Aptamer-Functionalized Platform for Selective Bacterial Isolation and Rapid RNA Purification Using Capture Pins
Abstract
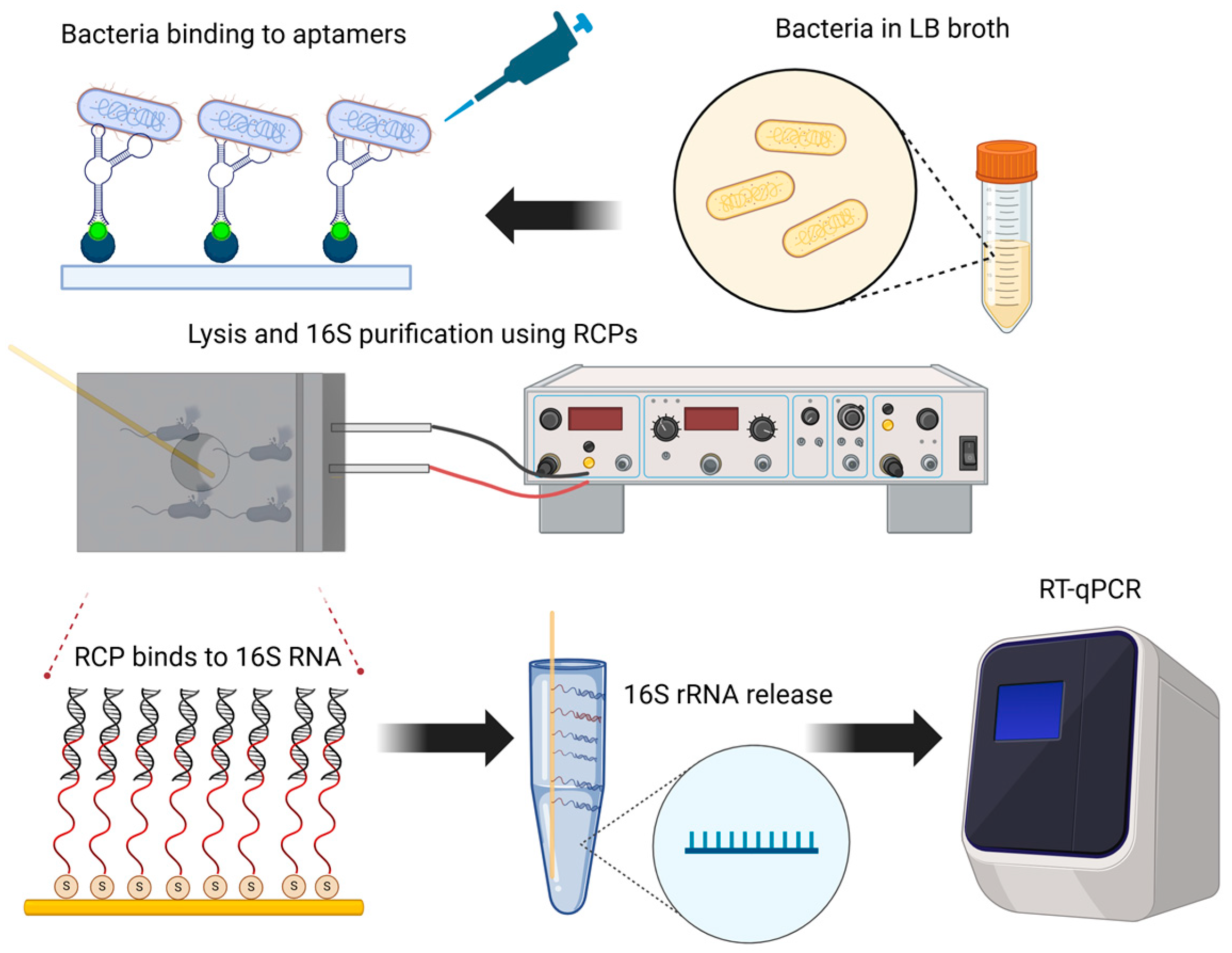
1. Introduction
2. Materials and Methods
2.1. Thiol-Conjugated 16S-Specific Oligos Immobilization to Gold-Plated RNA Capture Pins
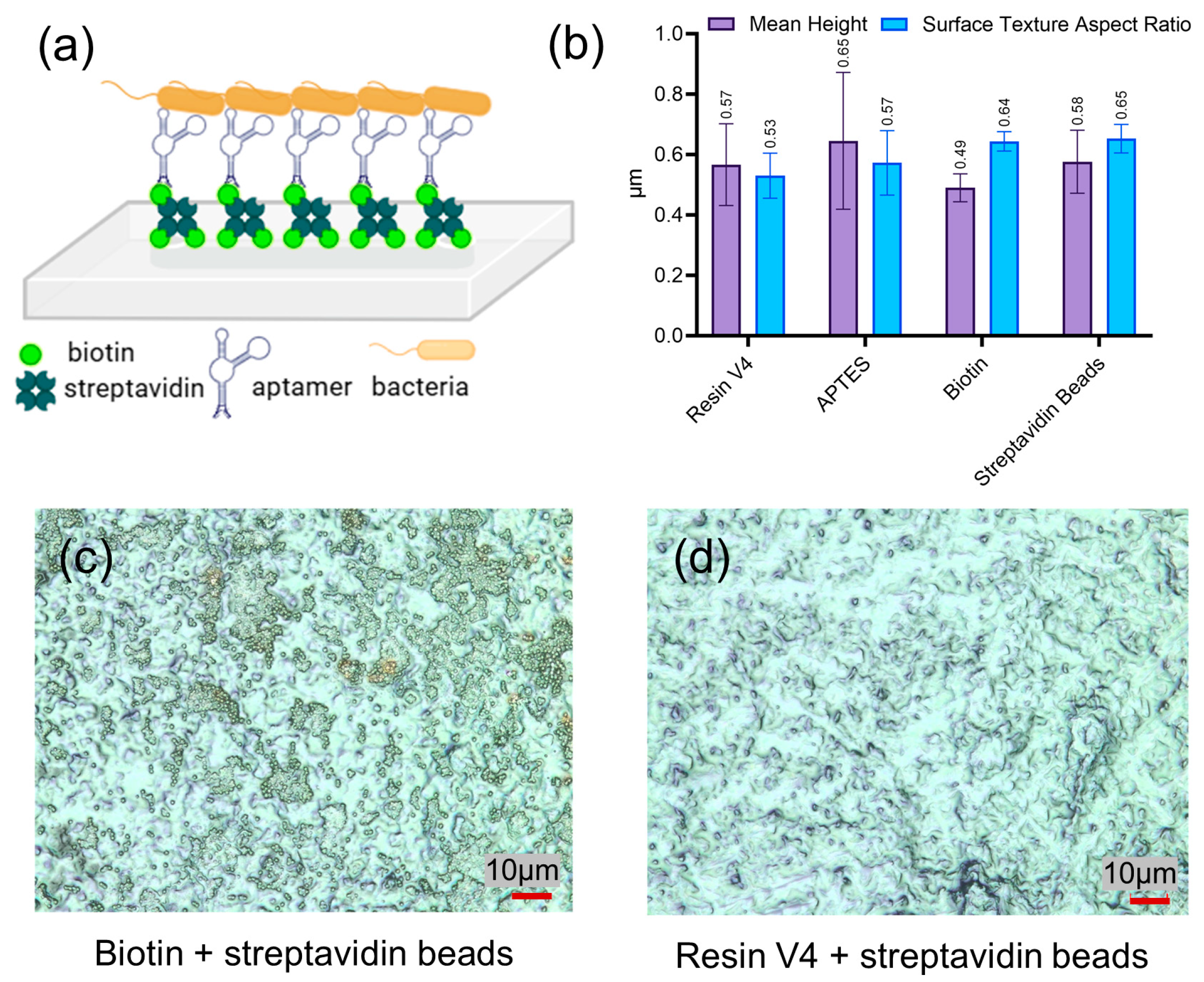
2.2. Scanning Electron Microscopy of RNA Capture Pin Functionalization
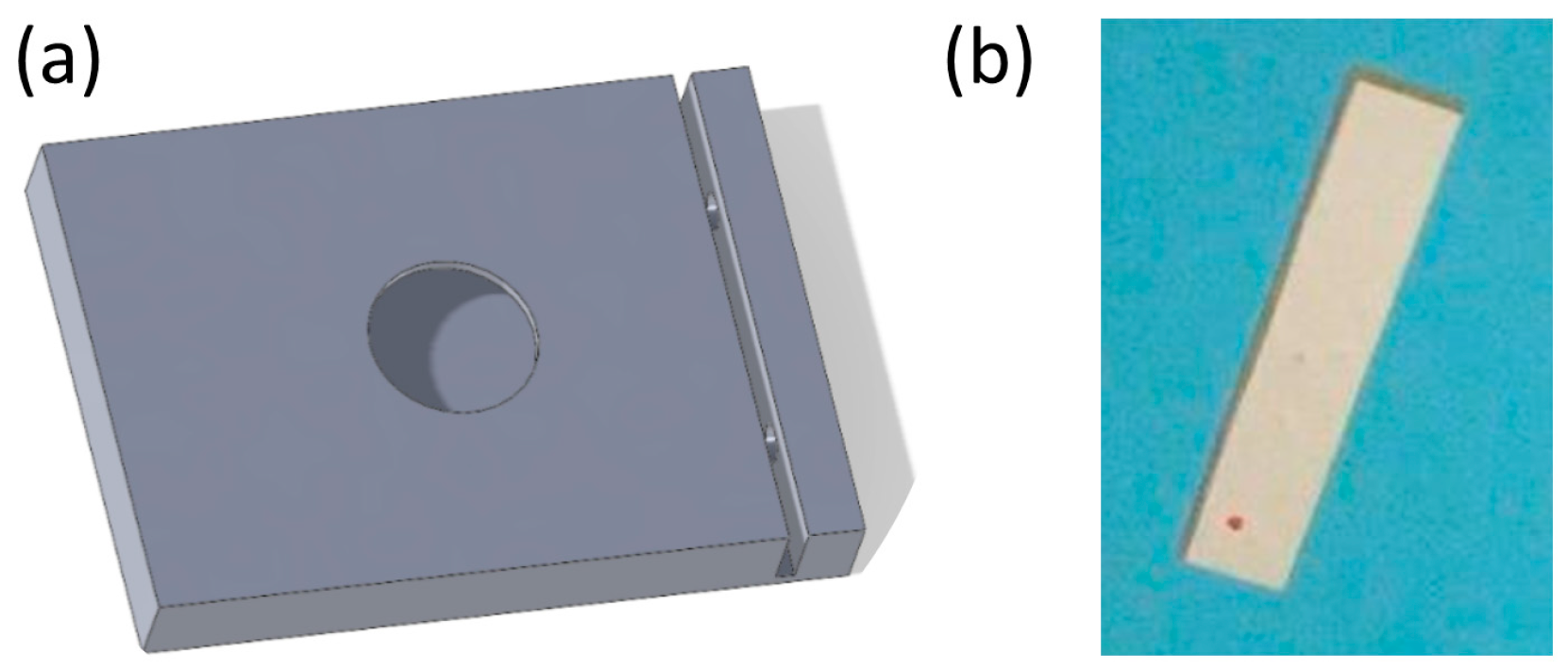
2.3. Fabrication of the Bacterial Lysis Platform with Integrated Piezoelectric Plates
2.4. Aptamers Functionalization of the Platform and Assessment of Bacterial Binding
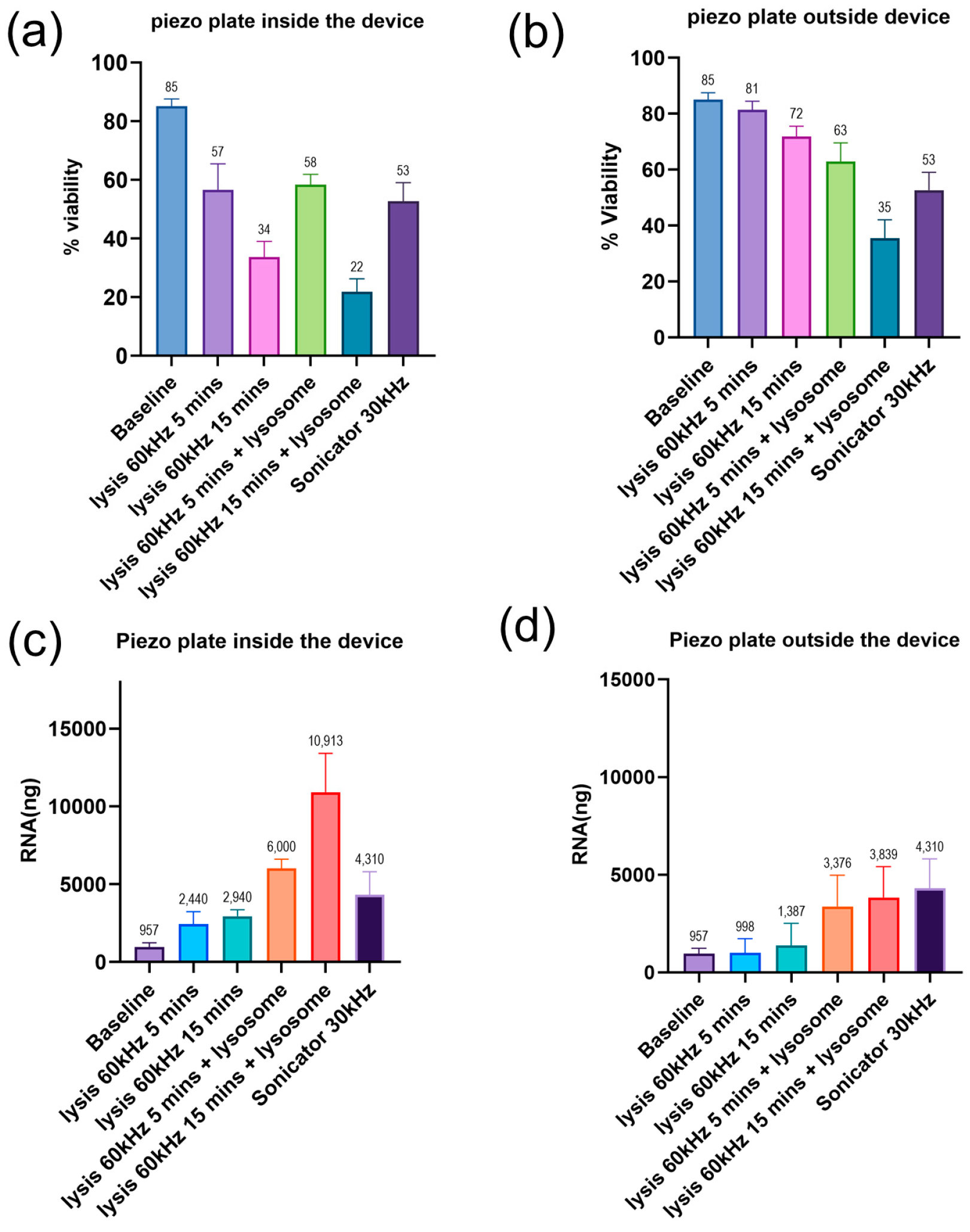
2.5. Assessment of Bacterial Lysis Efficiency and RNA Quantification
2.6. Agilent Bioanalyzer Assessment of RNA Capture Pin Specificity
2.7. RT-qPCR Using RNA Purified from RNA Capture Pins
3. Results and Discussion
3.1. Thiol-Conjugated Oligonucleotide Immobilization on the Gold-Plated RCPs
3.2. Device Design and Aptamer-Based Selective Capture of E. coli
3.3. Characterization of the Bacterial Lysis Efficiency of the Platform with an Integrated Piezoelectric Plate
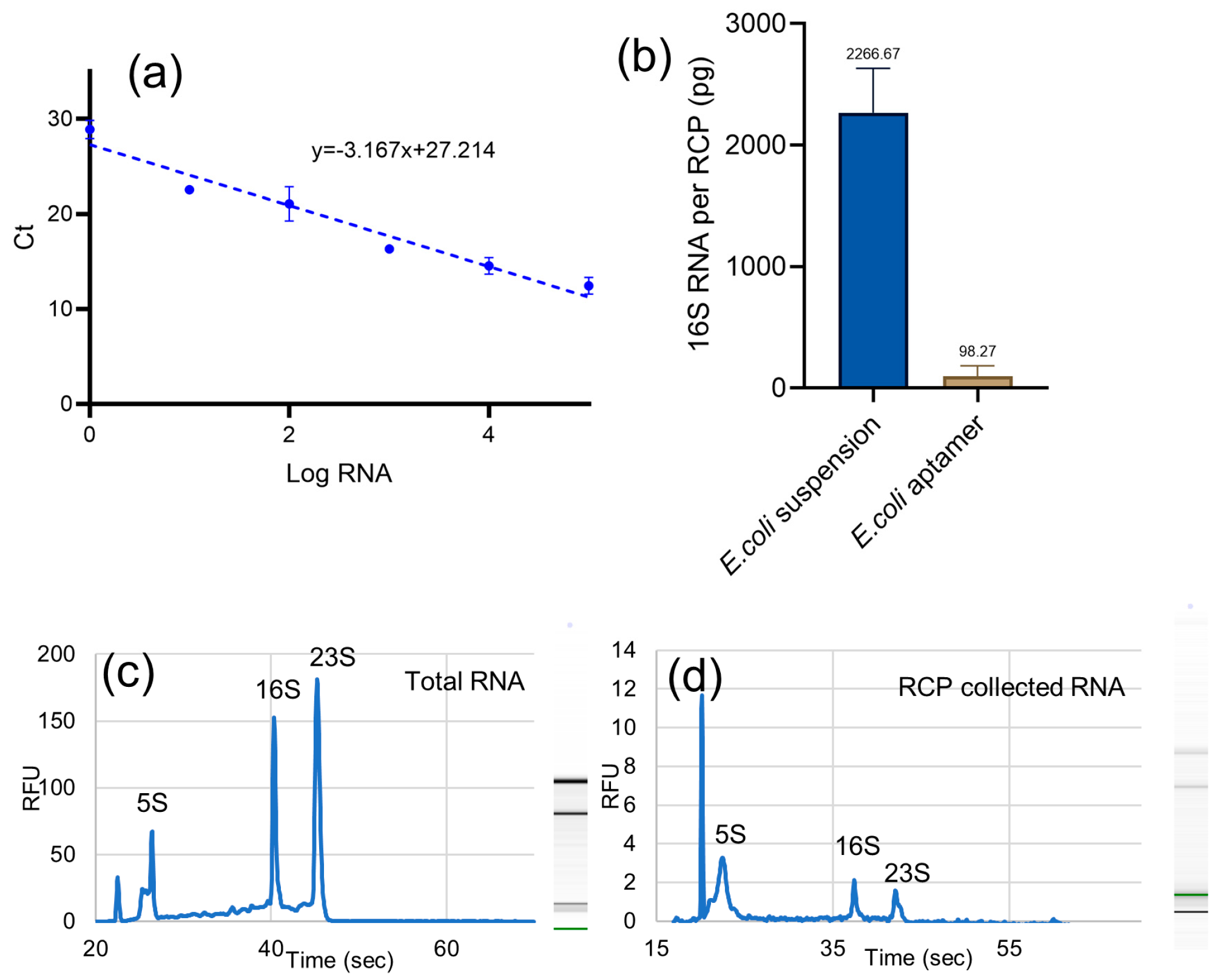
3.4. RNA Capture Pin Efficiency and Specificity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RCP | RNA capture pin |
| RT-qPCR | Reverse transcription-quantitative PCR |
| rRNA | Ribosomal RNA |
References
- Mothershed, E.A.; Whitney, A.M. Nucleic acid-based methods for the detection of bacterial pathogens: Present and future considerations for the clinical laboratory. Clin. Chim. Acta 2006, 363, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Rachakonda, S.; Cartee, L. Challenges in Antimicrobial Drug Discovery and the Potential of Nucleoside Antibiotics. Curr. Med. Chem. 2004, 11, 775–793. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Kim, H.U. Systems strategies for developing industrial microbial strains. Nat. Biotechnol. 2015, 33, 1061–1072. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.D.; Wright, G.D. New Targets and Screening Approaches in Antimicrobial Drug Discovery. Chem. Rev. 2005, 105, 759–774. [Google Scholar] [CrossRef]
- Cinti, S.; Singh, S.; Covone, G.; Tonietti, L.; Ricciardelli, A.; Cordone, A.; Iacono, R.; Mazzoli, A.; Moracci, M.; Rotundi, A.; et al. Reviewing the state of biosensors and lab-on-a-chip technologies: Opportunities for extreme environments and space exploration. Front. Microbiol. 2023, 14, 1215529. [Google Scholar] [CrossRef]
- Krakos, A. Lab-on-chip technologies for space research—Current trends and prospects. Microchim. Acta 2024, 191, 31. [Google Scholar] [CrossRef]
- Gomes, T.A.; Zanette, C.M.; Spier, M.R. An overview of cell disruption methods for intracellular biomolecules recovery. Prep. Biochem. Biotechnol. 2020, 50, 635–654. [Google Scholar] [CrossRef]
- Abolmaaty, A.; El-Shemy, M.G.; Khallaf, M.F.; Levin, R.E. Effect of lysing methods and their variables on the yield of Escherichia coli O157: H7 DNA and its PCR amplification. J. Microbiol. Methods 1998, 34, 133–141. [Google Scholar] [CrossRef]
- Vandeventer, P.E.; Weigel, K.M.; Salazar, J.; Erwin, B.; Irvine, B.; Doebler, R.; Nadim, A.; Cangelosi, G.A.; Niemz, A. Mechanical Disruption of Lysis-Resistant Bacterial Cells by Use of a Miniature, Low-Power, Disposable Device. J. Clin. Microbiol. 2011, 49, 2533–2539. [Google Scholar] [CrossRef]
- Zhang, L.; Foxman, B.; Gilsdorf, J.R.; Marrs, C.F. Bacterial Genomic DNA Isolation Using Sonication for Microarray Analysis. Biotechniques 2005, 39, 640–644. [Google Scholar] [CrossRef]
- Danaeifar, M. New horizons in developing cell lysis methods: A review. Biotechnol. Bioeng. 2022, 119, 3007–3021. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Lv, X.; Su, Y.; Fan, Z.; Fang, W.; Duan, J.; Zhang, S.; Ma, B.; Liu, F.; Chen, H.; et al. Piezoelectric Microchip for Cell Lysis through Cell–Microparticle Collision within a Microdroplet Driven by Surface Acoustic Wave Oscillation. Small 2019, 15, 1804593. [Google Scholar] [CrossRef] [PubMed]
- Boom, R.; Sol, C.J.; Salimans, M.M.; Jansen, C.L.; Wertheim-van Dillen, P.M.; van der Noordaa, J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990, 28, 495–503. [Google Scholar] [CrossRef]
- Pang, X.; Zhou, D.; Song, Y.; Pei, D.; Wang, J.; Guo, Z.; Yang, R. Bacterial mRNA Purification by Magnetic Capture-Hybridization Method. Microbiol. Immunol. 2004, 48, 91–96. [Google Scholar] [CrossRef]
- Scherp, P.; Hasenstein, K.H. Solid Phase Gene Extraction Isolates mRNA at High Spatial and Temporal Resolution. Biotechniques 2008, 45, 172–178. [Google Scholar] [CrossRef]
- Nestorova, G.G.; Hasenstein, K.; Nguyen, N.; DeCoster, M.A.; Crews, N.D. Lab-on-a-chip mRNA purification and reverse transcription via a solid-phase gene extraction technique. Lab Chip 2017, 17, 1128–1136. [Google Scholar] [CrossRef]
- Nestorova, G.G.; Crews, N.; Schramm, A.K.; Aquilina, R.A.; Parra, M.; Chin, M.; Chinn, T.; Hee, L. Spaceflight validation of one-step Gene Sampling tool for genetic analysis on the International Space Station. Acta Astronaut. 2022, 198, 225–232. [Google Scholar] [CrossRef]
- Gaines, D.; Brodsky, E.; Kaur, H.; Nestorova, G.G. RNA capture pin technology: Investigating long-term stability and mRNA purification specificity of oligonucleotide immobilization on gold and streptavidin surfaces. Anal. Bioanal. Chem. 2023, 415, 6077–6089. [Google Scholar] [CrossRef]
- Liedberg, B.; Cooper, J.M. Bioanalytical applications of self-assembled monolayers. In Immobilized Biomolecules in Analysis; Oxford University Press: Oxford, UK, 1999; pp. 55–78. [Google Scholar] [CrossRef]
- Deka, J.; Měch, R.; Ianeselli, L.; Amenitsch, H.; Cacho-Nerin, F.; Parisse, P.; Casalis, L. Surface Passivation Improves the Synthesis of Highly Stable and Specific DNA-Functionalized Gold Nanoparticles with Variable DNA Density. ACS Appl. Mater. Interfaces 2015, 7, 7033–7040. [Google Scholar] [CrossRef]
- Sypabekova, M.; Hagemann, A.; Rho, D.; Kim, S. Review: 3-Aminopropyltriethoxysilane (APTES) Deposition Methods on Oxide Surfaces in Solution and Vapor Phases for Biosensing Applications. Biosensors 2023, 13, 36. [Google Scholar] [CrossRef]
- Abdelrasoul, G.N.; Anwar, A.; MacKay, S.; Tamura, M.; Shah, M.A.; Khasa, D.P.; Montgomery, R.R.; Ko, A.I.; Chen, J. DNA aptamer-based non-faradaic impedance biosensor for detecting E. coli. Anal. Chim. Acta 2020, 1107, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Mutafopulos, K.; Heyman, J.A.; Spink, P.; Shen, L.; Wang, C.; Franke, T.; Weitz, D.A. Rapid additive-free bacteria lysis using traveling surface acoustic waves in microfluidic channels. Lab Chip 2019, 19, 4064–4070. [Google Scholar] [CrossRef] [PubMed]
- Tandiono, T.; Ow, D.S.-W.; Driessen, L.; Chin, C.S.-H.; Klaseboer, E.; Choo, A.B.-H.; Ohl, S.-W.; Ohl, C.-D. Sonolysis of Escherichia coli and Pichia pastoris in microfluidics. Lab Chip 2012, 12, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Gong, H.Q.; Zeng, X.; Lou, C.; Sze, C. A microfluidic liquid phase nucleic acid purification chip to selectively isolate DNA or RNA from low copy/single bacterial cells in minute sample volume followed by direct on-chip quantitative PCR assay. Anal. Chem. 2013, 85, 1484–1491. [Google Scholar] [CrossRef]
- Warner, C.L.; Bruckner-Lea, C.J.; Grate, J.W.; Straub, T.; Posakony, G.J.; Valentine, N.; Ozanich, R.; Bond, L.J.; Matzke, M.M.; Dockendorff, B.; et al. A Flow-Through Ultrasonic Lysis Module for the Disruption of Bacterial Spores. JALA J. Assoc. Lab. Autom. 2009, 14, 277–284. [Google Scholar] [CrossRef]
- Grigorov, E.; Kirov, B.; Marinov, M.B.; Galabov, V. Review of Microfluidic Methods for Cellular Lysis. Micromachines 2021, 12, 498. [Google Scholar] [CrossRef]
- Di Carlo, D.; Jeong, K.-H.; Lee, L.P. Reagentless mechanical cell lysis by nanoscale barbs in microchannels for sample preparation. Lab Chip 2003, 3, 287. [Google Scholar] [CrossRef]
- Petrov, A.S.; Bernier, C.R.; Hershkovits, E.; Xue, Y.; Waterbury, C.C.; Hsiao, C.; Stepanov, V.G.; Gaucher, E.A.; Grover, M.A.; Harvey, S.C.; et al. Secondary structure and domain architecture of the 23S and 5S rRNAs. Nucleic Acids Res. 2013, 41, 7522–7535. [Google Scholar] [CrossRef]
- Fabre, A.-L.; Colotte, M.; Luis, A.; Tuffet, S.; Bonnet, J. An efficient method for long-term room temperature storage of RNA. Eur. J. Hum. Genet. 2014, 22, 379–385. [Google Scholar] [CrossRef]
- Jones, K.L.; Drane, D.; Gowans, E.J. Long-Term Storage of DNA-Free RNA for use in Vaccine Studies. Biotechniques 2007, 43, 675–681. [Google Scholar] [CrossRef]
- Huang, T.; Tzeng, Y.; Dickson, R.M. FAST: Rapid determinations of antibiotic susceptibility phenotypes using label-free cytometry. Cytom. Part A 2018, 93, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Wojno, K.J.; Baunoch, D.; Luke, N.; Opel, M.; Korman, H.; Kelly, C.; Jafri, S.M.A.; Keating, P.; Hazelton, D.; Hindu, S.; et al. Multiplex PCR Based Urinary Tract Infection (UTI) Analysis Compared to Traditional Urine Culture in Identifying Significant Pathogens in Symptomatic Patients. Urology 2020, 136, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Lorena, S.M.; Cynthia, P.C.; Isabela, A.R.M.; Pamela, V.M.; Gabriela, R.H.; Jorge, B.; Mariano, G. Nucleic Acids Isolation for Molecular Diagnostics: Present and Future of the Silica-based DNA/RNA Purification Technologies. Sep. Purif. Rev. 2023, 52, 193–204. [Google Scholar] [CrossRef]


| Feature | RNA Capture Pin | Column-Based Purification | Magnetic Beads-Based Purification |
|---|---|---|---|
| Liquid Handling | Minimal or none | Requires multiple liquid-handling steps | Requires washing and elution steps |
| Processing Time | Fast (a few minutes) | Moderate (30–60 min) | Moderate (30–60 min) |
| Sample Volume Requirement | Very low | Moderate to high | Low to moderate |
| Contamination Risk | Low (direct capture reduces contaminants) | Moderate (genomic DNA, protein carryover) | Low (RNA-binding surfaces) |
| Selectivity | High (oligo modification allows selective RNA binding) | Moderate (require DNase treatment) | High (customizable bead coatings) |
| Ease of Use | Very simple, with no centrifugation or complex steps | Multiple wash/elution steps | Magnet setup and multiple wash steps |
| Scalability | High (adaptable for field applications) | Moderate (lab-dependent) | High (automation-friendly) |
| Limitations | Newer technology requires further optimization | Labor-intensive | Costly, requires optimization for different applications |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, M.A.; Giorno, R.; Nestorova, G.G. Aptamer-Functionalized Platform for Selective Bacterial Isolation and Rapid RNA Purification Using Capture Pins. Sensors 2025, 25, 1774. https://doi.org/10.3390/s25061774
Islam MA, Giorno R, Nestorova GG. Aptamer-Functionalized Platform for Selective Bacterial Isolation and Rapid RNA Purification Using Capture Pins. Sensors. 2025; 25(6):1774. https://doi.org/10.3390/s25061774
Chicago/Turabian StyleIslam, Md Aminul, Rebecca Giorno, and Gergana G. Nestorova. 2025. "Aptamer-Functionalized Platform for Selective Bacterial Isolation and Rapid RNA Purification Using Capture Pins" Sensors 25, no. 6: 1774. https://doi.org/10.3390/s25061774
APA StyleIslam, M. A., Giorno, R., & Nestorova, G. G. (2025). Aptamer-Functionalized Platform for Selective Bacterial Isolation and Rapid RNA Purification Using Capture Pins. Sensors, 25(6), 1774. https://doi.org/10.3390/s25061774






