Rapid Path Planning Algorithm for Percutaneous Rigid Needle Biopsy Based on Optical Illumination Principles
Abstract
1. Introduction
- (1)
- Image Acquisition and Target Identification: High-resolution CT images are acquired and imported into the system, where either the operator or dedicated software delineates the puncture target and maps the surrounding anatomical structures. This step includes assessing tissue traversability to ensure that the planned trajectory avoids regions that could trigger adverse or even fatal complications.
- (2)
- Trajectory Planning: Given the rigidity of the needle, the system identifies one or more safe entry points in three-dimensional space relative to the target. It then computes a straight-line trajectory that avoids critical structures (e.g., bones, blood vessels, airways, and fissures), thereby ensuring a safe path for needle insertion.
- (3)
- Physical Trajectory Establishment and Execution: The virtual path is transferred to the physical environment, where the needle is advanced along the pre-planned trajectory either manually by the operator or automatically via robotic control, ensuring precise needle placement.
- (1)
- Ultra-fast Path Computation: Leveraging optical principles and GPU acceleration, our method achieves ultra-fast path calculations by simulating light absorption and reflection, offering a computationally lightweight solution that circumvents complex geometric computations.
- (2)
- Simplified Vascular Modeling: Our approach simplifies vascular modeling by assigning light absorption parameters to vascular voxels, accumulating vascular thickness along the path without explicit vessel segmentation, thus balancing computational efficiency and vascular safety.
2. Materials and Methods
2.1. Constraints of Rigid Needle
2.1.1. Hard Constraints
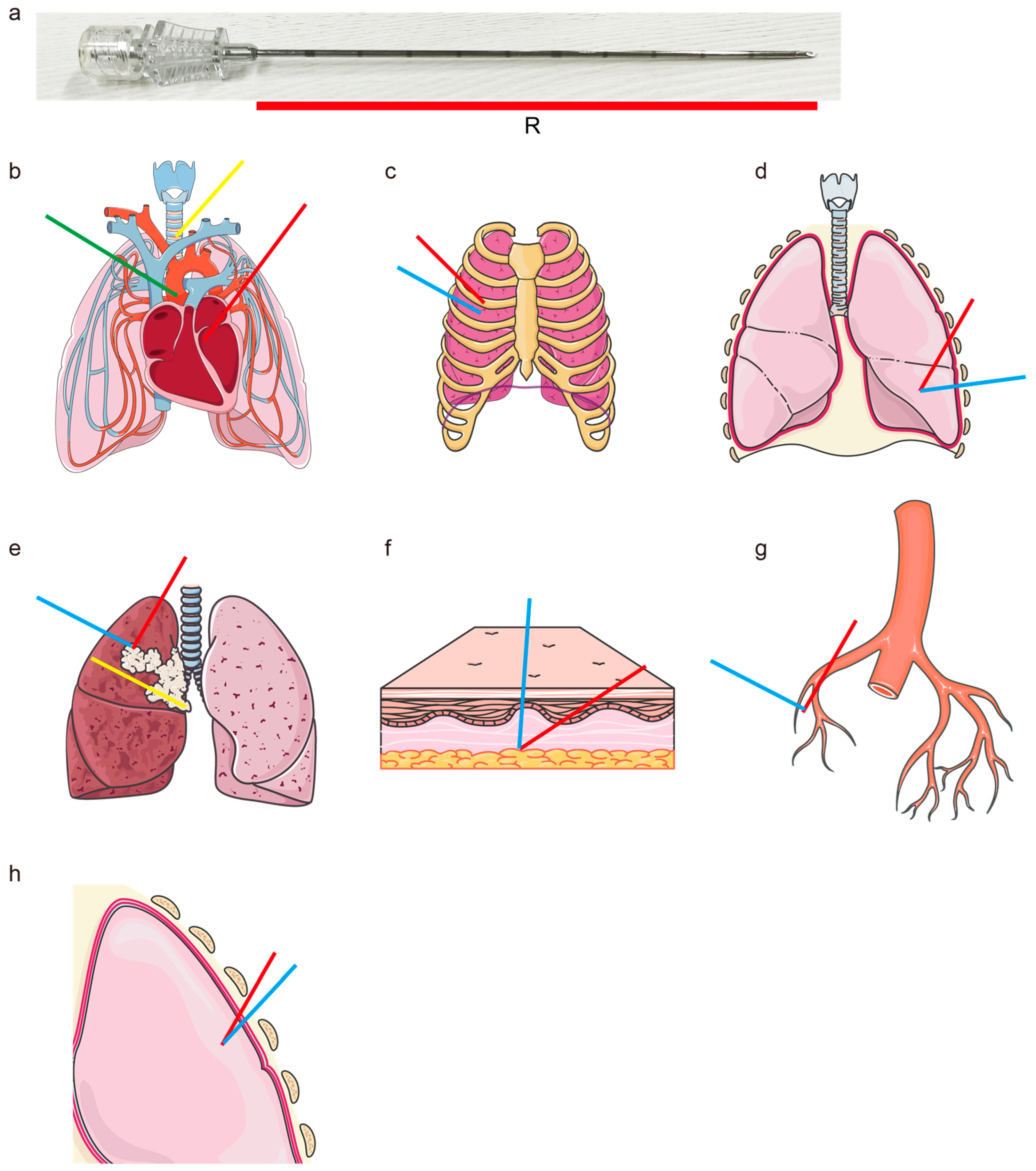
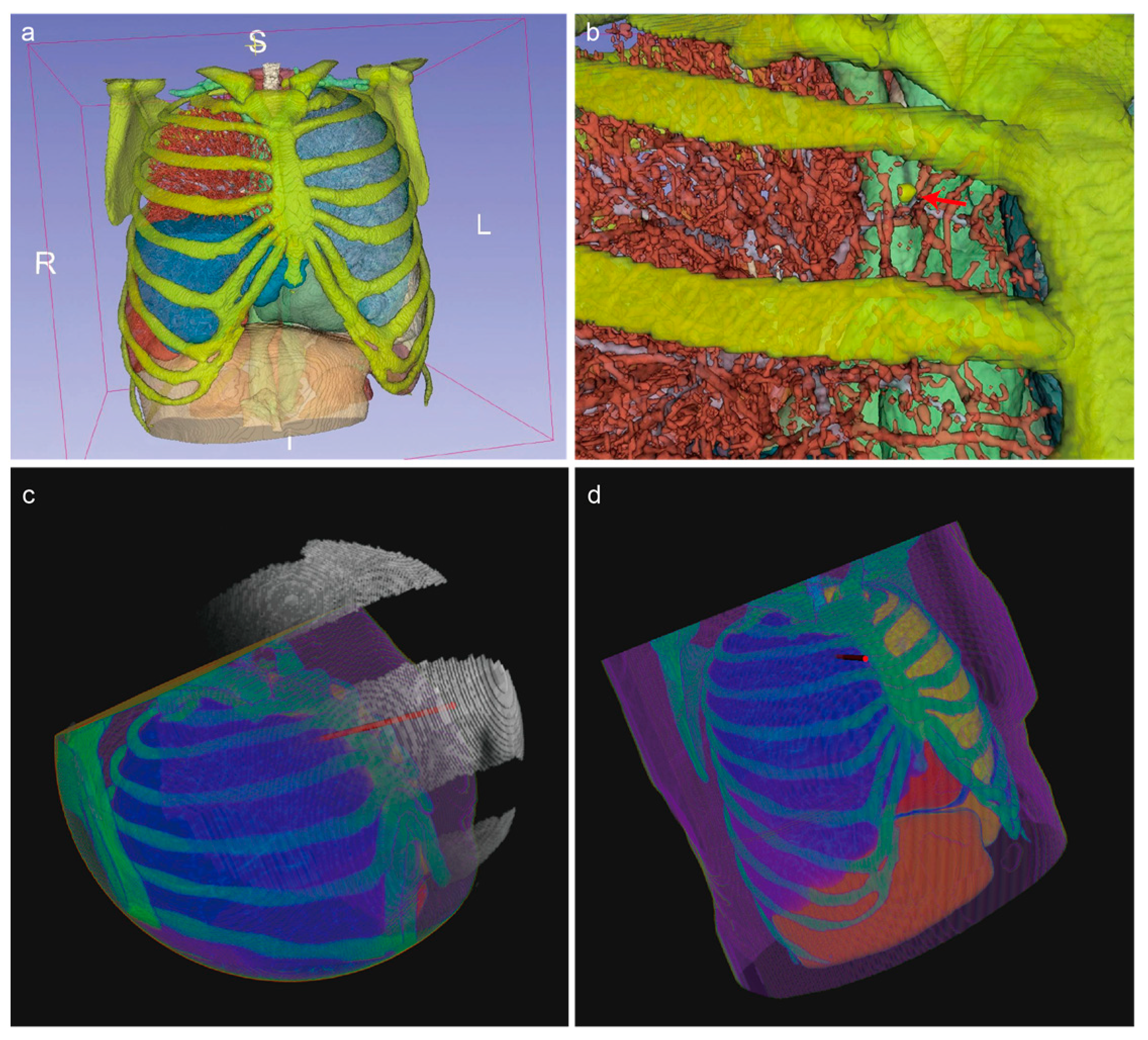
- Straight Path for Rigid Needle: The path must be straight to accommodate the rigidity of the needle (Figure 1a).
- Avoidance of Vital Organs/Tissues: The path must avoid the heart and major blood vessels to prevent severe complications (Figure 1b).
- Avoidance of Bones: The needle must not penetrate the ribs or other bones to prevent fractures or physical obstruction (Figure 1c).
- Only Pass Through Target Lung Lobe: The path must stay within the target lung lobe and avoid crossing into non-target lung lobes, ensuring no traversal of interlobar fissures (Figure 1d).
- Path Length Must Be Less Than Needle Length: The path length must be within the physical limits of the needle to ensure it can reach the target site effectively (Figure 1e).
2.1.2. Soft Constraints
- Minimize Path Length: Shorter paths are preferred to reduce the risk of complications (Figure 1e).
- Maximize Angle with Skin: The angle of insertion with the skin should be as large as possible for easier needle insertion (Figure 1f).
- Prefer Smaller and Fewer Vessels: If it is necessary to pass through vessels, the path should prefer smaller and fewer vessels to minimize the risk of bleeding or other complications (Figure 1g).
- Maximize Distance from Vital Organs/Tissues: The path should be optimized to maintain the maximum feasible distance from vital organs and tissues, thereby minimizing the risk of unintended damage (Figure 1h).
2.2. Segmentation of Organs, Tissues, and Lesions
2.2.1. Datasets
2.2.2. Data Annotation and Training
2.2.3. Validation of Segmentation Models
2.2.4. Robustness Test of Segmentation Models
2.3. Principles of an Optical Illumination-Based Path Planning Algorithm
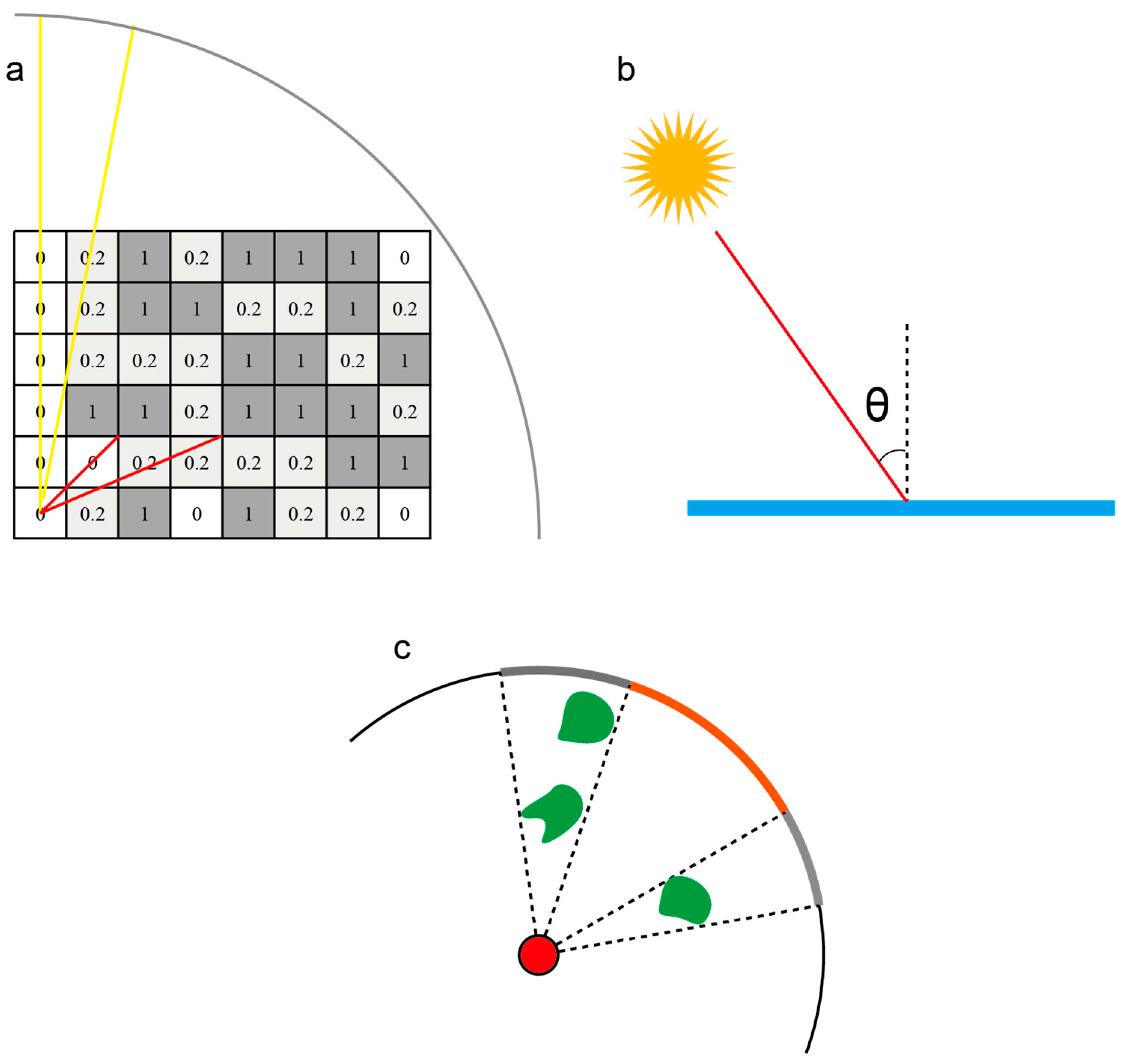
2.3.1. Basic Concept
2.3.2. Biopsy Needle Length Constraint
- 1.
- The tail is outside the skin: .
- 2.
- The line segment from the tail to intersects the skin surface .
- 3.
- The tail position is within the visible skin region of the CT scan.
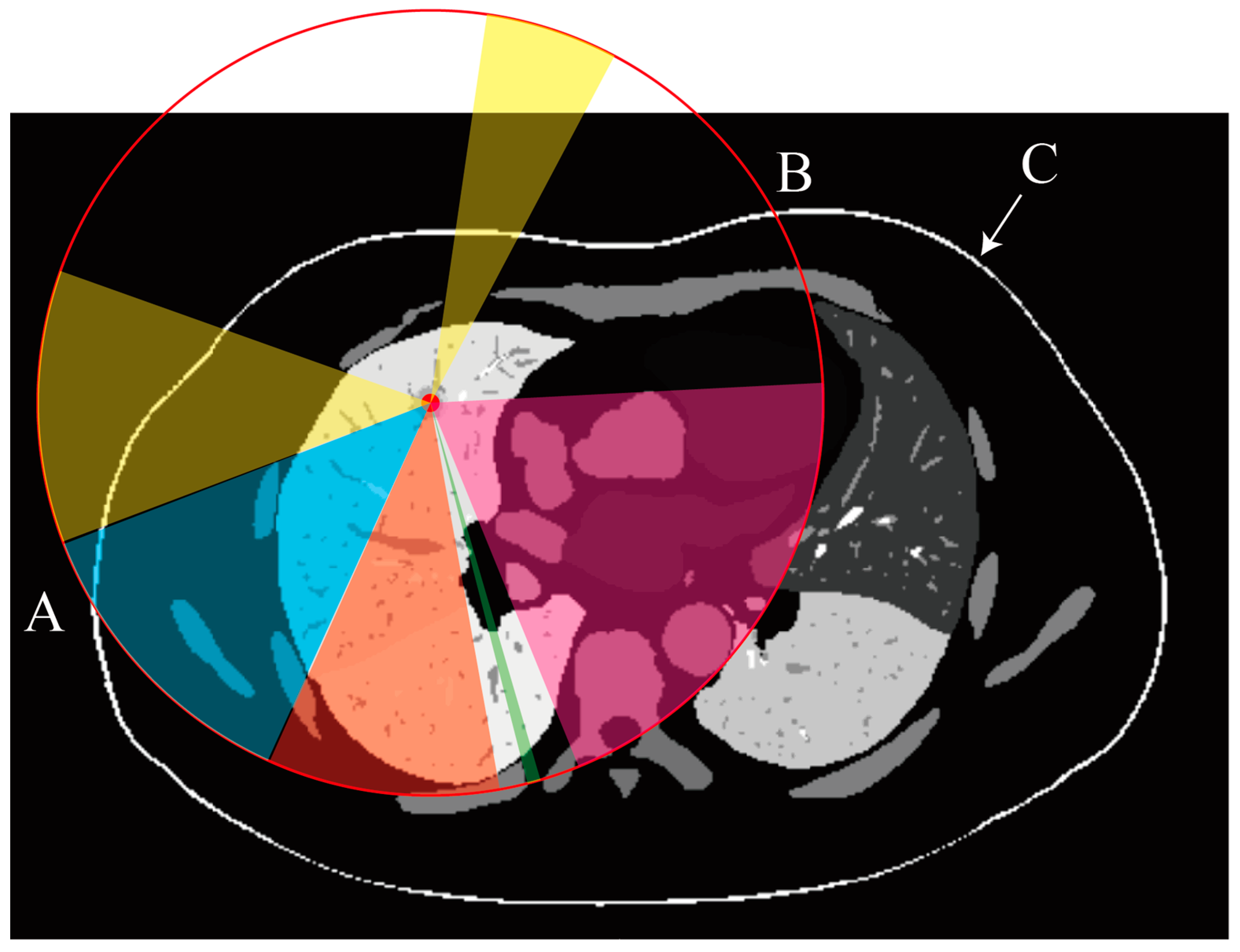
2.3.3. Organ and Vessel Avoidance and Depth Regulation
- a.
- Organ Avoidance
- b.
- Vessel Risk Assessment
- c.
- Puncture Depth Regulation
- d.
- Integration into Path Evaluation
2.3.4. Skin Angle Constraints
2.3.5. Constraints on Distance from Obstacles
2.3.6. Final Light Intensity Model
2.4. Implementation of the Optical Illumination-Inspired Path Planning Algorithm
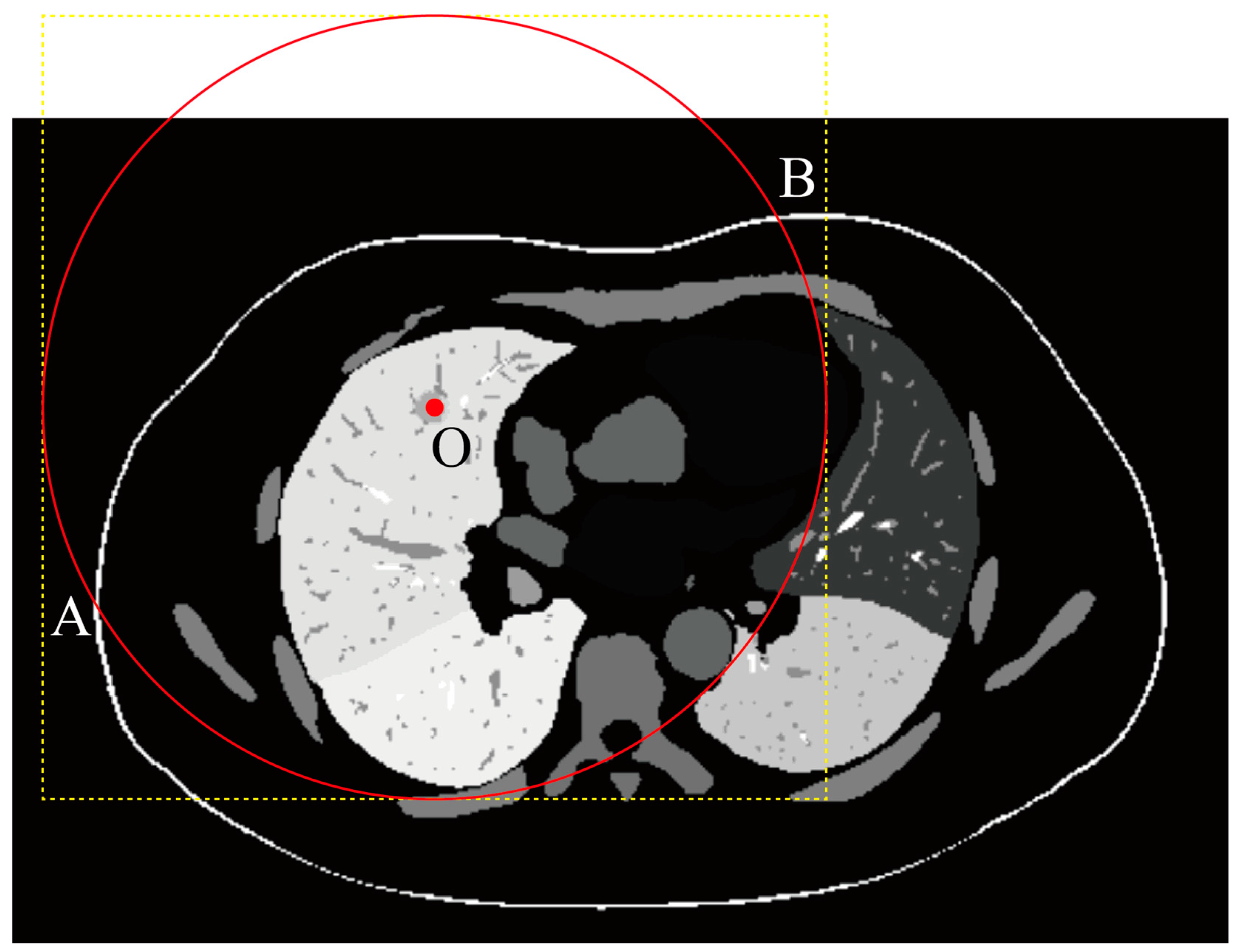
2.4.1. Segmentation Mask Acquisition
2.4.2. Mask Resampling
2.4.3. ROI Cropping
2.4.4. No-Skin Region Removal (Optional Step)
2.4.5. Intensity Calculation
2.4.6. Optimal Path Recommendation
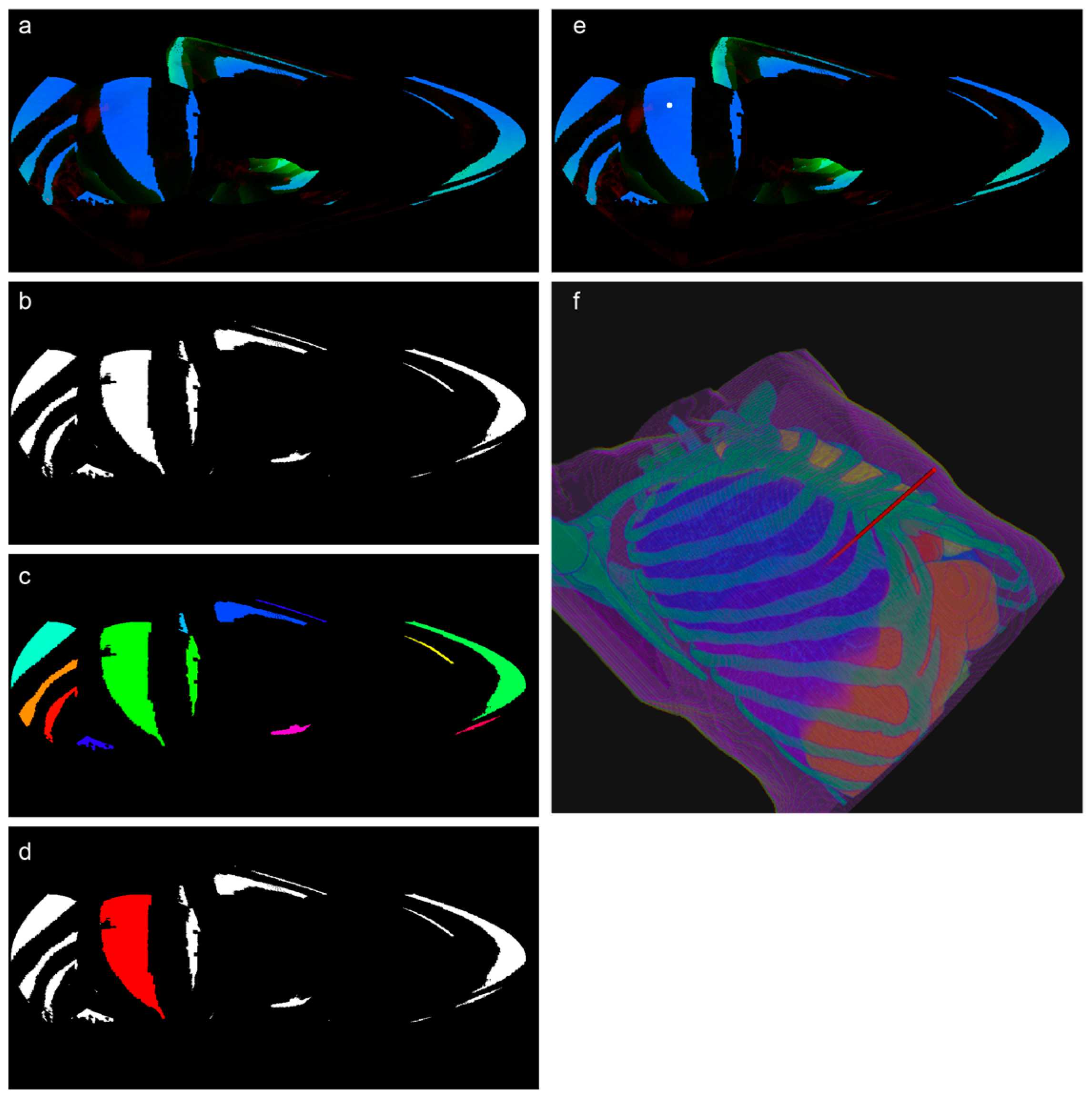
- a.
- Illumination Map Generation and RGB Projection
- b.
- Region Segmentation and Expert Annotation
- c.
- Feature Extraction and Dataset Preparation
- d.
- Machine Learning Model Training and Evaluation for Region Selection
- Random Forest: The random forest model was configured with 100 trees to ensure robust performance. The maximum depth of each tree was limited to 10 levels to prevent overfitting. To address class imbalance, the class_weight parameter was set to “balanced”, which automatically adjusts weights inversely proportional to class frequencies.
- XGBoost: The XGBoost model was trained using 100 boosting rounds. The maximum depth of each tree was set to 6 levels. To handle class imbalance, the scale_pos_weight parameter was used, which was set to the ratio of negative to positive samples in the resampled training data. The evaluation metric was set to log loss for binary classification.
- LightGBM: The LightGBM model was configured with 100 boosting rounds. To address class imbalance, the class_weight parameter was set to “balanced”.
- CatBoost: class imbalance was addressed using the class_weights parameter, with weights explicitly set to [1, ratio of negative to positive samples in resampled training data] to balance class influence.
- MLP Simple: Serving as a baseline shallow network, this model comprised a single hidden layer of 100 neurons. This architecture was designed to establish a performance reference point against which more complex models could be compared.
- MLP Deep: To investigate the influence of network depth, we implemented a two-hidden-layer MLP. The architecture consisted of a first hidden layer with 100 neurons, followed by a subsequent layer of 50 neurons. This configuration aimed to determine whether increased depth, while maintaining a constrained number of neurons per layer, could enhance feature representation and, consequently, model performance.
- MLP Wide: Complementing the exploration of depth, the “MLP Wide” model assessed the impact of network width. This architecture utilized a single hidden layer, but with an expanded capacity of 200 neurons. This design sought to evaluate whether a wider hidden layer, affording increased representational capacity within a single layer, would yield performance advantages.
- MLP Complex: A more elaborate architecture, “MLP Complex”, was implemented to integrate both increased depth and width. This model incorporated three hidden layers with a progressively refined neuron count: 150, 75, and 30 neurons in successive layers. This configuration was designed to examine the potential benefits of a more intricate network structure capable of hierarchical feature learning and capturing potentially nuanced relationships within the data.
- e.
- Final Path Recommendation Algorithm Testing
- Area Under the Receiver Operating Characteristic Curve (AUC-ROC): this assesses the overall classification ability, balancing sensitivity and specificity.
- Precision, Recall, and F1-score: these measure the quality of positive predictions and account for class imbalance.
- Specificity: this evaluates the ability to accurately identify negative regions.
- Sensitivity: this evaluates the ability to accurately identify positive regions.
- Confusion Matrix Components: this provides a detailed breakdown of the prediction outcomes, including TP, FP, TN, and FN.
3. Results
3.1. Segmentation Model Performance
3.1.1. Model Training and Setup
3.1.2. Quantitative Segmentation Accuracy Comparison
3.1.3. Ablation Analysis of Segmentation Experiments
3.1.4. Segmentation Robustness Analysis
3.2. Path Planning and Recommendation Algorithm Performance
3.2.1. Quantitative Evaluation of Path Planning and Machine Learning-Based Recommendation Algorithms
3.2.2. Evaluation of Biopsy Path Parameters and Clinical Feasibility
3.2.3. Ablation and Comparative Study
4. Discussion
4.1. Key Findings
4.2. Comparative Analysis with Existing Methods
4.2.1. Computation Speed
4.2.2. Accuracy and Performance
4.2.3. Novel Vascular Modeling
4.2.4. Multi-Objective Optimization
4.3. Limitations
- (1)
- Small Nodule Size: Lung nodules were considerably smaller in volume compared with other organs, rendering them highly susceptible to segmentation errors. Even a deviation of 1–2 voxels could lead to a substantial increase in both the ASD and HD95 metrics [46].
- (2)
- Boundary Ambiguity and Irregular Shapes: Lung nodules often present ambiguous boundaries and irregular shapes, posing a significant challenge for accurate segmentation. This was particularly pronounced for attached nodules and ground-glass nodules, where boundaries could be connected to surrounding structures such as blood vessels or lung lobes, thereby increasing segmentation errors [47,48].
- (3)
- Annotation Format of the LUNA16 Dataset: The annotation format inherent to the LUNA16 dataset introduced limitations in voxel-level accuracy. This dataset, used for training and evaluation, provides annotations in CSV format, defining nodules as spheres using center coordinates and diameters rather than precise voxel-level delineations [39]. This spherical CSV annotation can introduce false positive voxels when compared with a truly precise voxel-level ground truth.
4.4. Future Directions
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ROI | Region of Interest |
| DSC | Dice Similarity Coefficient |
| HD | Hausdorff Distance |
| ASD | Average Surface Distance |
References
- Lanouzière, M.; Varbédian, O.; Chevallier, O.; Griviau, L.; Guillen, K.; Popoff, R.; Aho-Glélé, S.-L.; Loffroy, R. Computed Tomography-Navigation™ Electromagnetic System Compared to Conventional Computed Tomography Guidance for Percutaneous Lung Biopsy: A Single-Center Experience. Diagnostics 2021, 11, 1532. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Song, B.; Zhang, Z.; Bo, B.; Xiong, A.; Ye, L.; Xie, D.; Li, J.; Zhao, S.; Cai, C.; et al. Evaluating efficacy and safety of a novel registration-free CT-guided needle biopsy navigation system (RC 120): A multicenter, prospective clinical trial. Lung Cancer 2024, 198, 108025. [Google Scholar] [CrossRef] [PubMed]
- Thandra, K.C.; Barsouk, A.; Saginala, K.; Aluru, J.S.; Barsouk, A. Epidemiology of lung cancer. Contemp. Oncol. 2021, 25, 45–52. [Google Scholar] [CrossRef]
- Klein, A.P. Pancreatic cancer epidemiology: Understanding the role of lifestyle and inherited risk factors. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 493–502. [Google Scholar] [CrossRef]
- Liu, Q.; Zhou, G.; Zhong, J.; Tang, L.; Lu, Y.; Qin, J.; He, L.; Zhang, J. Path planning for percutaneous lung biopsy based on the loose-Pareto and adaptive heptagonal optimization method. Med. Biol. Eng. Comput. 2023, 61, 1449–1472. [Google Scholar] [CrossRef] [PubMed]
- Too, C.W.; Fong, K.Y.; Hang, G.; Sato, T.; Nyam, C.Q.; Leong, S.H.; Ng, K.W.; Ng, W.L.; Kawai, T. Artificial Intelligence-Guided Segmentation and Path Planning Software for Transthoracic Lung Biopsy. J. Vasc. Interv. Radiol. 2024, 35, 780–789.E1. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wu, S.C.; Wu, W.W.; Gao, H.J.; Zhou, Z.H. Computer-assisted needle trajectory planning and mathematical modeling for liver tumor thermal ablation: A review. Math. Biosci. Eng. 2019, 16, 4846–4872. [Google Scholar] [CrossRef]
- Song, Z.; Ding, F.; Wu, W.; Zhou, Z.; Wu, S. Design of Path-Planning System for Interventional Thermal Ablation of Liver Tumors Based on CT Images. Sensors 2024, 24, 3537. [Google Scholar] [CrossRef]
- Bourgouin, P.P.; Rodriguez, K.J.; Fintelmann, F.J. Image-Guided Percutaneous Lung Needle Biopsy: How we do it. Tech. Vasc. Interv. Radiol. 2021, 24, 100770. [Google Scholar] [CrossRef]
- Du, K.; Liu, Y.; Wu, K.; Sun, Z.; Han, X.; Jiao, D. Percutaneous microwave ablation for lung tumors: A retrospective case-control study of conventional CT and C-arm CT guidance. Quant. Imaging Med. Surg. 2023, 13, 5737–5747. [Google Scholar] [CrossRef]
- Fan, H.; Xie, X.; Pang, Z.; Zhang, L.; Ding, R.; Wan, C.; Li, X.; Yang, Z.; Sun, J.; Kan, X.; et al. Risk assessment of pneumothorax in colorectal lung metastases treated by percutaneous thermal ablation: A multicenter retrospective cohort study. Int. J. Surg. 2024, 110, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.Y.; Tsai, S.C.; Wu, T.C.; Lin, F.C. Puncture frequency predicts pneumothorax in preoperative computed tomography-guided lung nodule localization for video-assisted thoracoscopic surgery. Thorac. Cancer 2022, 13, 1925–1932. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jiang, Y.; He, R.; Cui, F.; Lin, Y.; Xu, K.; Cai, W.; Hao, Z.; Zeng, Y.; Chen, X.; et al. Robotic-assisted navigation system for preoperative lung nodule localization: A pilot study. Transl. Lung Cancer Res. 2023, 12, 2283–2293. [Google Scholar] [CrossRef]
- Putzer, D.; Arco, D.; Schamberger, B.; Schanda, F.; Mahlknecht, J.; Widmann, G.; Schullian, P.; Jaschke, W.; Bale, R. Comparison of Two Electromagnetic Navigation Systems for CT-Guided Punctures: A Phantom Study. Rofo 2016, 188, 470–478. [Google Scholar] [CrossRef]
- Li, R.; An, C.; Wang, S.; Wang, G.; Zhao, L.; Yu, Y.; Wang, L. A heuristic method for rapid and automatic radiofrequency ablation planning of liver tumors. Int. J. Comput. Assist. Radiol. Surg. 2023, 18, 2213–2221. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Jiang, H.; Shi, T. Multi-stage puncture path planning algorithm of ablation needles for percutaneous radiofrequency ablation of liver tumors. Comput. Biol. Med. 2022, 145, 105506. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, Y.; Gong, Y.; Zhang, J.; He, L.; Zhong, J.; Tang, L. A lung biopsy path planning algorithm based on the double spherical constraint Pareto and indicators’ importance-correlation degree. Comput. Med. Imaging Graph. 2024, 117, 102426. [Google Scholar] [CrossRef]
- Seitel, A.; Engel, M.; Sommer, C.M.; Radeleff, B.A.; Essert-Villard, C.; Baegert, C.; Fangerau, M.; Fritzsche, K.H.; Yung, K.; Meinzer, H.P.; et al. Computer-assisted trajectory planning for percutaneous needle insertions. Med. Phys. 2011, 38, 3246–3259. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Campos-Nanez, E.; Yaniv, Z.; Banovac, F.; Abeledo, H.; Hata, N.; Cleary, K. Treatment planning and image guidance for radiofrequency ablation of large tumors. IEEE J. Biomed. Health Inform. 2014, 18, 920–928. [Google Scholar] [CrossRef]
- Jing, Y.; Jing, J.; Liu, J.; Zhang, J.; Jin, Y.; Bai, X. The clinical performance of robotic assisted navigation system versus conventional freehand technique for percutaneous transthoracic needle biopsy. Sci. Rep. 2025, 15, 5980. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, W.; Wu, S.; Zhou, Z.; Zhang, H. An Automatic Needle Puncture Path-Planning Method for Thermal Ablation of Lung Tumors. Diagnostics 2024, 14, 215. [Google Scholar] [CrossRef]
- Li, J.; Gao, H.; Shen, N.; Wu, D.; Feng, L.; Hu, P. High-security automatic path planning of radiofrequency ablation for liver tumors. Comput. Methods Programs Biomed. 2023, 242, 107769. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, Y.; Anvari, A.; Samir, A.E.; Arellano, R.S.; Prabhakar, A.M.; Uppot, R.N. Navigational Guidance and Ablation Planning Tools for Interventional Radiology. Curr. Probl. Diagn. Radiol. 2017, 46, 225–233. [Google Scholar] [CrossRef]
- Levin, A.A.; Klimov, D.D.; Nechunaev, A.A.; Vorotnikov, A.A.; Prokhorenko, L.S.; Grigorieva, E.V.; Astakhov, D.A.; Poduraev, Y.V.; Panchenkov, D.N. The comparison of the process of manual and robotic positioning of the electrode performing radiofrequency ablation under the control of a surgical navigation system. Sci. Rep. 2020, 10, 8612. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, J.; Han, P.; Chen, X.Z.; Zhang, Y.; Li, W.; Qin, J.; He, L. Path planning algorithm for percutaneous puncture lung mass biopsy procedure based on the multi-objective constraints and fuzzy optimization. Phys. Med. Biol. 2024, 69, 095006. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Cao, M.; Gu, F.; Gong, W.; Cai, Q. Method for puncture trajectory planning in liver tumors thermal ablation based on NSGA-III. Technol. Health Care 2022, 30, 1243–1256. [Google Scholar] [CrossRef]
- Scharll, Y.; Letrari, S.; Laimer, G.; Schullian, P.; Bale, R. Puncture accuracy of an optical tracked robotic aiming device-a phantom study. Eur. Radiol. 2022, 32, 6769–6776. [Google Scholar] [CrossRef]
- Wu, Q.; Cao, B.; Zheng, Y.; Liang, B.; Liu, M.; Wang, L.; Zhang, J.; Meng, L.; Luo, S.; He, X.; et al. Feasibility and safety of fine positioning needle-mediated breathing control in CT-guided percutaneous puncture of small lung/liver nodules adjacent to diaphragm. Sci. Rep. 2021, 11, 3411. [Google Scholar] [CrossRef]
- Hu, W.; Jiang, H.; Wang, M. Flexible needle puncture path planning for liver tumors based on deep reinforcement learning. Phys. Med. Biol. 2022, 67, 195008. [Google Scholar] [CrossRef]
- Lin, K.C.; Ko, W.C.; Tsai, Y.D.; Chang, C.Y.; Yang, Y.H.; Huang, Y.S.; Chang, Y.C. Hemorrhage risk prediction after computed tomography-guided lung biopsy: Clinical parameters and quantitative pulmonary vascular analysis. J. Formos. Med. Assoc. 2025, 124, 79–86. [Google Scholar] [CrossRef]
- Huang, Q.; Zhang, L.; Liu, L.; Cao, Y.; Ma, H.; Wang, L.; Zhou, C. 3D Visualization of Pulmonary Vessel Based on Low-Cost Segmentation and Fast Reconstruction. IEEE Access 2023, 11, 44866–44878. [Google Scholar] [CrossRef]
- Abe-Doi, M.; Murayama, R.; Takahashi, T.; Matsumoto, M.; Tamai, N.; Nakagami, G.; Sanada, H. Effects of ultrasound with an automatic vessel detection system using artificial intelligence on the selection of puncture points among ultrasound beginner clinical nurses. J. Vasc. Access 2024, 25, 1252–1260. [Google Scholar] [CrossRef] [PubMed]
- Baegert, C.; Villard, C.; Schreck, P.; Soler, L. Multi-criteria trajectory planning for hepatic radiofrequency ablation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019, Proceedings of the 22nd International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; Springer: Berlin/Heidelberg, Germany, 2007; Volume 10, pp. 676–684. [Google Scholar] [CrossRef]
- Belbachir, E.; Golkar, E.; Bayle, B.; Essert, C. Automatic planning of needle placement for robot-assisted percutaneous procedures. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 1429–1438. [Google Scholar] [CrossRef] [PubMed]
- Tashi, S.; Gogna, A.; Leong, S.; Venkatanarasimha, N.; Chandramohan, S. Vascular complications related to image-guided percutaneous thermal ablation of hepatic tumors. Diagn. Interv. Radiol. 2023, 29, 318–325. [Google Scholar] [CrossRef]
- Rhaiem, R.; Piardi, T.; Renard, Y.; Deguelte, S.; Kianmanesh, R. Laparoscopic thermal ablation of liver tumours. J. Visc. Surg. 2021, 158, 173–179. [Google Scholar] [CrossRef]
- Bhagavatula, S.K.; Graur, A.; Fintelmann, F.J. Lung Needle Biopsy and Lung Ablation: Indications, Patient Management, and Postprocedure Imaging Findings. Clin. Chest Med. 2024, 45, 307–323. [Google Scholar] [CrossRef]
- Keksel, A.; Schmidt, S.; Beck, D.; Seewig, J. Scientific modeling of optical 3D measuring devices based on GPU-accelerated ray tracing using the NVIDIA OptiX engine. In SPIE Optical Metrology, Proceedings of SPIE—Modeling Aspects in Optical Metrology IX–2023, Munich, Germany, 26–29 June 2023; SPIE: Bellingham, WA, USA; Volume 12619.
- The LUNA16 Dataset. Available online: https://luna16.grand-challenge.org/Home/ (accessed on 12 March 2025).
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- The Tianchi Dataset. Available online: https://tianchi.aliyun.com/competition/entrance/231601 (accessed on 12 March 2025).
- The LNDb Dataset. Available online: https://lndb.grand-challenge.org/ (accessed on 12 March 2025).
- Xie, R.L.; Wang, Y.; Zhao, Y.N.; Zhang, J.; Chen, G.B.; Fei, J.; Fu, Z. Lung nodule pre-diagnosis and insertion path planning for chest CT images. BMC Med. Imaging 2023, 23, 22. [Google Scholar] [CrossRef]
- He, L.; Meng, Y.; Zhong, J.; Tang, L.; Chui, C.; Zhang, J. Preoperative path planning algorithm for lung puncture biopsy based on path constraint and multidimensional space distance optimization. Biomed. Signal Process. Control 2023, 80, 104304. [Google Scholar] [CrossRef]
- Monfaredi, R.; Concepcion-Gonzalez, A.; Acosta Julbe, J.; Fischer, E.; Hernandez-Herrera, G.; Cleary, K.; Oluigbo, C. Automatic Path-Planning Techniques for Minimally Invasive Stereotactic Neurosurgical Procedures-A Systematic Review. Sensors 2024, 24, 5238. [Google Scholar] [CrossRef]
- Jiang, W.; Zhi, L.; Zhang, S.; Zhou, T. A Dual-Branch Framework with Prior Knowledge for Precise Segmentation of Lung Nodules in Challenging CT Scans. IEEE J. Biomed. Health Inform. 2024, 28, 1540–1551. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhou, M.; Liu, Z.; Liu, Z.; Gu, D.; Zang, Y.; Dong, D.; Gevaert, O.; Tian, J. Central focused convolutional neural networks: Developing a data-driven model for lung nodule segmentation. Med. Image Anal. 2017, 40, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Talbar, S.N. CSE-GAN: A 3D conditional generative adversarial network with concurrent squeeze-and-excitation blocks for lung nodule segmentation. Comput. Biol. Med. 2022, 147, 105781. [Google Scholar] [CrossRef]

| Target | Sens | Spec | DSC | RVE | HD95 | ASD |
|---|---|---|---|---|---|---|
| Airways | 0.9268 ± 0.0818 | 0.9999 ± 0.0000 | 0.9171 ± 0.0574 | 0.0707 ± 0.0678 | 1.0085 ± 0.0252 | 0.2884 ± 0.0899 |
| Body | 0.9683 ± 0.0056 | 0.9701 ± 0.0233 | 0.9418 ± 0.0127 | 0.0564 ± 0.0299 | 1.3660 ± 0.3660 | 0.8716 ± 0.0287 |
| Bones | 0.9139 ± 0.0124 | 0.9981 ± 0.0003 | 0.9144 ± 0.0118 | 0.0108 ± 0.0097 | 1.0009 ± 0.1153 | 0.4371 ± 0.0067 |
| Heart | 0.9694 ± 0.0081 | 0.9995 ± 0.0002 | 0.9684 ± 0.0098 | 0.0089 ± 0.0089 | 1.4142 ± 0.7915 | 0.5184 ± 0.1742 |
| Kidneys | 0.9444 ± 0.0387 | 0.9998 ± 0.0001 | 0.9419 ± 0.0488 | 0.0379 ± 0.0579 | 1.0170 ± 0.4502 | 0.3026 ± 0.1866 |
| Left Lower Lobe | 0.9663 ± 0.0305 | 0.9991 ± 0.0006 | 0.9660 ± 0.0227 | 0.0186 ± 0.0325 | 1.0001 ± 0.2558 | 0.2928 ± 0.1801 |
| Left Upper Lobe | 0.9673 ± 0.0329 | 0.9991 ± 0.0004 | 0.9697 ± 0.0210 | 0.0151 ± 0.0287 | 1.0000 ± 0.4294 | 0.2918 ± 0.0008 |
| Liver | 0.9840 ± 0.0045 | 0.9993 ± 0.0005 | 0.9830 ± 0.0077 | 0.0082 ± 0.0166 | 1.4142 ± 0.5166 | 0.4298 ± 0.0728 |
| Lung Nodules | 0.5830 ± 0.3157 | 0.9999 ± 0.0000 | 0.5328 ± 0.2596 | 0.7777 ± 1.0616 | 201.9190 ± 52.7719 | 7.18075 ± 1.4455 |
| Pancreas | 0.8481 ± 0.1448 | 0.9998 ± 0.0001 | 0.8512 ± 0.1328 | 0.1084 ± 0.1394 | 1.7320 ± 0.3647 | 0.3906 ± 0.0016 |
| Pulmonary Arteries | 0.8806 ± 0.0366 | 0.9998 ± 0.0000 | 0.8826 ± 0.0275 | 0.0378 ± 0.0312 | 1.0014 ± 0.4073 | 0.2080 ± 0.0870 |
| Pulmonary Veins | 0.8433 ± 0.0430 | 0.9994 ± 0.0001 | 0.8576 ± 0.0264 | 0.0715 ± 0.0568 | 1.0920 ± 0.6981 | 0.3399 ± 0.2280 |
| Right Lower Lobe | 0.9586 ± 0.0611 | 0.9990 ± 0.0007 | 0.9557 ± 0.0847 | 0.0347 ± 0.1325 | 1.0000 ± 0.1270 | 0.2962 ± 0.0124 |
| Right Middle Lobe | 0.9368 ± 0.0363 | 0.9990 ± 0.0007 | 0.9193 ± 0.0479 | 0.0774 ± 0.1411 | 8.9746 ± 3.9526 | 1.0472 ± 0.2161 |
| Right Upper Lobe | 0.9577 ± 0.0479 | 0.9988 ± 0.0007 | 0.9554 ± 0.0309 | 0.0350 ± 0.0526 | 2.0182 ± 0.6456 | 0.4376 ± 0.1180 |
| Skin | 0.6685 ± 0.0177 | 0.9863 ± 0.005 | 0.6720 ± 0.2131 | 0.0211 ± 0.0009 | 28.5059 ± 10.9514 | 3.8390 ± 1.2075 |
| Spine | 0.9442 ± 0.0072 | 0.9993 ± 0.0001 | 0.9432 ± 0.0048 | 0.0086 ± 0.0058 | 1.0000 ± 0.4110 | 0.3772 ± 0.0483 |
| Spleen | 0.9674 ± 0.0409 | 0.9998 ± 0.0000 | 0.9691 ± 0.0254 | 0.0158 ± 0.0426 | 1.0846 ± 0.2497 | 0.3664 ± 0.0105 |
| Vessels | 0.9281 ± 0.0202 | 0.9993 ± 0.0001 | 0.9302 ± 0.0174 | 0.0179 ± 0.0181 | 1.7320 ± 0.9526 | 0.5909 ± 0.2047 |
| Target | Sens | Spec | DSC | RVE | HD95 | ASD |
|---|---|---|---|---|---|---|
| Airways | 0.9321 ± 0.0885 | 0.9999 ± 0.0000 | 0.9205 ± 0.0587 | 0.0740 ± 0.0742 | 1.0000 ± 0.5919 | 0.3676 ± 0.1954 |
| Body | 0.9626 ± 0.0211 | 0.9713 ± 0.0080 | 0.9404 ± 0.0073 | 0.0471 ± 0.1248 | 1.3660 ± 0.3660 | 0.8838 ± 0.0239 |
| Bones | 0.9091 ± 0.0115 | 0.9983 ± 0.0003 | 0.9149 ± 0.0102 | 0.0149 ± 0.0104 | 1.0000 ± 0.0184 | 0.4314 ± 0.0065 |
| Heart | 0.9700 ± 0.0085 | 0.9995 ± 0.003 | 0.9680 ± 0.0116 | 0.0119 ± 0.0149 | 1.4142 ± 0.6882 | 0.5144 ± 0.0260 |
| Kidneys | 0.9515 ± 0.0314 | 0.9999 ± 0.0001 | 0.9391 ± 0.0547 | 0.0514 ± 0.1028 | 1.0000 ± 0.4149 | 0.3230 ± 0.0250 |
| Left Lower Lobe | 0.9690 ± 0.0201 | 0.9991 ± 0.0007 | 0.9666 ± 0.0210 | 0.0181 ± 0.0265 | 1.0208 ± 0.1559 | 0.3120 ± 0.1938 |
| Left Upper Lobe | 0.9688 ± 0.0317 | 0.9991 ± 0.0006 | 0.9699 ± 0.0209 | 0.0169 ± 0.0294 | 1.4142 ± 0.6981 | 0.3045 ± 0.0130 |
| Liver | 0.9847 ± 0.0041 | 0.9993 ± 0.0007 | 0.9826 ± 0.0085 | 0.0089 ± 0.0166 | 1.4142 ± 0.5974 | 0.4108 ± 0.0514 |
| Lung Nodules | 0.6775 ± 0.2702 | 0.9999 ± 0.0000 | 0.5756 ± 0.2477 | 1.3663 ± 4.0629 | 356.7180 ± 49.8263 | 103.8800 ± 48.1003 |
| Pancreas | 0.8673 ± 0.0981 | 0.9998 ± 0.0001 | 0.8544 ± 0.1025 | 0.0962 ± 0.1143 | 1.0085 ± 0.3120 | 0.3282 ± 0.0015 |
| Pulmonary Arteries | 0.8869 ± 0.0334 | 0.9999 ± 0.0000 | 0.8893 ± 0.0246 | 0.0420 ± 0.0323 | 1.0096 ± 0.5099 | 0.2396 ± 0.0667 |
| Pulmonary Veins | 0.8513 ± 0.0545 | 0.9995 ± 0.0002 | 0.8597 ± 0.0306 | 0.1005 ± 0.0708 | 1.0726 ± 0.3692 | 0.3868 ± 0.0216 |
| Right Lower Lobe | 0.9584 ± 0.0571 | 0.9991 ± 0.0009 | 0.9564 ± 0.0855 | 0.0395 ± 0.1658 | 21.5045 ± 7.8824 | 6.1628 ± 0.1149 |
| Right Middle Lobe | 0.9318 ± 0.0366 | 0.9992 ± 0.0005 | 0.9230 ± 0.0366 | 0.0641 ± 0.0737 | 10.1462 ± 3.7860 | 1.1372 ± 0.0052 |
| Right Upper Lobe | 0.9576 ± 0.0549 | 0.9989 ± 0.0007 | 0.9553 ± 0.0349 | 0.0365 ± 0.0602 | 2.0816 ± 0.9378 | 0.4522 ± 0.0091 |
| Skin | 0.6963 ± 0.0035 | 0.98534 ± 0.0000 | 0.6707 ± 0.2126 | 0.1546 ± 0.0019 | 33.0670 ± 15.8281 | 3.3824 ± 0.4396 |
| Spine | 0.9429 ± 0.0072 | 0.9994 ± 0.0001 | 0.9444 ± 0.0047 | 0.0076 ± 0.0064 | 1.0089 ± 0.1060 | 0.3795 ± 0.0450 |
| Spleen | 0.9742 ± 0.0133 | 0.9999 ± 0.0001 | 0.9723 ± 0.0111 | 0.0152 ± 0.0224 | 1.0086 ± 0.4226 | 0.3531 ± 0.0415 |
| Vessels | 0.9378 ± 0.0139 | 0.9993 ± 0.0003 | 0.9306 ± 0.0182 | 0.0249 ± 0.0265 | 1.7320 ± 0.6671 | 0.5475 ± 0.1030 |
| Target | Sens | Spec | DSC | RVE | HD95 | ASD |
|---|---|---|---|---|---|---|
| Airways | 0.9075 ± 0.0999 | 0.9998 ± 0.0000 | 0.8856 ± 0.0669 | 0.1018 ± 0.0994 | 1.0003 ± 0.4887 | 0.3280 ± 0.0319 |
| Body | 0.9620 ± 0.0087 | 0.9635 ± 0.0009 | 0.9296 ± 0.0310 | 0.0695 ± 0.0135 | 1.5770 ± 0.4230 | 1.0645 ± 0.1788 |
| Bones | 0.8908 ± 0.0203 | 0.9975 ± 0.0005 | 0.8885 ± 0.0183 | 0.0246 ± 0.0264 | 1.4142 ± 0.6456 | 0.5371 ± 0.0089 |
| Heart | 0.9509 ± 0.0249 | 0.9992 ± 0.0006 | 0.9475 ± 0.0249 | 0.0265 ± 0.0421 | 2.4142 ± 0.1949 | 0.7250 ± 0.0329 |
| Kidneys | 0.8595 ± 0.1603 | 0.9998 ± 0.0002 | 0.8760 ± 0.1331 | 0.1268 ± 0.1659 | 7.8783 ± 2.1186 | 1.0679 ± 0.3029 |
| Left Lower Lobe | 0.9568 ± 0.0340 | 0.9986 ± 0.0008 | 0.9525 ± 0.0325 | 0.0261 ± 0.0282 | 1.4142 ± 0.0524 | 0.3967 ± 0.0539 |
| Left Upper Lobe | 0.9501 ± 0.0389 | 0.9989 ± 0.0007 | 0.9571 ± 0.0257 | 0.0286 ± 0.0377 | 1.4142 ± 0.6274 | 0.4009 ± 0.0235 |
| Liver | 0.9753 ± 0.0112 | 0.9990 ± 0.0005 | 0.9740 ± 0.0084 | 0.0140 ± 0.0150 | 2.4142 ± 0.4351 | 0.6933 ± 0.1203 |
| Lung Nodules | 0.3910 ± 0.3582 | 0.9999 ± 0.0000 | 0.3675 ± 0.3239 | 1.8281 ± 4.5187 | 334.1962 ± 48.8511 | 329.6033 ± 18.2415 |
| Pancreas | 0.6357 ± 0.2247 | 0.9996 ± 0.0003 | 0.6391 ± 0.2082 | 0.4054 ± 0.8568 | 5.0096 ± 1.0732 | 0.7854 ± 0.0575 |
| Pulmonary Arteries | 0.6651 ± 0.0517 | 0.9995 ± 0.0001 | 0.6460 ± 0.0376 | 0.1043 ± 0.0945 | 14.2426 ± 5.9513 | 2.4166 ± 0.2596 |
| Pulmonary Veins | 0.7690 ± 0.0426 | 0.9989 ± 0.0003 | 0.7635 ± 0.0397 | 0.1115 ± 0.0926 | 2.7320 ± 0.7813 | 0.4889 ± 0.2309 |
| Right Lower Lobe | 0.9438 ± 0.0689 | 0.9985 ± 0.0010 | 0.9403 ± 0.0954 | 0.0430 ± 0.1838 | 1.7320 ± 0.4073 | 0.3946 ± 0.2175 |
| Right Middle Lobe | 0.8891 ± 0.0581 | 0.9988 ± 0.0005 | 0.8840 ± 0.0559 | 0.0715 ± 0.0744 | 7.8783 ± 1.8511 | 1.0683 ± 0.5540 |
| Right Upper Lobe | 0.9417 ± 0.0519 | 0.9983 ± 0.0010 | 0.9382 ± 0.0376 | 0.0428 ± 0.0569 | 2.7320 ± 0.7296 | 0.5497 ± 0.0423 |
| Skin | 0.6832 ± 0.0046 | 0.9870 ± 0.0020 | 0.6840 ± 0.1409 | 0.0043 ± 0.0015 | 45.3406 ± 0.2558 | 5.0630 ± 2.1102 |
| Spine | 0.9324 ± 0.0124 | 0.9991 ± 0.0002 | 0.9254 ± 0.0100 | 0.0199 ± 0.0150 | 1.4142 ± 0.9324 | 0.5290 ± 0.0732 |
| Spleen | 0.9444 ± 0.0525 | 0.9997 ± 0.0001 | 0.9486 ± 0.0393 | 0.0300 ± 0.0432 | 1.0010 ± 0.5243 | 0.5558 ± 0.0145 |
| Vessels | 0.8753 ± 0.0372 | 0.9989 ± 0.0004 | 0.8806 ± 0.0333 | 0.0413 ± 0.0325 | 7.9282 ± 3.141 | 1.4424 ± 0.2505 |
| Model Variant | Average DSC | Infer Times (s) | GPU Mem (MB) |
|---|---|---|---|
| U-Net + Base Conv (3D U-Net) | 0.8921 ± 0.0045 | 60.288 ± 14.656 | 3960 ± 12 |
| U-Net + Res Block (SegResNet) | 0.9122 ± 0.0096 | 63.601 ± 19.671 | 4321 ± 16 |
| U-Net + Transformer Block (UNETR) | 0.8609 ± 0.0029 | 80.214 ± 40.882 | 6410 ± 51 |
| Dataset | Average Sens | Average Spec | Average DSC | Average RVE | Average HD95 | Average ASD |
|---|---|---|---|---|---|---|
| LUNA16 | 0.9031 | 0.9983 | 0.9122 | 0.0668 | 12.7881 | 0.7806 |
| Tianchi | 0.9018 | 0.9968 | 0.8749 | 0.1616 | 8.5356 | 1.7746 |
| LNDb | 0.8952 | 0.9965 | 0.8710 | 0.1921 | 7.7988 | 2.1419 |
| Clinical | 0.8905 | 0.9969 | 0.8563 | 0.1985 | 8.1350 | 1.7342 |
| Model | AUC-ROC | Precision | Recall | F1 | Specificity | TP | TN | FP | FN |
|---|---|---|---|---|---|---|---|---|---|
| RandomForest | 0.948 | 0.706 | 0.766 | 0.735 | 0.956 | 36 | 324 | 15 | 11 |
| XGBoost | 0.942 | 0.717 | 0.702 | 0.710 | 0.962 | 33 | 326 | 13 | 14 |
| LightGBM | 0.944 | 0.717 | 0.702 | 0.710 | 0.962 | 33 | 326 | 13 | 14 |
| CatBoost | 0.951 | 0.643 | 0.766 | 0.699 | 0.941 | 36 | 319 | 20 | 11 |
| MLP_Simple | 0.950 | 0.694 | 0.723 | 0.708 | 0.956 | 34 | 324 | 15 | 13 |
| MLP_Deep | 0.957 | 0.694 | 0.723 | 0.708 | 0.956 | 34 | 324 | 15 | 13 |
| MLP_Wide | 0.950 | 0.708 | 0.723 | 0.716 | 0.959 | 34 | 325 | 14 | 13 |
| MLP_Complex | 0.949 | 0.654 | 0.723 | 0.687 | 0.947 | 34 | 321 | 18 | 13 |
| Manual | / | 0.660 | 0.500 | 0.569 | 0.949 | 33 | 319 | 17 | 33 |
| Method | Average Depth (mm) | Average Angle (°) | Average Distance (mm) | Average Time Cost (s) |
|---|---|---|---|---|
| Generation of RGB Images | / | / | / | 1.758 ± 0.061 |
| RandomForest | 70.774 ± 29.320 | 63.353 ± 11.216 | 3.326 ± 3.512 | 0.072 ± 0.022 |
| XGBoost | 72.651 ± 29.511 | 62.703 ± 11.177 | 3.154 ± 3.547 | 0.081 ± 0.022 |
| LightGBM | 72.247 ± 29.530 | 63.191 ± 11.054 | 3.183 ± 3.527 | 0.106 ± 0.028 |
| CatBoost | 73.189 ± 30.838 | 63.085 ± 11.487 | 3.292 ± 3.552 | 0.082 ± 0.028 |
| MLP_Simple | 76.697 ± 31.278 | 60.809 ± 11.826 | 2.892 ± 2.861 | 0.055 ± 0.021 |
| MLP_Deep | 73.619 ± 30.956 | 62.146 ± 12.175 | 3.115 ± 3.375 | 0.053 ± 0.021 |
| MLP_Wide | 74.520 ± 32.881 | 62.278 ± 12.600 | 3.027 ± 3.377 | 0.054 ± 0.021 |
| MLP_Complex | 74.638 ± 30.476 | 61.592 ± 11.985 | 3.323 ± 3.350 | 0.054 ± 0.022 |
| Manual | 74.956 ± 29.570 | 62.435 ± 9.511 | 2.883 ± 3.133 | 1.968 ± 0.802 |
| Ground Truth | 71.640 ± 31.556 | 62.696 ± 11.794 | 3.142 ± 3.590 | / |
| Study | Existing Methods and Limitations | Proposed Improvements | Vascular Handling | Acceleration Strategy | Multi-Objective Optimization | Time Cost | Includes Organ Segmentation? |
|---|---|---|---|---|---|---|---|
| Claire Baegert (2007) [33] | Manual constraint of optimization; artificial minima risk. | Safe entry region calculation, refined via constrained triangle elimination/subdivision. | Collision detection to avoid organs, bones, and vessels. | Coarse angle discretization + local optimization. | Weighted sum: tumor volume, organ distance, and insertion depth. | ~30 s | Not specified |
| Qi Dong (2022) [26] | Sphere models; manual weight setting; limited flexibility. | NSGA-III with adaptive morphology. | 3 mm vessel dilation; boundary adjustments. | DICOM resampling + HashMap for collision detection. | NSGA-III balances efficiency and treatment. | Not specified | Not specified |
| Min Luo (2022) [16] | Multi-criteria and electrode planning; high cost, few clinical factors. | Multi-stage planning with risk index and potential fields. | Risk maps to avoid critical vessels. | Not specified | Not specified | <200 ms | Liver reconstruction: 5–20 min |
| Ruikun Li (2023) [15] | Deep learning for insertion/ablation; high cost, partial solutions. | Heuristic RFA planning with 3D-to-2D projection. | Adjusts vessel positions via projection. | Heuristic rules within projection framework. | Not specified | <3 min | Not specified |
| Jing Li (2023) [22] | Single/multi-needle planning and Pareto optimization; manual threshold and limited clinical constraints. | Fixed-point, ROI-based multi-needle planning. | Strict constraints to avoid vessels. | Triangular mesh reduces search space. | NSGA-II for needle placement vs. tumor coverage. | ~41.4 s | Not specified |
| Rong-Li Xie (2023) [43] | Lung segmentation with boundary repair; repair failures, suboptimal paths. | Adaptive thresholding, morphological repair, shortest path. | Advanced segmentation defines safe zones. | Not specified | Distance-based optimization to shorten paths. | ~5 s | Not specified |
| Qi Liu (2023) [5] | Ablation/biopsy planning; manual tasks, ignored constraints. | Relaxed Pareto with adaptive heptagonal constraints. | Geometric constraints to avoid nerves and vessels. | Voxel downsampling (1 mm) boosts efficiency. | Relaxed Pareto enables flexible decisions. | 2045.3~4518.3 s | Not specified |
| Zhengshuai Wang (2024) [21] | Semi-automatic planning; manual interaction, sampling issues. | Cube mapping with Pareto optimization. | Cube mapping optimizes vessel avoidance. | Parallel generation of constraint maps. | Weighted Pareto improves trajectory planning. | ~35 s | Not specified |
| Hui Yang (2024) [17] | Rigid needle planning; suboptimal targets, few clinical factors. | Dual-sphere Pareto with importance-correlation scoring. | Limits vessel intersections (≤10). | Not specified | Dual-sphere Pareto for refined planning. | ~179.11 s | Not specified |
| Chow Wei Too (2024) [6] | Manual nodule marking, inverse kinematics; heavy manual intervention. | CNN-based lesion detection with Bayesian planning. | Dynamic vessel avoidance via Bayesian optimization. | Bayesian optimization adjusts trajectory in real time. | Not specified | <5 min | Not specified |
| Ziwei Song (2024) [8] | 3D visualization; semi-automatic planning struggles with complex cases. | Fully automated planning with Pareto and weighted sum. | Ensures safe paths with effective vessel avoidance. | Not specified. | Combined weighted sum and Pareto optimization. | ~4 min | Not specified |
| Jiayu Zhang (2024) [25] | Semi-automatic, centroid targeting; manual input, ignores respiratory motion. | Multi-objective fuzzy optimization. | Fuzzy logic yields zero vessel intersections. | Not specified. | Fuzzy-based Pareto for flexible decision-making. | 2669.4~2477.1 s | Not specified |
| Ling He (2023) [44] | Constraint-based with weighted sum and Pareto; subjective weights, manual selection. | Multi-dimensional Pareto optimization. | Distance-based vessel avoidance minimizes risk. | Not specified. | Multi-dimensional Pareto for nuanced decisions. | Not specified | Not specified |
| Ours | Brute-force methods are slow; downsampling lowers resolution; detailed risk assessment needed. | Optical algorithm-based environment modeling. | Optical absorption simulates vessel risk. | GPU-accelerated optical algorithms for rapid computation. | Machine learning models mimic clinicians’ preferences. | 1.905 ± 0.089 s | Segmentation: 63.601 ± 19.671 s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Kang, S.; Ren, J.; Zhang, D.; Niu, B.; Xu, K. Rapid Path Planning Algorithm for Percutaneous Rigid Needle Biopsy Based on Optical Illumination Principles. Sensors 2025, 25, 2137. https://doi.org/10.3390/s25072137
Liu J, Kang S, Ren J, Zhang D, Niu B, Xu K. Rapid Path Planning Algorithm for Percutaneous Rigid Needle Biopsy Based on Optical Illumination Principles. Sensors. 2025; 25(7):2137. https://doi.org/10.3390/s25072137
Chicago/Turabian StyleLiu, Jian, Shuai Kang, Juan Ren, Dongxia Zhang, Bing Niu, and Kai Xu. 2025. "Rapid Path Planning Algorithm for Percutaneous Rigid Needle Biopsy Based on Optical Illumination Principles" Sensors 25, no. 7: 2137. https://doi.org/10.3390/s25072137
APA StyleLiu, J., Kang, S., Ren, J., Zhang, D., Niu, B., & Xu, K. (2025). Rapid Path Planning Algorithm for Percutaneous Rigid Needle Biopsy Based on Optical Illumination Principles. Sensors, 25(7), 2137. https://doi.org/10.3390/s25072137






