Enhanced Prediction of Soil Carbon via Encoder-Decoder Neural Networks for a Boreal Study Area in Northern Ontario
Abstract
1. Introduction
2. Materials and Methods
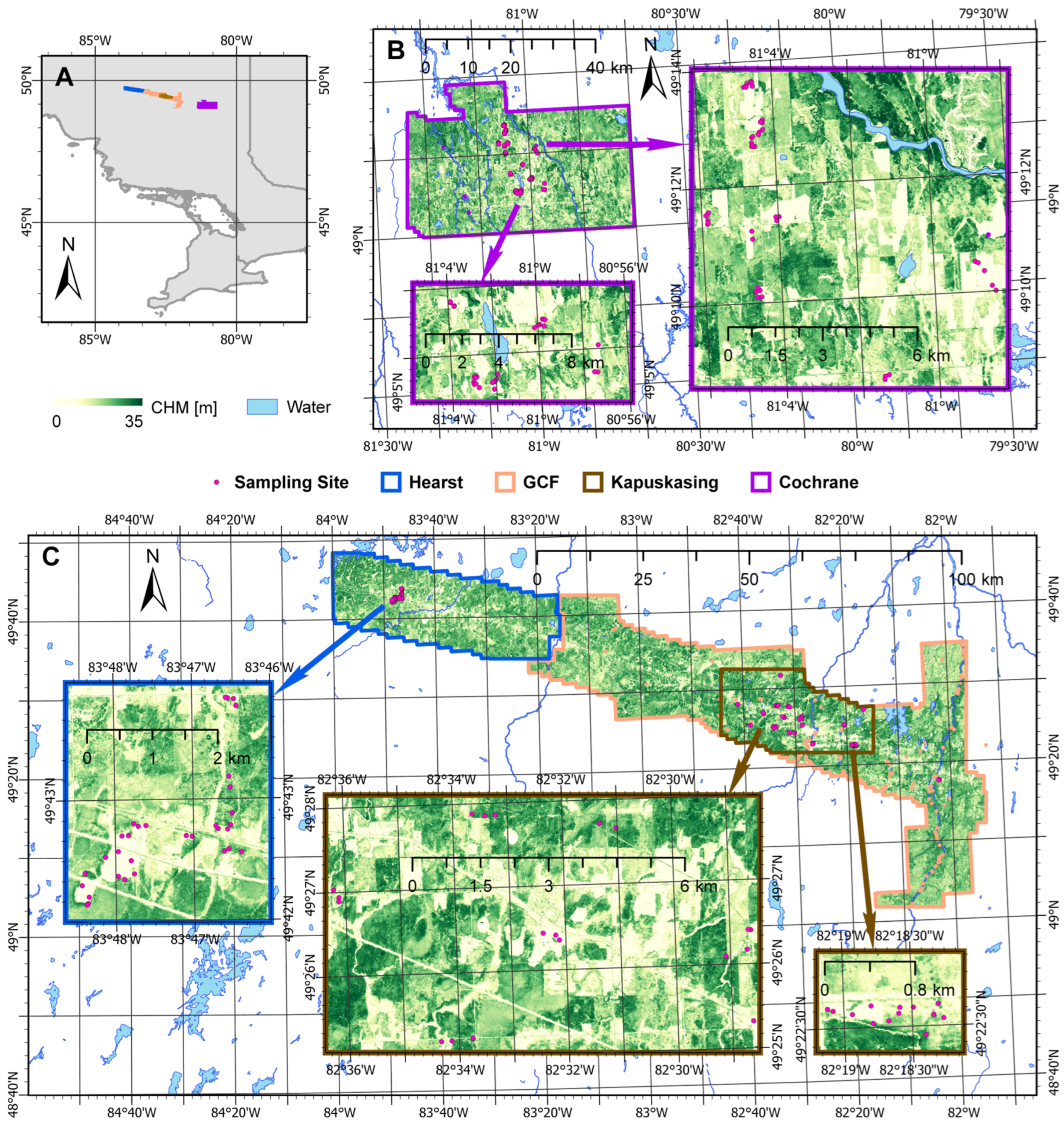
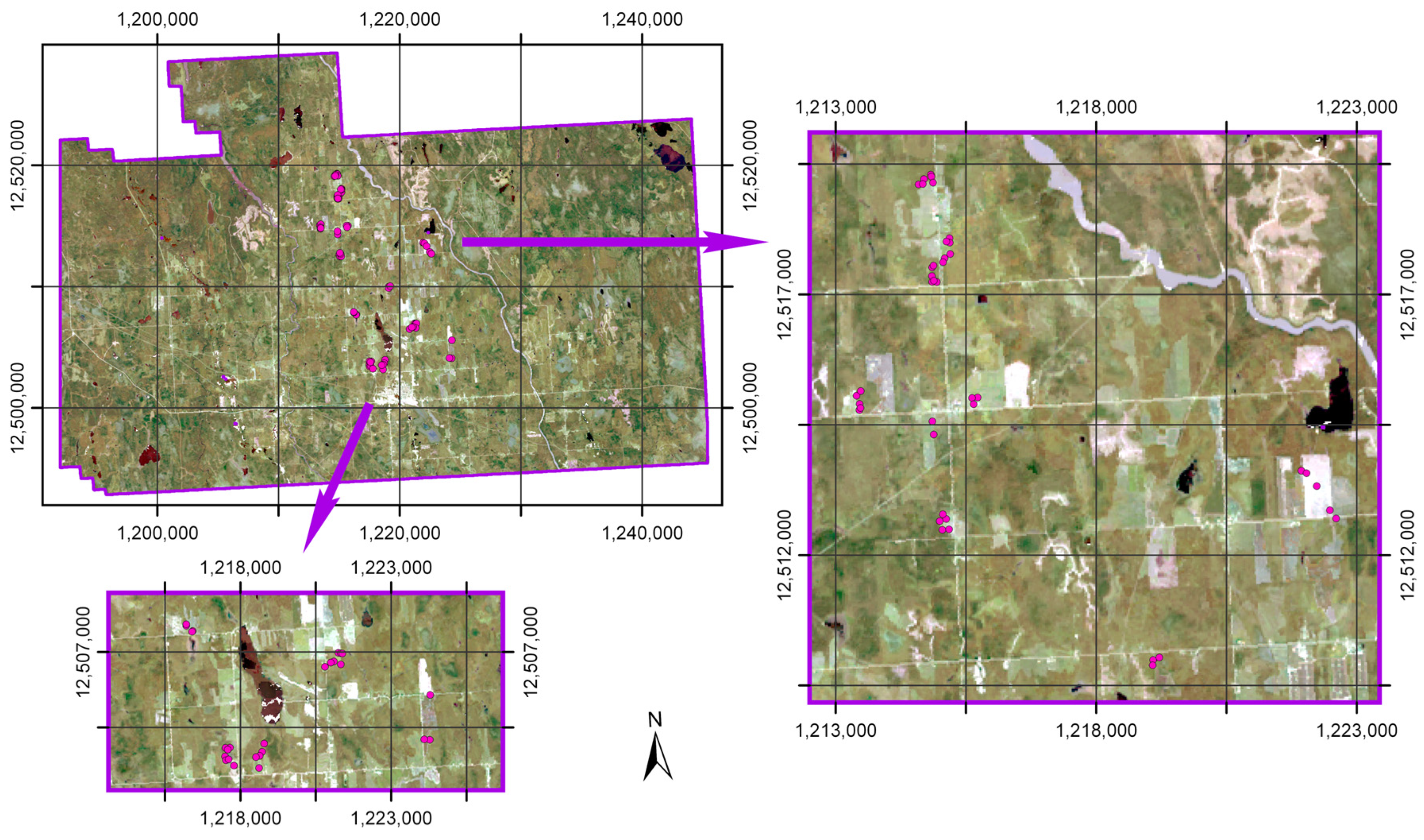
2.1. Study Region
2.2. Soil Data
2.3. Environmental Covariates
2.4. Modeling
2.4.1. Normalization
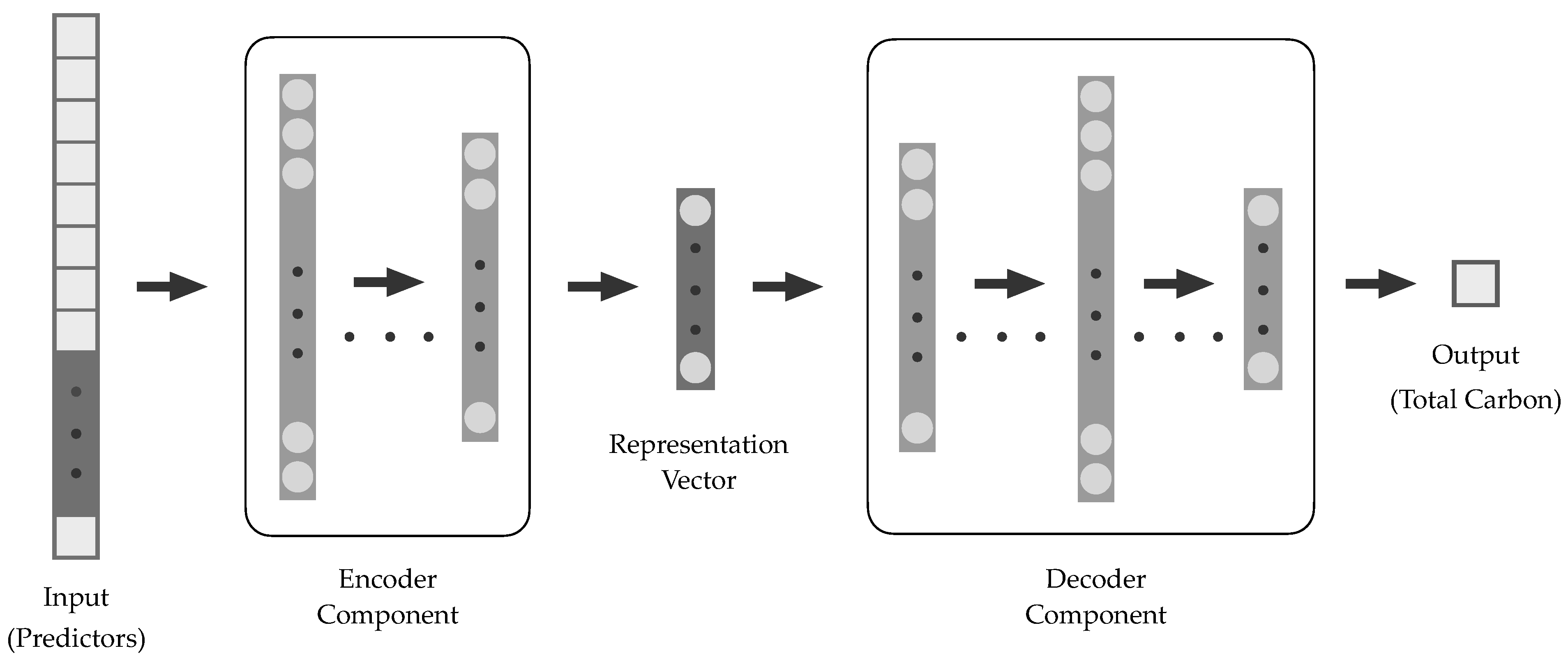
2.4.2. Encoder-Decoder Neural Networks
2.4.3. Other Models
2.4.4. Model Evaluation
2.5. Uncertainty Quantification
3. Results
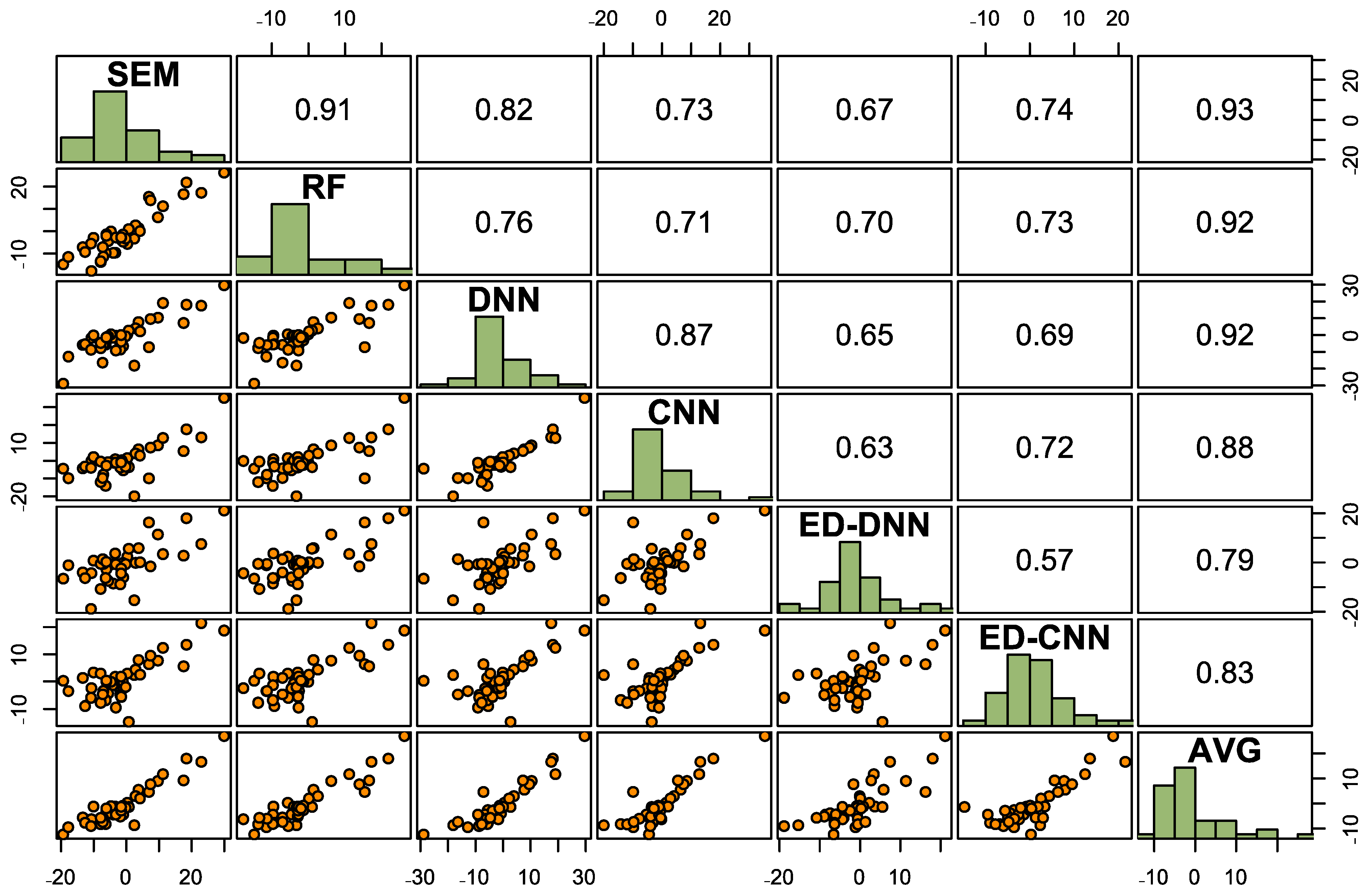
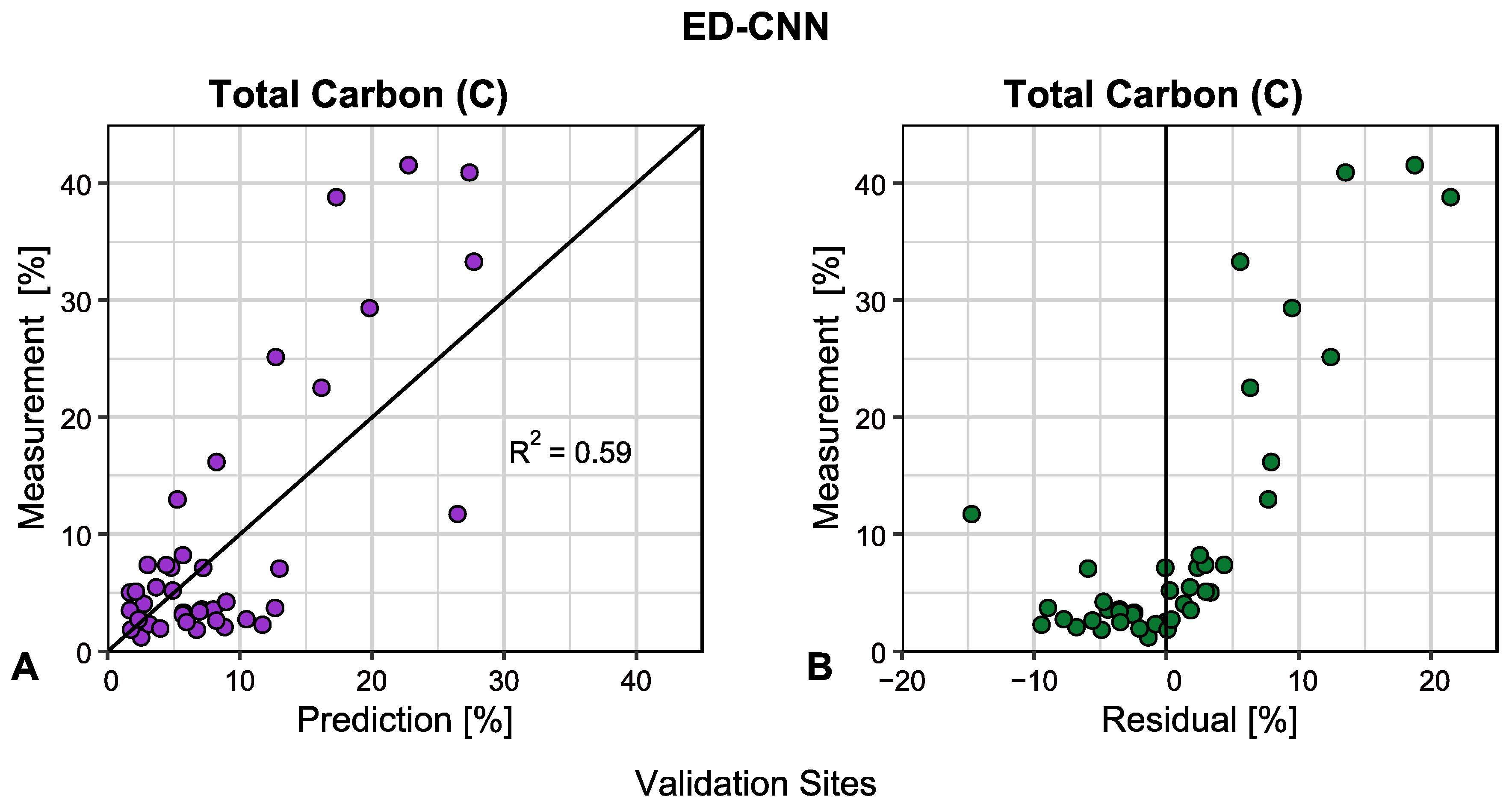
3.1. Modeling Accuracies
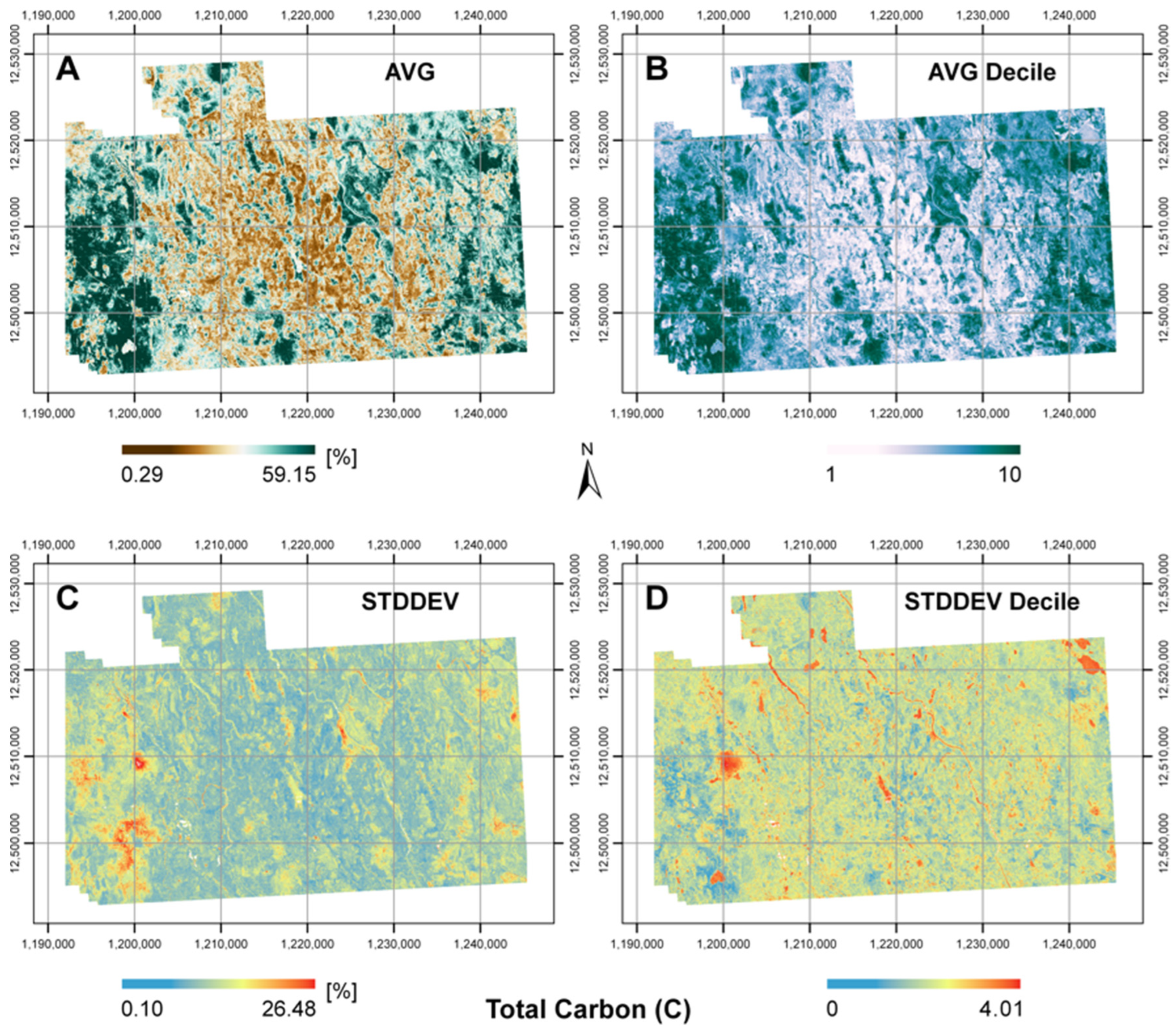
3.2. Prediction Maps
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gao, B.; Taylor, A.R.; Searle, E.B.; Kumar, P.; Ma, Z.; Hume, A.M.; Chen, H.Y.H. Carbon Storage Declines in Old Boreal Forests Irrespective of Succession Pathway. Ecosystems 2018, 21, 1168–1182. [Google Scholar] [CrossRef]
- Laganière, J.; Paré, D.; Bergeron, Y.; Chen, H.Y.H.; Brassard, B.W.; Cavard, X. Stability of Soil Carbon Stocks Varies with Forest Composition in the Canadian Boreal Biome. Ecosystems 2013, 16, 852–865. [Google Scholar] [CrossRef]
- Boulanger, Y.; Taylor, A.R.; Price, D.T.; Cyr, D.; McGarrigle, E.; Rammer, W.; Sainte-Marie, G.; Beaudoin, A.; Guindon, L.; Mansuy, N. Climate Change Impacts on Forest Landscapes along the Canadian Southern Boreal Forest Transition Zone. Landsc. Ecol. 2017, 32, 1415–1431. [Google Scholar] [CrossRef]
- Minasny, B.; Berglund, Ö.; Connolly, J.; Hedley, C.; de Vries, F.; Gimona, A.; Kempen, B.; Kidd, D.; Lilja, H.; Malone, B.; et al. Digital Mapping of Peatlands—A Critical Review. Earth Sci. Rev. 2019, 196, 102870. [Google Scholar] [CrossRef]
- Heung, B.; Ho, H.C.; Zhang, J.; Knudby, A.; Bulmer, C.E.; Schmidt, M.G. An Overview and Comparison of Machine-Learning Techniques for Classification Purposes in Digital Soil Mapping. Geoderma 2016, 265, 62–77. [Google Scholar] [CrossRef]
- Nussbaum, M.; Spiess, K.; Baltensweiler, A.; Grob, U.; Keller, A.; Greiner, L.; Schaepman, M.E.; Papritz, A. Evaluation of Digital Soil Mapping Approaches with Large Sets of Environmental Covariates. Soil 2018, 4, 1–22. [Google Scholar] [CrossRef]
- McBratney, A.B.; Mendonça Santos, M.L.; Minasny, B. On Digital Soil Mapping. Geoderma 2003, 117, 3–52. [Google Scholar] [CrossRef]
- Mulder, V.L.; de Bruin, S.; Schaepman, M.E.; Mayr, T.R. The Use of Remote Sensing in Soil and Terrain Mapping—A Review. Geoderma 2011, 162, 1–19. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Brungard, C.W.; Boettinger, J.L.; Duniway, M.C.; Wills, S.A.; Edwards, T.C. Machine Learning for Predicting Soil Classes in Three Semi-Arid Landscapes. Geoderma 2015, 239, 68–83. [Google Scholar] [CrossRef]
- Yang, R.M.; Zhang, G.L.; Liu, F.; Lu, Y.Y.; Yang, F.; Yang, F.; Yang, M.; Zhao, Y.G.; Li, D.C. Comparison of Boosted Regression Tree and Random Forest Models for Mapping Topsoil Organic Carbon Concentration in an Alpine Ecosystem. Ecol. Indic. 2016, 60, 870–878. [Google Scholar] [CrossRef]
- Were, K.; Bui, D.T.; Dick, Ø.B.; Singh, B.R. A Comparative Assessment of Support Vector Regression, Artificial Neural Networks, and Random Forests for Predicting and Mapping Soil Organic Carbon Stocks across an Afromontane Landscape. Ecol. Indic. 2015, 52, 394–403. [Google Scholar] [CrossRef]
- Heung, B.; Hodúl, M.; Schmidt, M.G. Comparing the Use of Training Data Derived from Legacy Soil Pits and Soil Survey Polygons for Mapping Soil Classes. Geoderma 2017, 290, 51–68. [Google Scholar] [CrossRef]
- Mansuy, N.; Thiffault, E.; Paré, D.; Bernier, P.; Guindon, L.; Villemaire, P.; Poirier, V.; Beaudoin, A. Digital Mapping of Soil Properties in Canadian Managed Forests at 250m of Resolution Using the K-Nearest Neighbor Method. Geoderma 2014, 235–236, 59–73. [Google Scholar] [CrossRef]
- Beguin, J.; Fuglstad, G.A.; Mansuy, N.; Paré, D. Predicting Soil Properties in the Canadian Boreal Forest with Limited Data: Comparison of Spatial and Non-Spatial Statistical Approaches. Geoderma 2017, 306, 195–205. [Google Scholar] [CrossRef]
- Wang, B.; Waters, C.; Orgill, S.; Gray, J.; Cowie, A.; Clark, A.; Liu, D.L. High Resolution Mapping of Soil Organic Carbon Stocks Using Remote Sensing Variables in the Semi-Arid Rangelands of Eastern Australia. Sci. Total Environ. 2018, 630, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M.; Wing, J.; Weston, S.; Williams, A.; Keefer, C.; Engelhardt, A.; Cooper, T.; Mayer, Z.; Ziem, A.; Scrucca, L.; et al. Package ’caret’: Classification and Regression Training. 2020. Available online: https://cran.r-project.org/web/packages/caret/caret.pdf (accessed on 16 April 2025).
- Lee, H.; Song, J. Introduction to Convolutional Neural Network Using Keras; An Understanding from a Statistician. Commun. Stat. Appl. Methods 2019, 26, 591–610. [Google Scholar] [CrossRef]
- Natesan, S.; Armenakis, C.; Vepakomma, U. Individual Tree Species Identification Using Dense Convolutional Network (Densenet) on Multitemporal RGB Images from UAV. J. Unmanned Veh. Syst. 2020, 8, 310–333. [Google Scholar] [CrossRef]
- Chauhan, R.; Ghanshala, K.K.; Joshi, R.C. Convolutional Neural Network (CNN) for Image Detection and Recognition. In Proceedings of the ICSCCC 2018—1st International Conference on Secure Cyber Computing and Communications, Jalandhar, India, 15–17 December 2018; pp. 278–282. [Google Scholar] [CrossRef]
- Simard, P.Y.; Steinkraus, D.; Platt, J.C. Best Practices for Convolutional Neural Networks Applied to Visual Document Analysis. In Proceedings of the International Conference on Document Analysis and Recognition, ICDAR 2003, Edinburgh, UK, 3–6 August 2003; pp. 958–963. [Google Scholar] [CrossRef]
- Ji, Y.; Zhang, H.; Zhang, Z.; Liu, M. CNN-Based Encoder-Decoder Networks for Salient Object Detection: A Comprehensive Review and Recent Advances. Inf. Sci. 2021, 546, 835–857. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, Y.; Kalra, M.K.; Lin, F.; Chen, Y.; Liao, P.; Zhou, J.; Wang, G. Low-Dose CT with a Residual Encoder-Decoder Convolutional Neural Network. IEEE Trans. Med. Imaging 2017, 36, 2524–2535. [Google Scholar] [CrossRef]
- Li, Q.; Li, Z.; Shangguan, W.; Wang, X.; Li, L.; Yu, F. Improving Soil Moisture Prediction Using a Novel Encoder-Decoder Model with Residual Learning. Comput. Electron. Agric. 2022, 195, 106816. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Z.; Li, Q.; Zhu, J. Enhancing Soil Moisture Forecasting Accuracy with REDF-LSTM: Integrating Residual En-Decoding and Feature Attention Mechanisms. Water 2024, 16, 1376. [Google Scholar] [CrossRef]
- Li, X.; Zhu, Y.; Li, Q.; Zhao, H.; Zhu, J.; Zhang, C. Interpretable Spatio-Temporal Modeling for Soil Temperature Prediction. Front. For. Glob. Chang. 2023, 6, 1295731. [Google Scholar] [CrossRef]
- Ke, Z.; Ren, S.; Yin, L. Advancing Soil Property Prediction with Encoder-Decoder Structures Integrating Traditional Deep Learning Methods in Vis-NIR Spectroscopy. Geoderma 2024, 449, 117006. [Google Scholar] [CrossRef]
- Liu, L.; Yang, B.; Zhang, Y. Inverting Magnetotelluric Data Using a Physics-Guided Auto-Encoder with Scaling Laws Extension. Front. Earth Sci. 2024, 12, 1510962. [Google Scholar] [CrossRef]
- Bai, G.; Liao, C.; Liu, Y.; Cheng, Y.F. Far-Field Phaseless Diagnosis for Impaired Arrays Based on Artificial Neural Networks and Compressed Sensing. IEEE Trans. Antennas Propag. 2024, 72, 1581–1592. [Google Scholar] [CrossRef]
- Hu, B.; Jung, W.M.; Liu, J.; Shang, J. RETRIEVAL OF LEAF AREA INDEX AND LEAF CHLOROPHYLL CONTENT FROM HYPERSPECTRAL DATA USING DEEP LEARNING NETWORKS. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2022, 43, 397–404. [Google Scholar] [CrossRef]
- Engler, R.; Waser, L.T.; Zimmermann, N.E.; Schaub, M.; Berdos, S.; Ginzler, C.; Psomas, A. Combining Ensemble Modeling and Remote Sensing for Mapping Individual Tree Species at High Spatial Resolution. For. Ecol. Manag. 2013, 310, 64–73. [Google Scholar] [CrossRef]
- Huang, J.; Gao, J. An Ensemble Simulation Approach for Artificial Neural Network: An Example from Chlorophyll a Simulation in Lake Poyang, China. Ecol. Inform. 2017, 37, 52–58. [Google Scholar] [CrossRef]
- Maraun, D. Bias Correction, Quantile Mapping, and Downscaling: Revisiting the Inflation Issue. J. Clim. 2013, 26, 2137–2143. [Google Scholar] [CrossRef]
- Airborne Imaging. Final Report for Project Cochrane LiDAR; Airborne Imaging: Calgary, AB, Canada, 2018. [Google Scholar]
- Environment Canada Canadian Climate Normals 1981–2010 Station Data: Mattice TCPL Ontario. Available online: https://climate.weather.gc.ca/climate_normals/results_e.html?searchType=stnProx&txtRadius=100&selCity=&selPark=&optProxType=custom&txtCentralLatDeg=49&txtCentralLatMin=30&txtCentralLatSec=00&txtCentralLongDeg=84&txtCentralLongMin=00&txtCentralLongSec=00&t (accessed on 26 June 2023).
- Environment Canada Canadian Climate Normals 1981–2010 Station Data: Kapuskasing A. Available online: https://climate.weather.gc.ca/climate_normals/results_1981_2010_e.html?searchType=stnName&txtStationName=Kapuskasing&searchMethod=contains&txtCentralLatMin=0&txtCentralLatSec=0&txtCentralLongMin=0&txtCentralLongSec=0&stnID=4157&dispBack=0 (accessed on 1 July 2020).
- Environment Canada Canadian Climate Normals 1981–2010 Station Data: Cochrane Ontario. Available online: https://climate.weather.gc.ca/climate_normals/results_e.html?searchType=stnProv&lstProvince=ON&txtCentralLatMin=0&txtCentralLatSec=0&txtCentralLongMin=0&txtCentralLongSec=0&stnID=4142&dispBack=0 (accessed on 26 June 2023).
- Kroetsch, D.J.; Geng, X.; Chang, S.X.; Saurette, D.D. Organic Soils of Canada: Part 1. Wetland Organic Soils. Can. J. Soil. Sci. 2011, 91, 807–822. [Google Scholar] [CrossRef]
- Pittman, R.; Hu, B. Soil Sampling Protocol for York University Field Campaigns; Department of Earth and Space Science and Engineering, York University: Toronto, ON, Canada, 2024. [Google Scholar]
- Pittman, R.; Hu, B.; Pittman, T.; Webster, K.L.; Shang, J.; Nelson, S.A. Inferential Approach for Evaluating the Association Between Land Cover and Soil Carbon in Northern Ontario. Earth 2025, 6, 1. [Google Scholar] [CrossRef]
- Pittman, R.; Hu, B. Constructing Rasterized Covariates from LiDAR Point Cloud Data via Structured Query Language. Proceedings 2024, 110, 1. [Google Scholar] [CrossRef]
- Conrad, O.; Bechtel, B.; Bock, M.; Dietrich, H.; Fischer, E.; Gerlitz, L.; Wehberg, J.; Wichmann, V.; Böhner, J. System for Automated Geoscientific Analyses (SAGA) v. 2.1.4. Geosci. Model. Dev. 2015, 8, 1991–2007. [Google Scholar] [CrossRef]
- Fradette, O.; Marty, C.; Faubert, P.; Dessureault, P.L.; Paré, M.; Bouchard, S.; Villeneuve, C. Additional Carbon Sequestration Potential of Abandoned Agricultural Land Afforestation in the Boreal Zone: A Modelling Approach. For. Ecol. Manag. 2021, 499, 119565. [Google Scholar] [CrossRef]
- Beaudoin, A.; Bernier, P.Y.; Villemaire, P.; Guindon, L.; Guo, X.J. Tracking Forest Attributes across Canada between 2001 and 2011 Using a k Nearest Neighbors Mapping Approach Applied to MODIS Imagery. Can. J. For. Res. 2018, 48, 85–93. [Google Scholar] [CrossRef]
- Chollet, F. Keras 2015. Available online: https://github.com/keras-team/keras (accessed on 16 April 2025).
- Li, D.; Li, L. Detection of Water PH Using Visible Near-Infrared Spectroscopy and One-Dimensional Convolutional Neural Network. Sensors 2022, 22, 5809. [Google Scholar] [CrossRef]
- Sindi, H.; Nour, M.; Rawa, M.; Öztürk, Ş.; Polat, K. Random Fully Connected Layered 1D CNN for Solving the Z-Bus Loss Allocation Problem. Measurement 2021, 171, 108794. [Google Scholar] [CrossRef]
- Ajayi, O.G.; Ashi, J. Effect of Varying Training Epochs of a Faster Region-Based Convolutional Neural Network on the Accuracy of an Automatic Weed Classification Scheme. Smart Agric. Technol. 2023, 3, 100128. [Google Scholar] [CrossRef]
- Kim, M.; Lee, W. Deep Spread Multiplexing and Study of Training Methods for DNN-Based Encoder and Decoder. Sensors 2023, 23, 3848. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by RandomForest. R News 2002, 2, 18–22. [Google Scholar]
- Rosseel, Y. Lavaan: An R Package for Structural Equation Modeling. J. Stat. Softw. 2012, 48, 1–36. [Google Scholar] [CrossRef]
- Nakagawa, S.; Schielzeth, H. A General and Simple Method for Obtaining R2 from Generalized Linear Mixed-Effects Models. Methods Ecol. Evol. 2013, 4, 133–142. [Google Scholar] [CrossRef]
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. Dplyr: A Grammar of Data Manipulation. 2023. Available online: https://dplyr.tidyverse.org/ (accessed on 16 April 2025).
- Tao, Z.; Huiling, L.; Wenwen, W.; Xia, Y. GA-SVM Based Feature Selection and Parameter Optimization in Hospitalization Expense Modeling. Appl. Soft Comput. J. 2019, 75, 323–332. [Google Scholar] [CrossRef]
- Wadoux, A.M.J.C.; Heuvelink, G.B.M.; de Bruin, S.; Brus, D.J. Spatial Cross-Validation Is Not the Right Way to Evaluate Map Accuracy. Ecol. Modell. 2021, 457, 109692. [Google Scholar] [CrossRef]
- Adin, A.; Krainski, E.T.; Lenzi, A.; Liu, Z.; Martínez-Minaya, J.; Rue, H. Automatic Cross-Validation in Structured Models: Is It Time to Leave out Leave-One-Out? Spat. Stat. 2024, 62, 100843. [Google Scholar] [CrossRef]
- El Abidine, A.Z.; Bernier, P.Y.; Stewart, J.D.; Plamondon, A.P. Water Stress Preconditioning of Black Spruce Seedlings from Lowland and Upland Sites. Can. J. Bot. 1994, 72, 1511–1518. [Google Scholar] [CrossRef]
- Zhang, Q.; Min, B.; Hang, Y.; Chen, H.; Qiu, J. A Full-Scale Lung Image Segmentation Algorithm Based on Hybrid Skip Connection and Attention Mechanism. Sci. Rep. 2024, 14, 23233. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, C.; Ding, W.; Sun, S.; Yue, X.; Fujita, H. Target-Aware U-Net with Fuzzy Skip Connections for Refined Pancreas Segmentation. Appl. Soft Comput. 2022, 131, 109818. [Google Scholar] [CrossRef]


| Predictor | Source | Soil Formation Factor |
|---|---|---|
| Digital elevation model (DEM) * | LiDAR | Relief |
| Canopy height model (CHM) | LiDAR | Vegetation |
| Gap fraction | LiDAR | Vegetation |
| Aspect | DEM (LiDAR) | Relief |
| Convergence index | DEM (LiDAR) | Relief |
| Mid-slope position * | DEM (LiDAR) | Relief |
| Multi-resolution ridge top flatness (MRRTF) | DEM (LiDAR) | Relief |
| Multi-resolution valley bottom flatness (MRVBF) | DEM (LiDAR) | Relief |
| SAGA topographic wetness index (SAGA TWI) | DEM (LiDAR) | Relief |
| Slope * | DEM (LiDAR) | Relief |
| Slope height * | DEM (LiDAR) | Relief |
| Slope length | DEM (LiDAR) | Relief |
| Stream power index | DEM (LiDAR) | Relief |
| Terrain ruggedness index (TRI) | DEM (LiDAR) | Relief |
| Topographic wetness index (TWI) | DEM (LiDAR) | Relief |
| Total curvature | DEM (LiDAR) | Relief |
| Valley depth | DEM (LiDAR) | Relief |
| Visible sky | DEM (LiDAR) | Relief |
| B1 summer 2017 | SR | Vegetation |
| B2 summer 2017 | SR | Vegetation |
| B3 summer 2017 | SR | Vegetation |
| B4 summer 2017 | SR | Vegetation |
| B5 summer 2017 | SR | Vegetation |
| B6 summer 2017 | SR | Vegetation |
| B7 summer 2017 | SR | Vegetation |
| B10 summer 2017 | SR | Vegetation |
| Modified normalized difference water index (MNDWI) summer 2017 | SR | Relief (water) |
| Normalized difference vegetation index (NDVI) summer 2017 | SR | Vegetation |
| Change magnitude B3 B4 B5 summer 1984–2005 * | SR | Time |
| SAR C VH May 2017 | SAR | Relief (water) |
| SAR C VV May 2017 * | SAR | Relief (water) |
| Gravity anomaly 2016 | Aeromagnetic | Parent material |
| Magnetic residual November 2018 | Aeromagnetic | Parent material |
| NFI black spruce 2011 * | k-NN model | Vegetation |
| ED-DNN | ED-CNN | |||||
|---|---|---|---|---|---|---|
| Layer | Output Shape | # Parameters | Layer | Output Shape | # Parameters | |
| Encoder: | Encoder: | |||||
| Input layer | 34 | 0 | Input layer | (1, 34) | 0 | |
| Dense | 32 | 1120 | Convolution 1-D | (1, 16) | 560 | |
| Dense | 16 | 528 | Max pooling 1-D | (1, 16) | 0 | |
| Dense | 8 | 136 | Convolution 1-D | (1, 32) | 544 | |
| Decoder: | Flatten | 32 | 0 | |||
| Input layer | 8 | 0 | Dense | 8 | 264 | |
| Dense | 32 | 288 | Decoder: | |||
| Dense | 16 | 528 | Input layer | 8 | 0 | |
| Dense | 1 | 17 | Dense | 64 | 576 | |
| Reshape | (1, 64) | 0 | ||||
| Total parameters: | 2617 | Convolution 1-D | (1, 32) | 2080 | ||
| Upsampling 1-D | (2, 32) | 0 | ||||
| Convolution 1-D | (2, 16) | 528 | ||||
| Flatten | 32 | 0 | ||||
| Dense | 16 | 528 | ||||
| Dense | 1 | 17 | ||||
| Total parameters: | 5097 |
| Model | R2 | RMSE | MAE | |
|---|---|---|---|---|
| [%] | [%] | |||
| Structural equation model | SEM | 0.21 | 10.18 | 7.63 |
| Random forest | RF | 0.22 | 10.09 | 7.83 |
| Dense neural network | DNN | 0.18 | 10.38 | 7.35 |
| Convolutional neural network 1-D | CNN | 0.37 | 9.07 | 6.05 |
| Encoder-decoder DNN | ED-DNN | 0.54 | 7.78 | 5.40 |
| Encoder-decoder CNN 1-D | ED-CNN | 0.59 | 7.30 | 5.38 |
| Ensemble * | AVG | 0.50 | 8.06 | 6.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pittman, R.; Hu, B. Enhanced Prediction of Soil Carbon via Encoder-Decoder Neural Networks for a Boreal Study Area in Northern Ontario. Sensors 2025, 25, 2583. https://doi.org/10.3390/s25082583
Pittman R, Hu B. Enhanced Prediction of Soil Carbon via Encoder-Decoder Neural Networks for a Boreal Study Area in Northern Ontario. Sensors. 2025; 25(8):2583. https://doi.org/10.3390/s25082583
Chicago/Turabian StylePittman, Rory, and Baoxin Hu. 2025. "Enhanced Prediction of Soil Carbon via Encoder-Decoder Neural Networks for a Boreal Study Area in Northern Ontario" Sensors 25, no. 8: 2583. https://doi.org/10.3390/s25082583
APA StylePittman, R., & Hu, B. (2025). Enhanced Prediction of Soil Carbon via Encoder-Decoder Neural Networks for a Boreal Study Area in Northern Ontario. Sensors, 25(8), 2583. https://doi.org/10.3390/s25082583







