Evaluating Sparse Inertial Measurement Unit Configurations for Inferring Treadmill Running Motion
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset
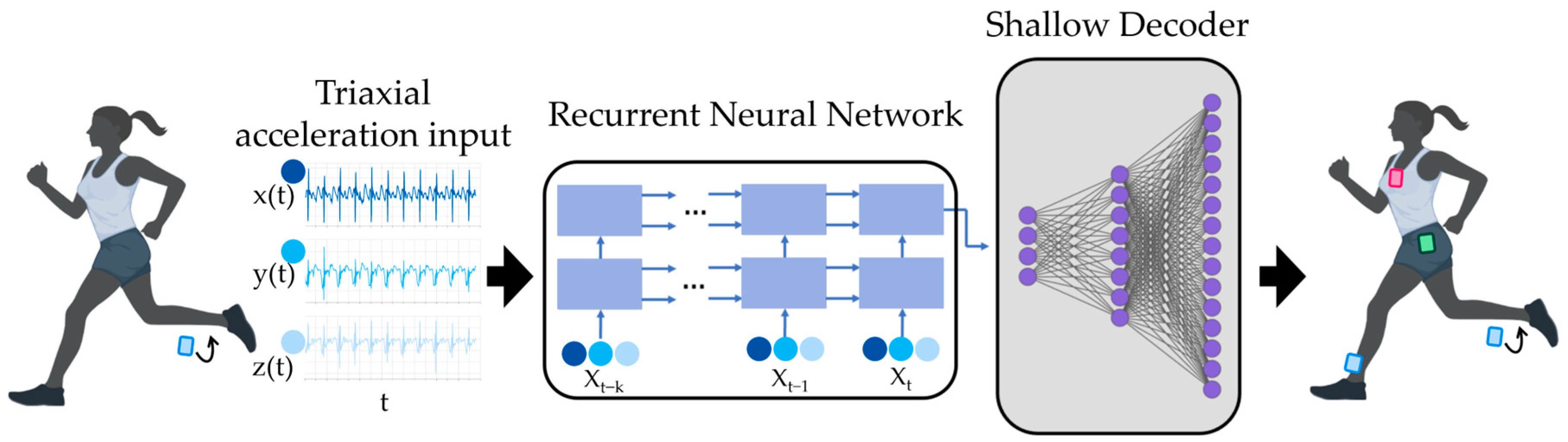
2.2. Shallow Recurrent Decoder Parameters
2.3. Experimental Parameters
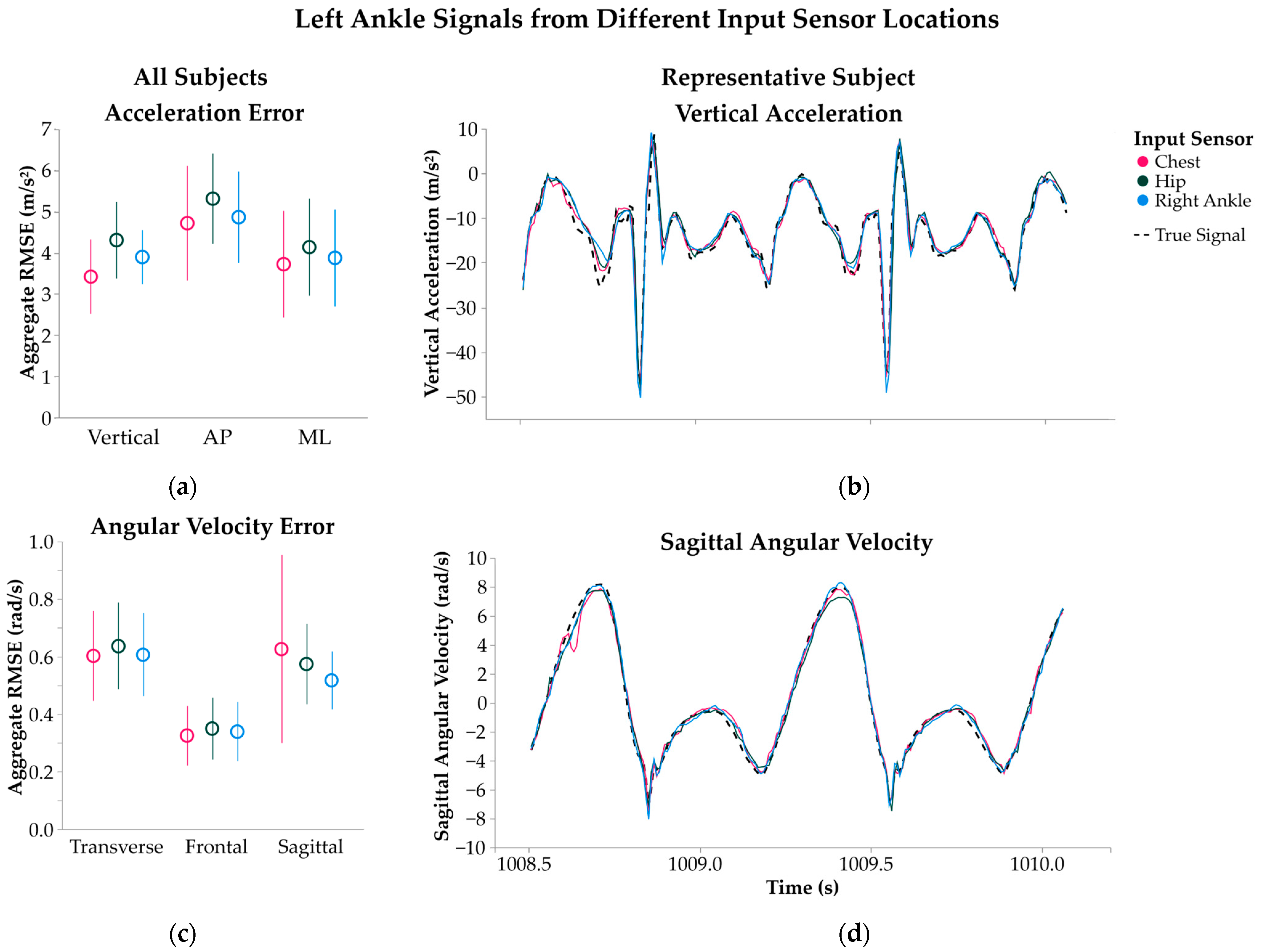
2.3.1. Input Sensor Location
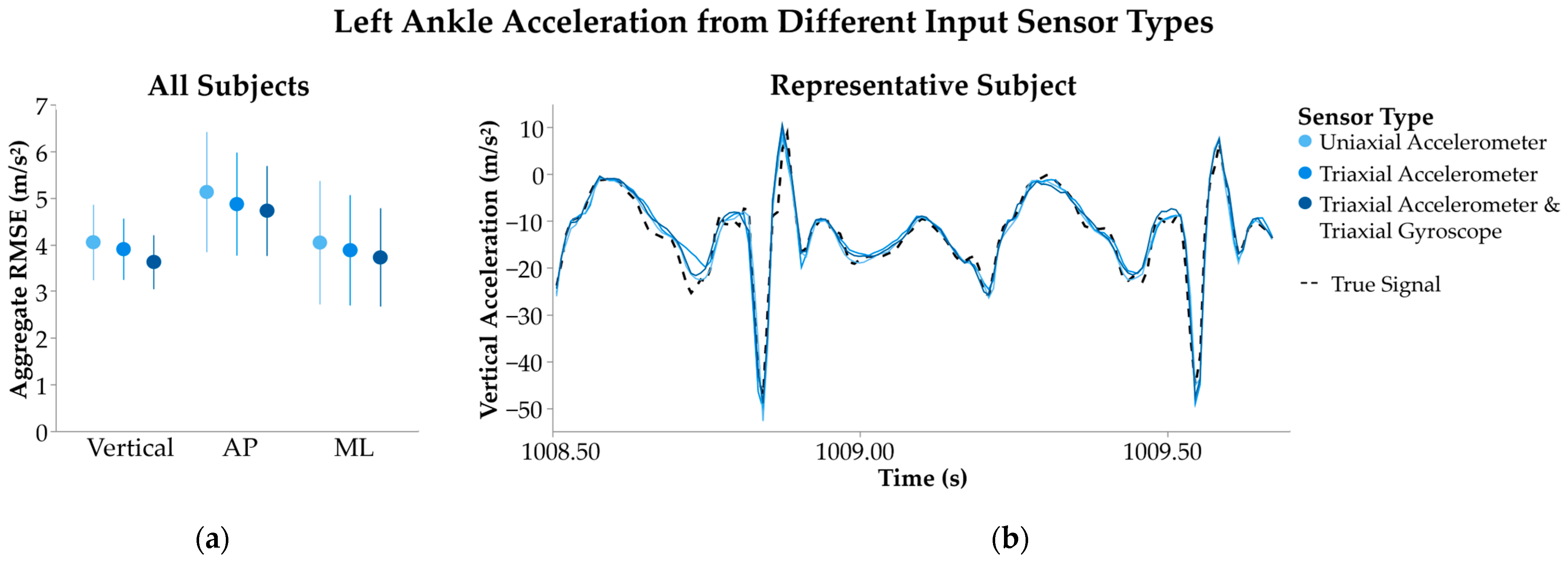
2.3.2. Input Sensor Type
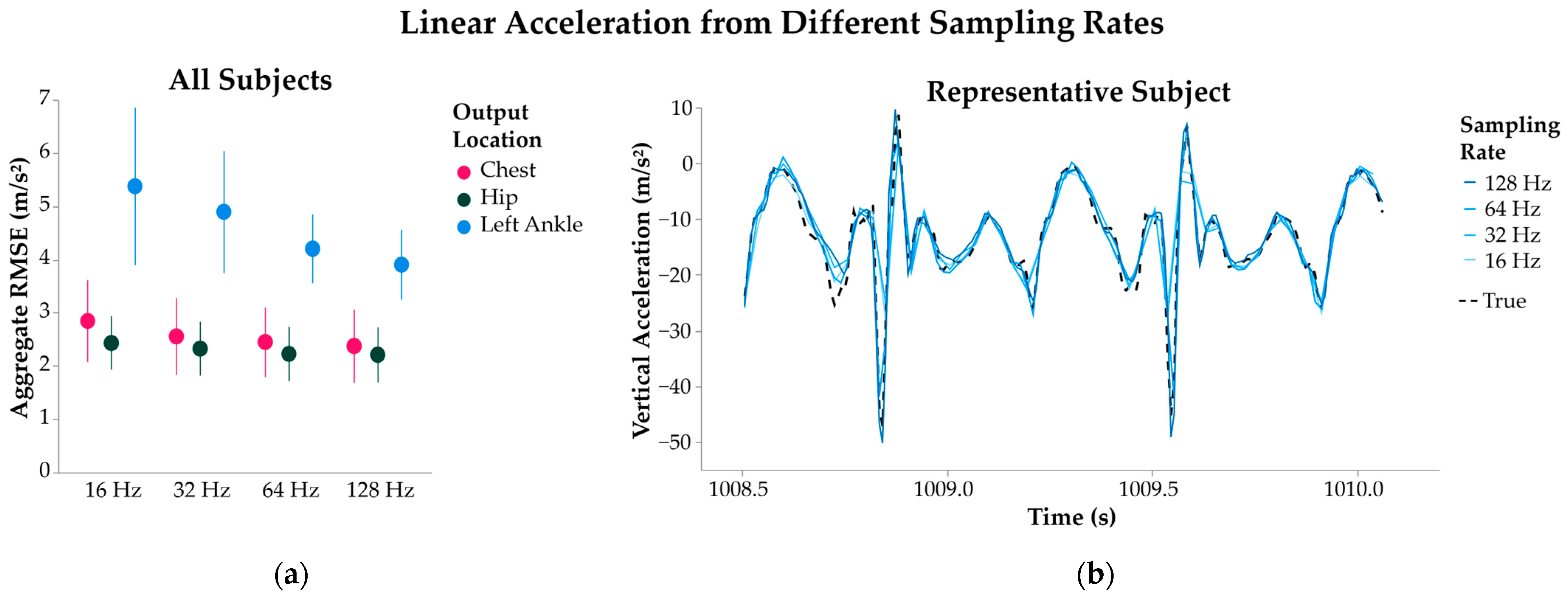
2.3.3. Sampling Rate
2.3.4. Running Speed
2.4. Data Analysis
3. Results
3.1. Single Speed
3.1.1. Input Sensor Location
3.1.2. Input Sensor Type
3.1.3. Sampling Rate
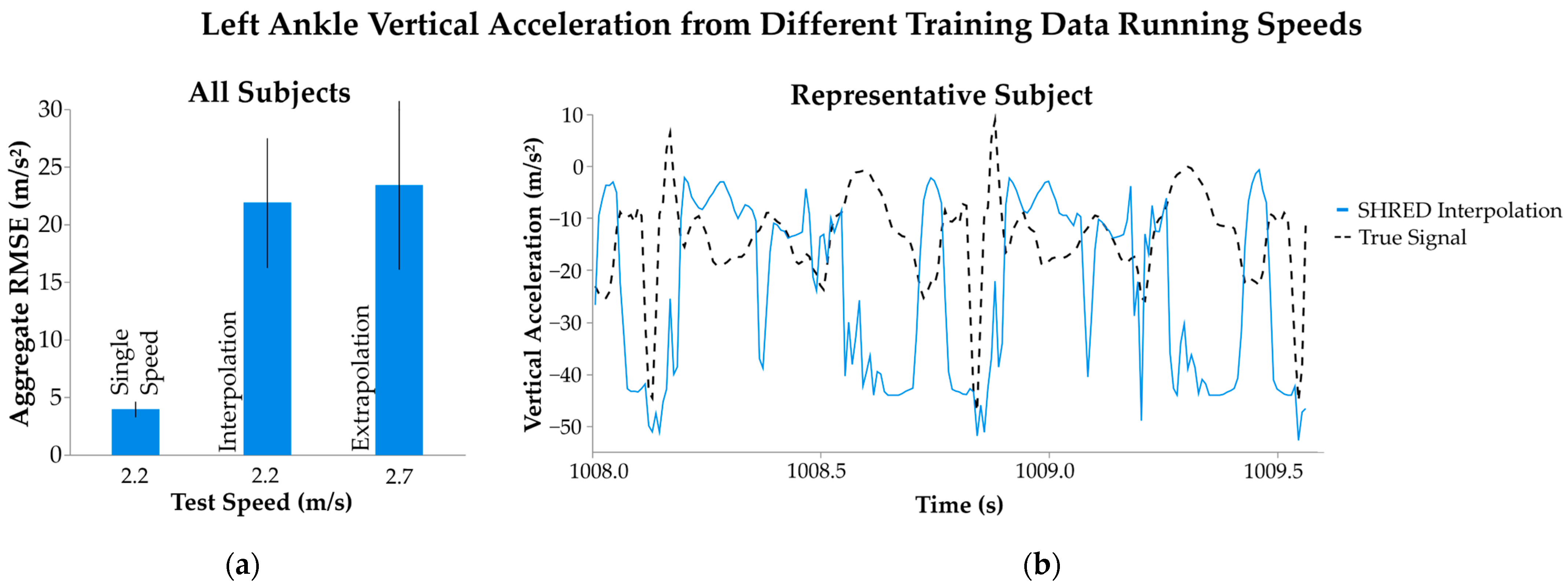
3.2. Multiple Speeds
4. Discussion
4.1. Inference Accuracy
4.1.1. Input Sensor Location
4.1.2. Input Sensor Type
4.1.3. Sampling Rate
4.1.4. Running Speed
4.2. Significance
4.3. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| IMU | Inertial measurement unit |
| SHRED | Shallow recurrent decoder network |
| LSTM | Long short-term memory network |
| AP | Anteroposterior |
| ML | Mediolateral |
| RMSE | Root mean squared error |
| MAE | Mean absolute error |
| MBE | Mean bias error |
| MDC | Minimal detectable change |
References
- Mason, R.; Pearson, L.T.; Barry, G.; Young, F.; Lennon, O.; Godfrey, A.; Stuart, S. Wearables for Running Gait Analysis: A Systematic Review. Sports Med. 2023, 53, 241–268. [Google Scholar] [CrossRef] [PubMed]
- Preatoni, E.; Bergamini, E.; Fantozzi, S.; Giraud, L.I.; Orejel Bustos, A.S.; Vannozzi, G.; Camomilla, V. The Use of Wearable Sensors for Preventing, Assessing, and Informing Recovery from Sport-Related Musculoskeletal Injuries: A Systematic Scoping Review. Sensors 2022, 22, 3225. [Google Scholar] [CrossRef] [PubMed]
- Benson, L.C.; Räisänen, A.M.; Clermont, C.A.; Ferber, R. Is This the Real Life, or Is This Just Laboratory? A Scoping Review of IMU-Based Running Gait Analysis. Sensors 2022, 22, 1722. [Google Scholar] [CrossRef]
- Neal, B.S.; Bramah, C.; McCarthy-Ryan, M.F.; Moore, I.S.; Napier, C.; Paquette, M.R.; Gruber, A.H. Using Wearable Technology Data to Explain Recreational Running Injury: A Prospective Longitudinal Feasibility Study. Phys. Ther. Sport 2024, 65, 130–136. [Google Scholar] [CrossRef]
- Carter, J.; Chen, X.; Cazzola, D.; Trewartha, G.; Preatoni, E. Consumer-Priced Wearable Sensors Combined with Deep Learning Can Be Used to Accurately Predict Ground Reaction Forces during Various Treadmill Running Conditions. PeerJ 2024, 12, e17896. [Google Scholar] [CrossRef]
- Elstub, L.J.; Nurse, C.A.; Grohowski, L.M.; Volgyesi, P.; Wolf, D.N.; Zelik, K.E. Tibial Bone Forces Can Be Monitored Using Shoe-Worn Wearable Sensors during Running. J. Sports Sci. 2022, 40, 1741–1749. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Paolieri, M.; Stewart, H.E.; Golubchik, L.; McNitt-Gray, J.L.; Misra, V.; Shah, D. Estimating Ground Reaction Forces from Inertial Sensors. IEEE Trans. Biomed. Eng. 2024, 72, 595–608. [Google Scholar] [CrossRef]
- Tan, T.; Strout, Z.A.; Shull, P.B. Accurate Impact Loading Rate Estimation During Running via a Subject-Independent Convolutional Neural Network Model and Optimal IMU Placement. IEEE J. Biomed. Health Inform. 2021, 25, 1215–1222. [Google Scholar] [CrossRef]
- Dorschky, E.; Nitschke, M.; Mayer, M.; Weygers, I.; Gassner, H.; Seel, T.; Eskofier, B.M.; Koelewijn, A.D. Comparing Sparse Inertial Sensor Setups for Sagittal-Plane Walking and Running Reconstructions. Front. Bioeng. Biotechnol. 2025, 13, 1507162. [Google Scholar]
- Wolski, L.; Halaki, M.; Hiller, C.E.; Pappas, E.; Fong Yan, A. Validity of an Inertial Measurement Unit System to Measure Lower Limb Kinematics at Point of Contact during Incremental High-Speed Running. Sensors 2024, 24, 5718. [Google Scholar] [CrossRef]
- Jeon, T.H.; Lee, J.K. IMU-Based Joint Angle Estimation Under Various Walking and Running Conditions. J. Korean Soc. Precis. Eng. 2018, 35, 1199–1204. [Google Scholar] [CrossRef]
- Baker, L.M.; Yawar, A.; Lieberman, D.E.; Walsh, C.J. Predicting Overstriding with Wearable IMUs during Treadmill and Overground Running. Sci. Rep. 2024, 14, 6347. [Google Scholar] [CrossRef]
- Faisal, A.I.; Majumder, S.; Mondal, T.; Cowan, D.; Naseh, S.; Deen, M.J. Monitoring Methods of Human Body Joints: State-of-the-Art and Research Challenges. Sensors 2019, 19, 2629. [Google Scholar] [CrossRef] [PubMed]
- Dorschky, E.; Nitschke, M.; Martindale, C.F.; Van Den Bogert, A.J.; Koelewijn, A.D.; Eskofier, B.M. CNN-Based Estimation of Sagittal Plane Walking and Running Biomechanics From Measured and Simulated Inertial Sensor Data. Front. Bioeng. Biotechnol. 2020, 8, 604. [Google Scholar] [CrossRef]
- Wouda, F.J.; Giuberti, M.; Bellusci, G.; Maartens, E.; Reenalda, J.; Van Beijnum, B.-J.F.; Veltink, P.H. Estimation of Vertical Ground Reaction Forces and Sagittal Knee Kinematics During Running Using Three Inertial Sensors. Front. Physiol. 2018, 9, 218. [Google Scholar] [CrossRef]
- Havens, K.L.; Cohen, S.C.; Pratt, K.A.; Sigward, S.M. Accelerations from Wearable Accelerometers Reflect Knee Loading during Running after Anterior Cruciate Ligament Reconstruction. Clin. Biomech. 2018, 58, 57–61. [Google Scholar] [CrossRef]
- Von Marcard, T.; Rosenhahn, B.; Black, M.J.; Pons-Moll, G. Sparse Inertial Poser: Automatic 3D Human Pose Estimation from Sparse IMUs. Comput. Graph. Forum 2017, 36, 349–360. [Google Scholar] [CrossRef]
- Huang, Y.; Kaufmann, M.; Aksan, E.; Black, M.J.; Hilliges, O.; Pons-Moll, G. Deep Inertial Poser: Learning to Reconstruct Human Pose from Sparse Inertial Measurements in Real Time. ACM Trans. Graph. 2018, 37, 1–15. [Google Scholar] [CrossRef]
- Yi, X.; Zhou, Y.; Habermann, M.; Shimada, S.; Golyanik, V.; Theobalt, C.; Xu, F. Physical Inertial Poser (PIP): Physics-Aware Real-Time Human Motion Tracking from Sparse Inertial Sensors. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 19–24 June 2022; pp. 13157–13168. [Google Scholar]
- Wouwe, T.V.; Lee, S.; Falisse, A.; Delp, S.; Liu, C.K. DiffusionPoser: Real-Time Human Motion Reconstruction From Arbitrary Sparse Sensors Using Autoregressive Diffusion. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2024, Seattle, WA, USA, 17–21 June 2024. [Google Scholar]
- Williams, J.P.; Zahn, O.; Kutz, J.N. Sensing with Shallow Recurrent Decoder Networks. Proc. R. Soc. A 2024, 480, 20240054. [Google Scholar] [CrossRef]
- Ebers, M.R.; Williams, J.P.; Steele, K.M.; Kutz, J.N. Leveraging Arbitrary Mobile Sensor Trajectories with Shallow Recurrent Decoder Networks for Full-State Reconstruction. IEEE Access 2024, 12, 97428–97439. [Google Scholar]
- Ebers, M.R.; Pitts, M.; Steele, K.M.; Kutz, J.N. Human Motion Data Expansion from Arbitrary Sparse Sensors with Shallow Recurrent Decoders. bioRxiv 2024. [Google Scholar] [CrossRef]
- Riva, S.; Introini, C.; Cammi, A.; Kutz, J.N. Robust State Estimation from Partial Out-Core Measurements with Shallow Recurrent Decoder for Nuclear Reactors. arXiv 2024, arXiv:2409.12550. [Google Scholar]
- Ingraham, K.A.; Ferris, D.P.; Remy, C.D. Using Portable Physiological Sensors to Estimate Energy Cost for ‘Body-in-the-Loop’ Optimization of Assistive Robotic Devices. In Proceedings of the 2017 IEEE Global Conference on Signal and Information Processing (GlobalSIP), Montreal, QC, Canada, 14–16 November 2017; pp. 413–417. [Google Scholar]
- Buchheit, M.; Gray, A.; Morin, J.-B. Assessing Stride Variables and Vertical Stiffness with GPS-Embedded Accelerometers: Preliminary Insights for the Monitoring of Neuromuscular Fatigue on the Field. J. Sports Sci. Med. 2015, 14, 698–701. [Google Scholar] [PubMed]
- Lawson, M.; Naemi, R.; Needham, R.A.; Chockalingam, N. Can Machine Learning Predict Running Kinematics Based on Upper Trunk GPS-Based IMU Acceleration? A Novel Method of Conducting Biomechanical Analysis in the Field Using Artificial Neural Networks. Appl. Sci. 2024, 14, 1730. [Google Scholar] [CrossRef]
- Neville, J.G.; Rowlands, D.D.; Lee, J.B.; James, D.A. A Model for Comparing Over-Ground Running Speed and Accelerometer Derived Step Rate in Elite Level Athletes. IEEE Sens. J. 2016, 16, 185–191. [Google Scholar] [CrossRef]
- Hughes, T.; Jones, R.K.; Starbuck, C.; Sergeant, J.C.; Callaghan, M.J. The Value of Tibial Mounted Inertial Measurement Units to Quantify Running Kinetics in Elite Football (Soccer) Players. A Reliability and Agreement Study Using a Research Orientated and a Clinically Orientated System. J. Electromyogr. Kinesiol. 2019, 44, 156–164. [Google Scholar] [CrossRef]
- O’Day, J.; Lee, M.; Seagers, K.; Hoffman, S.; Jih-Schiff, A.; Kidziński, Ł.; Delp, S.; Bronte-Stewart, H. Assessing Inertial Measurement Unit Locations for Freezing of Gait Detection and Patient Preference. J NeuroEngineering Rehabil. 2022, 19, 20. [Google Scholar] [CrossRef]
- Sheerin, K.R.; Reid, D.; Besier, T.F. The Measurement of Tibial Acceleration in Runners—A Review of the Factors That Can Affect Tibial Acceleration during Running and Evidence-Based Guidelines for Its Use. Gait Posture 2019, 67, 12–24. [Google Scholar] [CrossRef]
- Phan, V.; Song, K.; Silva, R.S.; Silbernagel, K.G.; Baxter, J.R.; Halilaj, E. Seven Things to Know about Exercise Classification with Inertial Sensing Wearables. IEEE J. Biomed. Health Inform. 2024, 28, 3411–3421. [Google Scholar] [CrossRef]
- Yoshida, T.; Nozaki, J.; Urano, K.; Hiroi, K.; Kaji, K.; Yonezawa, T.; Kawaguchi, N. Sampling Rate Dependency in Pedestrian Walking Speed Estimation Using DualCNN-LSTM. In Proceedings of the Adjunct Proceedings of the 2019 ACM International Joint Conference on Pervasive and Ubiquitous Computing and Proceedings of the 2019 ACM International Symposium on Wearable Computers, London, UK, 9–13 September 2019; pp. 862–868. [Google Scholar]
- Halilaj, E.; Rajagopal, A.; Fiterau, M.; Hicks, J.L.; Hastie, T.J.; Delp, S.L. Machine Learning in Human Movement Biomechanics: Best Practices, Common Pitfalls, and New Opportunities. J. Biomech. 2018, 81, 1–11. [Google Scholar] [CrossRef]
- Clouthier, A.L.; Ross, G.B.; Graham, R.B. Sensor Data Required for Automatic Recognition of Athletic Tasks Using Deep Neural Networks. Front. Bioeng. Biotechnol. 2020, 7, 473. [Google Scholar] [CrossRef] [PubMed]
- Al Borno, M.; O’Day, J.; Ibarra, V.; Dunne, J.; Seth, A.; Habib, A.; Ong, C.; Hicks, J.; Uhlrich, S.; Delp, S. OpenSense: An Open-Source Toolbox for Inertial-Measurement-Unit-Based Measurement of Lower Extremity Kinematics over Long Durations. J. Neuroeng. Rehabil. 2022, 19, 22. [Google Scholar] [CrossRef] [PubMed]
| Experiment | Training Parameter | Running Speed (m/s) | RMSE | MAE | MBE |
|---|---|---|---|---|---|
| Input Sensor Location | Chest | 2.2 | 3.4 ± 0.90 | 2.1 ± 0.52 | −0.07 ± 0.27 |
| Hip | 4.3 ± 0.93 | 2.4 ± 0.44 | −0.32 ± 0.33 | ||
| Right Ankle | 3.9 ± 0.66 | 2.2 ± 0.38 | −0.14 ± 0.23 | ||
| Input Sensor Type | Uniaxial Accelerometer | 2.2 | 4.0 ± 0.81 | 2.3 ± 0.43 | −0.12 ± 0.17 |
| Triaxial Accelerometer | 3.9 ± 0.66 | 2.2 ± 0.38 | −0.14 ± 0.23 | ||
| Triaxial Accelerometer and Triaxial Gyroscope | 3.6 ± 0.58 | 2.1 ± 0.32 | −0.16 ± 0.29 | ||
| Sampling Rate | 16 Hz * | 2.2 | 5.4 ± 1.5 | 2.8 ± 0.54 | 0.07 ± 0.49 |
| 32 Hz * | 4.9 ± 1.1 | 2.5 ± 0.44 | −0.15 ± 0.28 | ||
| 64 Hz * | 4.2 ± 0.65 | 2.3 ± 0.35 | 0.01 ± 0.25 | ||
| 128 Hz | 3.9 ± 0.66 | 2.2 ± 0.38 | −0.14 ± 0.23 | ||
| Multiple Speeds | Interpolation | Train: 1.8, 2.7; Test: 2.2 | 21.8 ± 5.6 | 17.1 ± 4.7 | 9.3 ± 3.8 |
| Extrapolation | Train: 1.8, 2.2; Test: 2.7 | 23.4 ± 7.3 | 18.0 ± 5.7 | 11.9 ± 6.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pitts, M.N.; Ebers, M.R.; Agresta, C.E.; Steele, K.M. Evaluating Sparse Inertial Measurement Unit Configurations for Inferring Treadmill Running Motion. Sensors 2025, 25, 2105. https://doi.org/10.3390/s25072105
Pitts MN, Ebers MR, Agresta CE, Steele KM. Evaluating Sparse Inertial Measurement Unit Configurations for Inferring Treadmill Running Motion. Sensors. 2025; 25(7):2105. https://doi.org/10.3390/s25072105
Chicago/Turabian StylePitts, Mackenzie N., Megan R. Ebers, Cristine E. Agresta, and Katherine M. Steele. 2025. "Evaluating Sparse Inertial Measurement Unit Configurations for Inferring Treadmill Running Motion" Sensors 25, no. 7: 2105. https://doi.org/10.3390/s25072105
APA StylePitts, M. N., Ebers, M. R., Agresta, C. E., & Steele, K. M. (2025). Evaluating Sparse Inertial Measurement Unit Configurations for Inferring Treadmill Running Motion. Sensors, 25(7), 2105. https://doi.org/10.3390/s25072105






