P.; Li, Y.; Sun, F. A lightweight attention-based CNN model for efficient gait recognition with wearable IMU sensors. Sensors 2021, 21, 2866. [Google Scholar] [CrossRef]
- Ruan, J.; Xie, M.; Gao, J.; Liu, T.; Fu, Y. Ege-unet: An efficient group enhanced unet for skin lesion segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 7–11 October 2024; Springer: Berlin/Heidelberg, Germany, 2023; pp. 481–490. [Google Scholar]
- Li, X.; Qin, X.; Huang, C.; Lu, Y.; Cheng, J.; Wang, L.; Liu, O.; Shuai, J.; Yuan, C.A. SUnet: A multi-organ segmentation network based on multiple attention. Comput. Biol. Med. 2023, 167, 107596. [Google Scholar] [CrossRef]
- Jia, Y.; Chen, G.; Chi, H. Retinal fundus image super-resolution based on generative adversarial network guided with vascular structure prior. Sci. Rep. 2024, 14, 22786. [Google Scholar] [CrossRef]


| Params | Flops | RITE | PanNuKe | |||||
|---|---|---|---|---|---|---|---|---|
| Method | M | G | mIoU | Acc (%) | Recall (%) | mIoU | Acc (%) | Recall (%) |
| U-Net | 1.56 | 4.08 | 83.77 | 98.17 | 87.81 | 80.01 | 92.67 | 87.23 |
| Res-Unet | 4.11 | 2.56 | 80.67 | 97.98 | 82.04 | 82.53 | 93.07 | 88.31 |
| UNet++ | 13.41 | 31.13 | 87.40 | 98.67 | 88.37 | 85.42 | 94.19 | 91.06 |
| Att-Unet | 13.75 | 32.23 | 85.21 | 98.43 | 87.07 | 85.94 | 94.41 | 90.97 |
| SUnet | 23.01 | 6.31 | 86.74 | 98.53 | 90.24 | 88.00 | 95.30 | 92.82 |
| MCANET | 5.56 | 16.44 | 90.08 | 99.04 | 91.80 | 91.16 | - | - |
| MDEU-Net | 17.18 | 44.11 | 92.32 | 99.19 | 93.45 | 91.53 | 96.18 | 93.42 |
| Params | Flops | RITE | PanNuKe | |||||
|---|---|---|---|---|---|---|---|---|
| Method | M | G | mIoU | Acc (%) | Recall (%) | mIoU | Acc (%) | Recall (%) |
| MDEU2-NET | 17.62 | 44.11 | 91.87 | 99.15 | 93.12 | 90.91 | 95.97 | 93.51 |
| MDEU4-NET | 17.52 | 44.11 | 91.04 | 99.05 | 92.72 | 91.46 | 95.13 | 93.78 |
| MDEU8-NET | 17.18 | 44.11 | 92.32 | 99.19 | 93.45 | 91.53 | 96.18 | 93.42 |
| MDEU16-NET | 17.46 | 44.11 | 91.59 | 99.11 | 93.25 | 90.86 | 95.62 | 93.04 |
| MDEU32-NET | 17.44 | 44.11 | 90.71 | 99.03 | 91.72 | 89.49 | 95.86 | 92.31 |
| MDMSC | EF | mIoU | Acc (%) | Recall (%) |
|---|---|---|---|---|
| √ | √ | 92.32 | 99.19 | 93.45 |
| × | √ | 91.12 | 99.07 | 92.15 |
| √ | × | 91.05 | 99.06 | 92.17 |
| MDMSC | EF | mIoU | Acc (%) | Recall (%) |
|---|---|---|---|---|
| √ | √ | 91.53 | 96.18 | 93.42 |
| × | √ | 87.90 | 95.26 | 92.47 |
| √ | × | 88.59 | 95.53 | 93.12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, S.; Lei, Y.; Zhang, J.; Gao, X.; Li, X.; Wang, P.; Cao, H. MDEU-Net: Medical Image Segmentation Network Based on Multi-Head Multi-Scale Cross-Axis. Sensors 2025, 25, 2917. https://doi.org/10.3390/s25092917
Yan S, Lei Y, Zhang J, Gao X, Li X, Wang P, Cao H. MDEU-Net: Medical Image Segmentation Network Based on Multi-Head Multi-Scale Cross-Axis. Sensors. 2025; 25(9):2917. https://doi.org/10.3390/s25092917
Chicago/Turabian StyleYan, Shengxian, Yuyang Lei, Jing Zhang, Xiao Gao, Xiang Li, Penghui Wang, and Hui Cao. 2025. "MDEU-Net: Medical Image Segmentation Network Based on Multi-Head Multi-Scale Cross-Axis" Sensors 25, no. 9: 2917. https://doi.org/10.3390/s25092917
APA StyleYan, S., Lei, Y., Zhang, J., Gao, X., Li, X., Wang, P., & Cao, H. (2025). MDEU-Net: Medical Image Segmentation Network Based on Multi-Head Multi-Scale Cross-Axis. Sensors, 25(9), 2917. https://doi.org/10.3390/s25092917






