Identification of People in a Household Using Ballistocardiography Signals Through Deep Learning
Abstract
1. Introduction
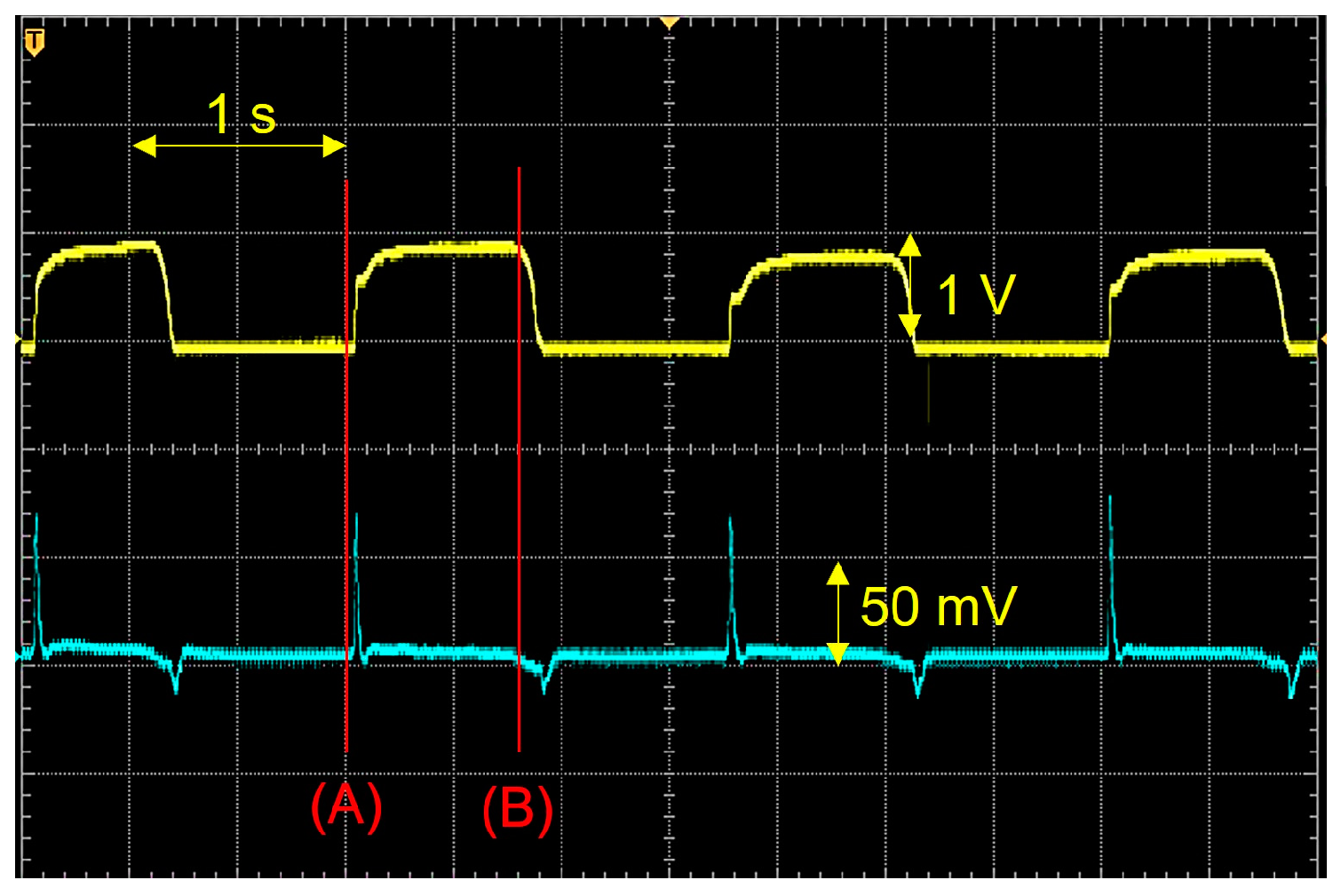
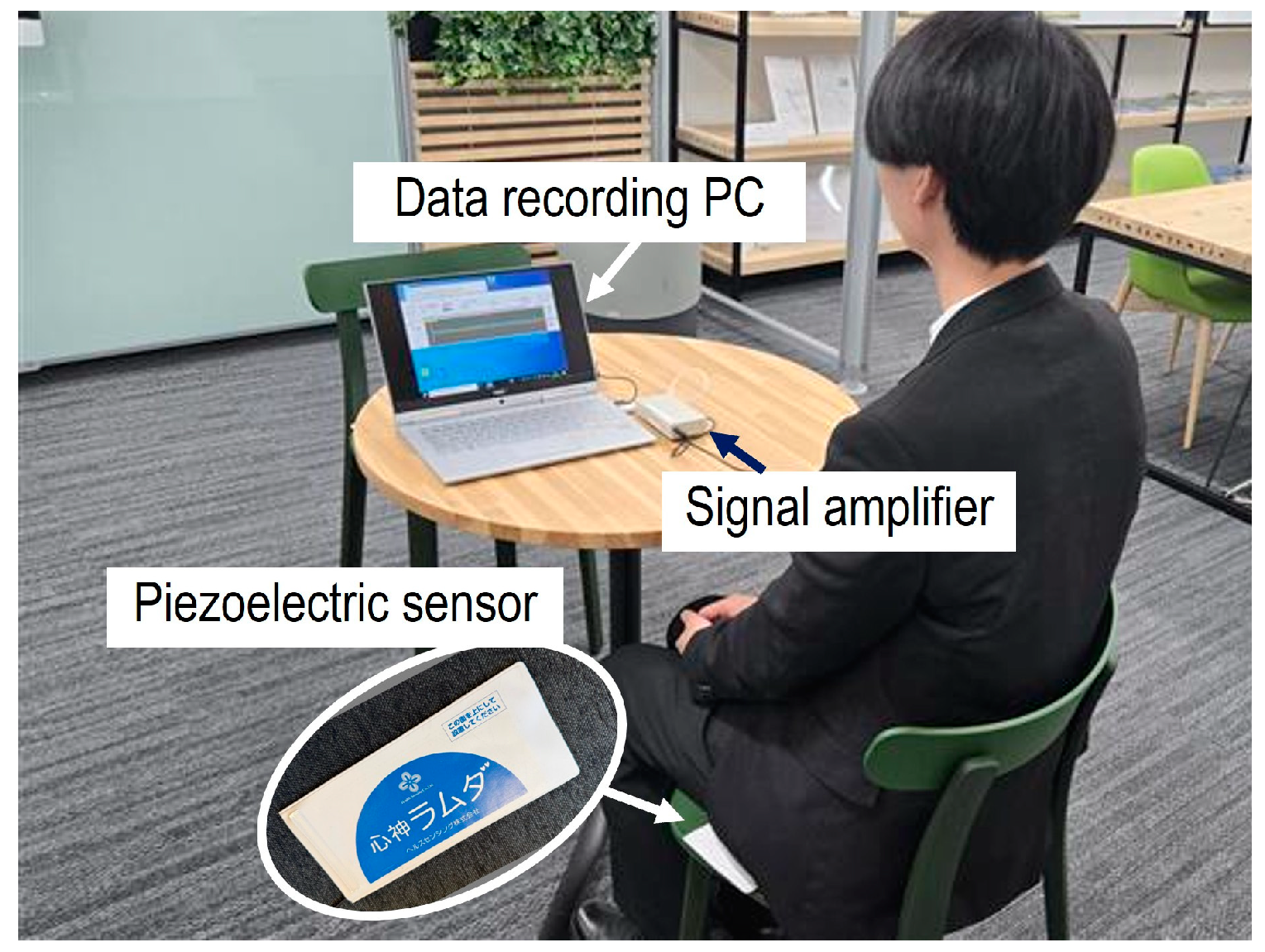
2. Sensor Selection and Data Acquisition
2.1. Sensor Selection
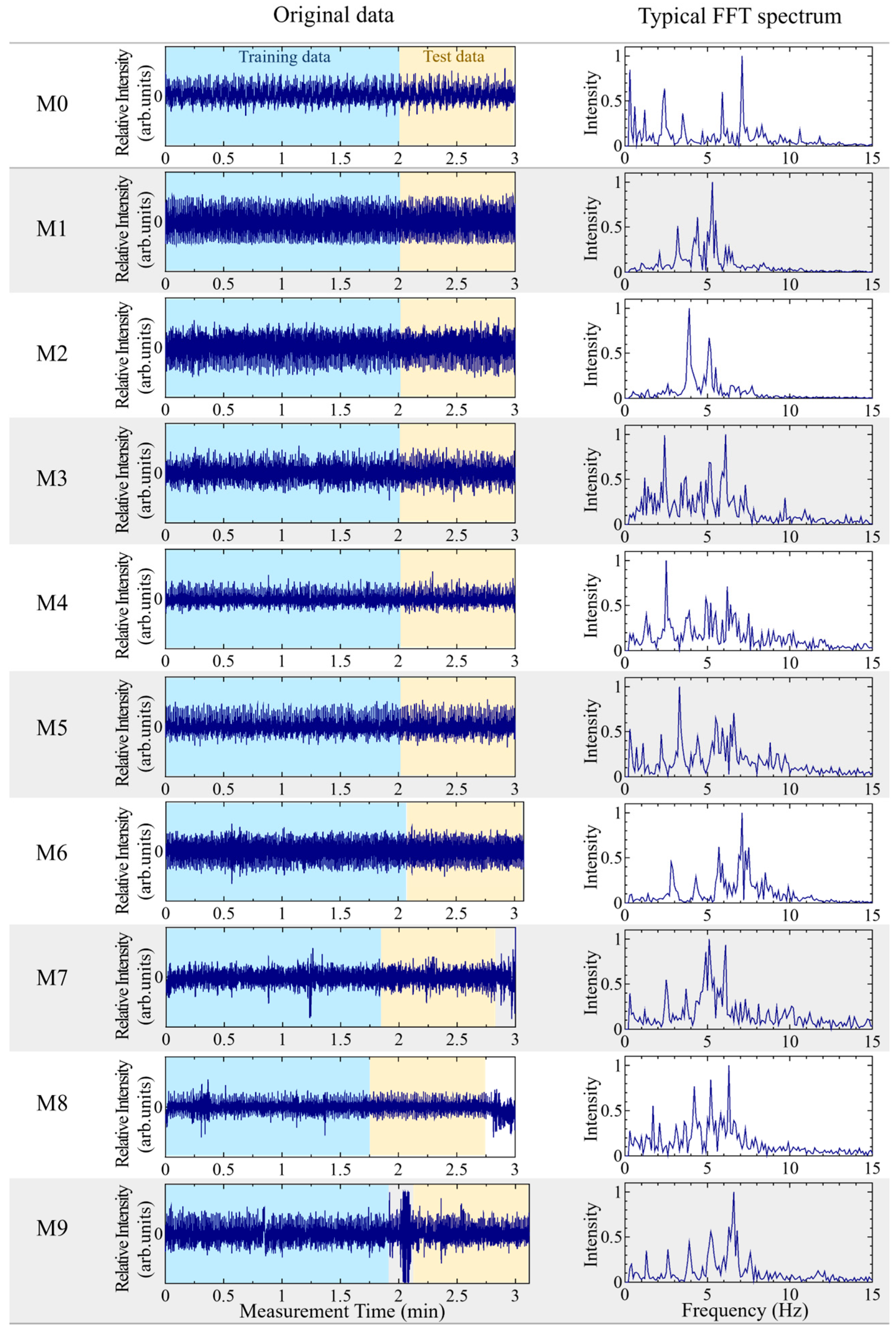
2.2. Sample Dataset
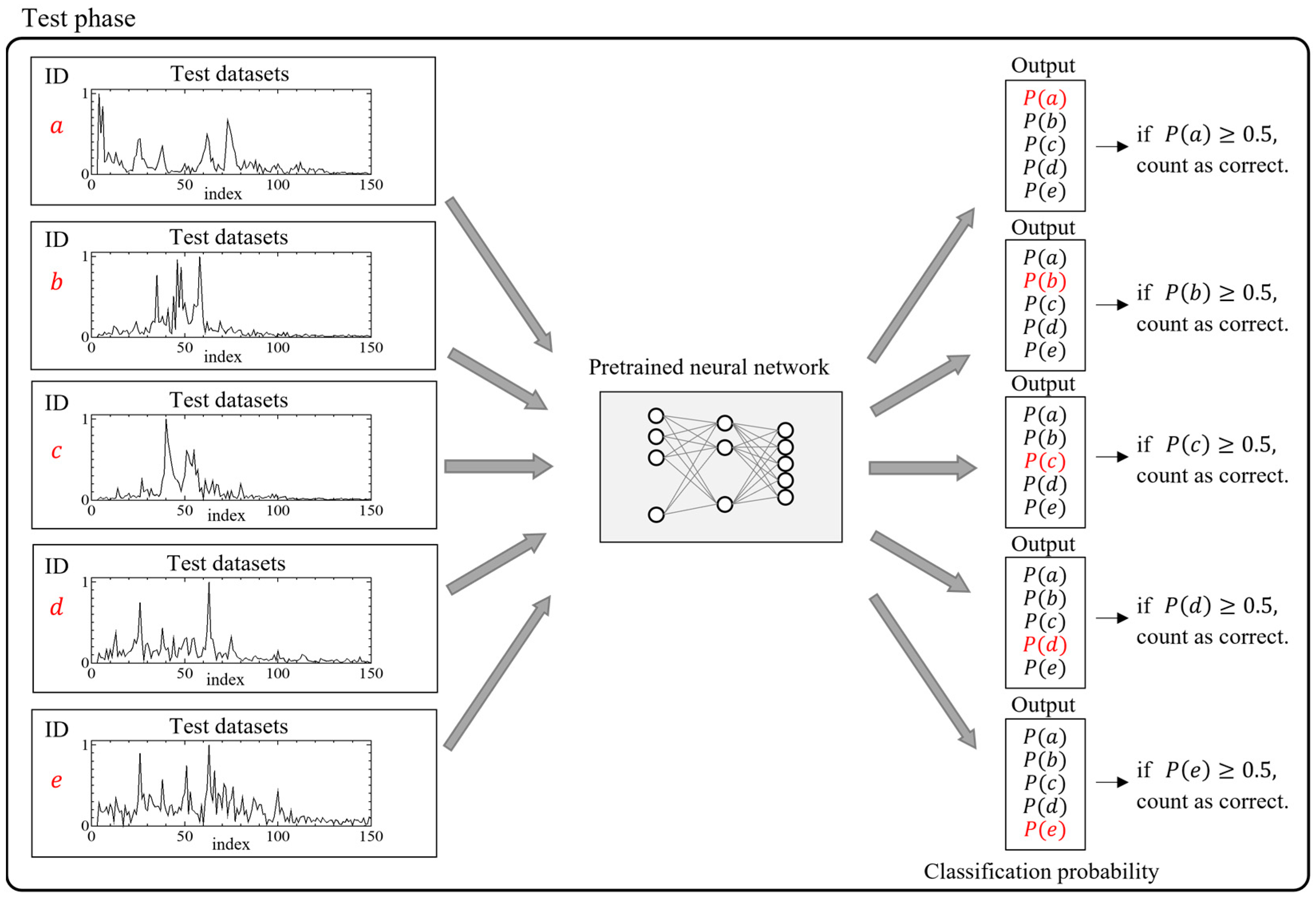
2.3. Five-Classification Analysis
2.3.1. Creation of Training and Test Dataset
2.3.2. Structure of the Neural Network
3. Results
3.1. Processing Time
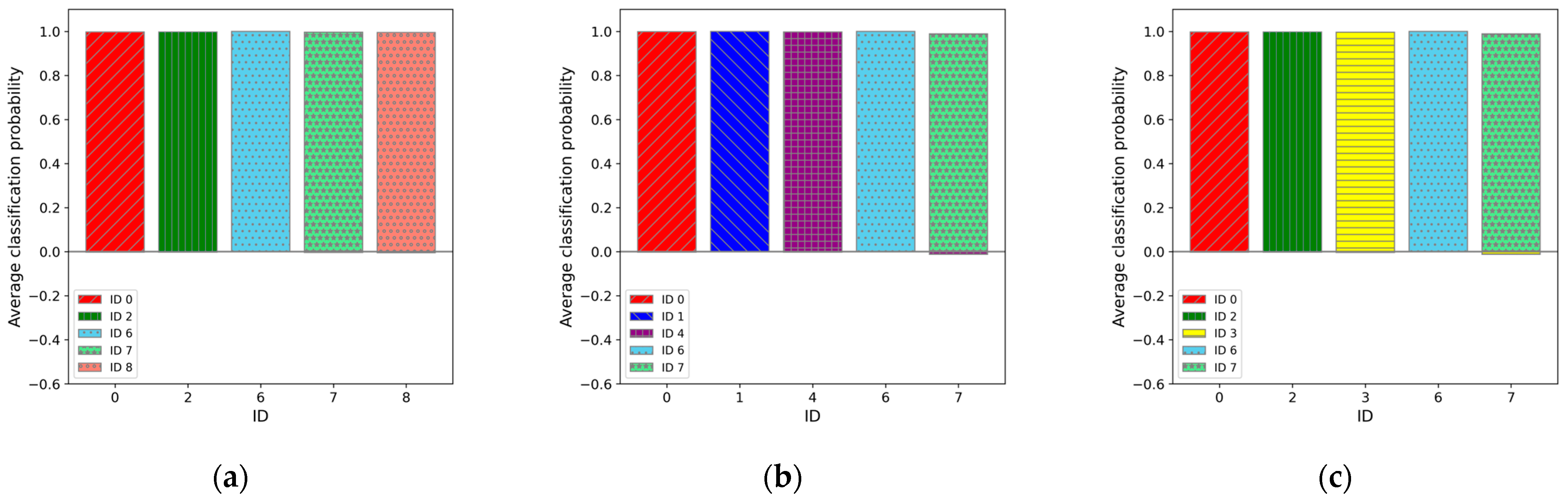
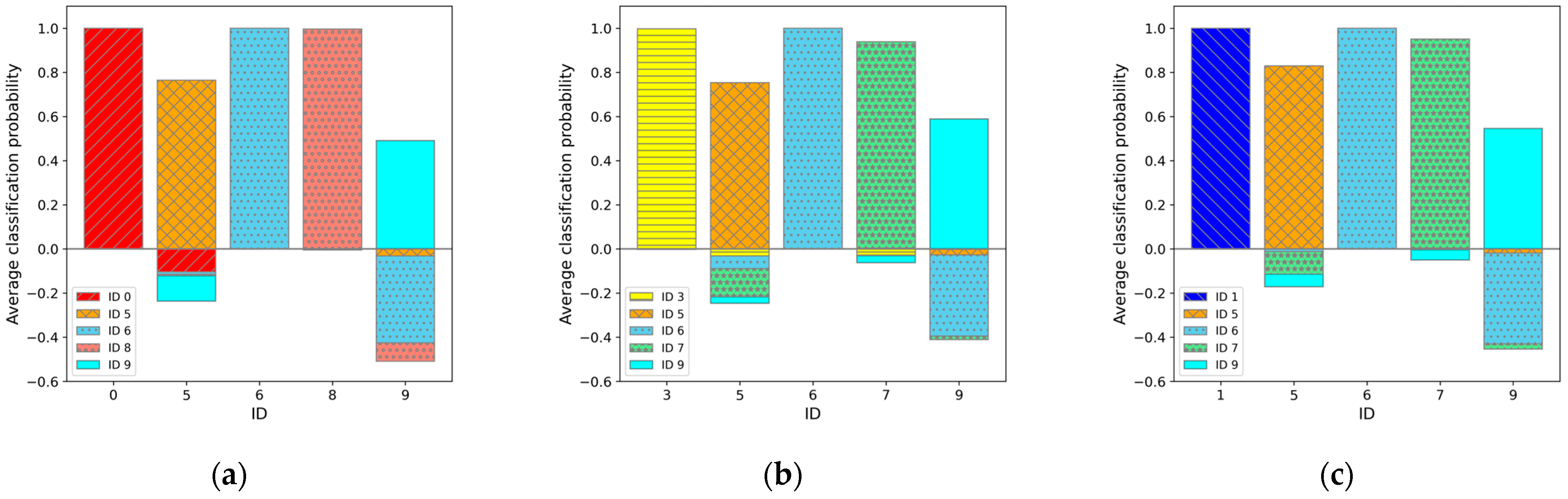
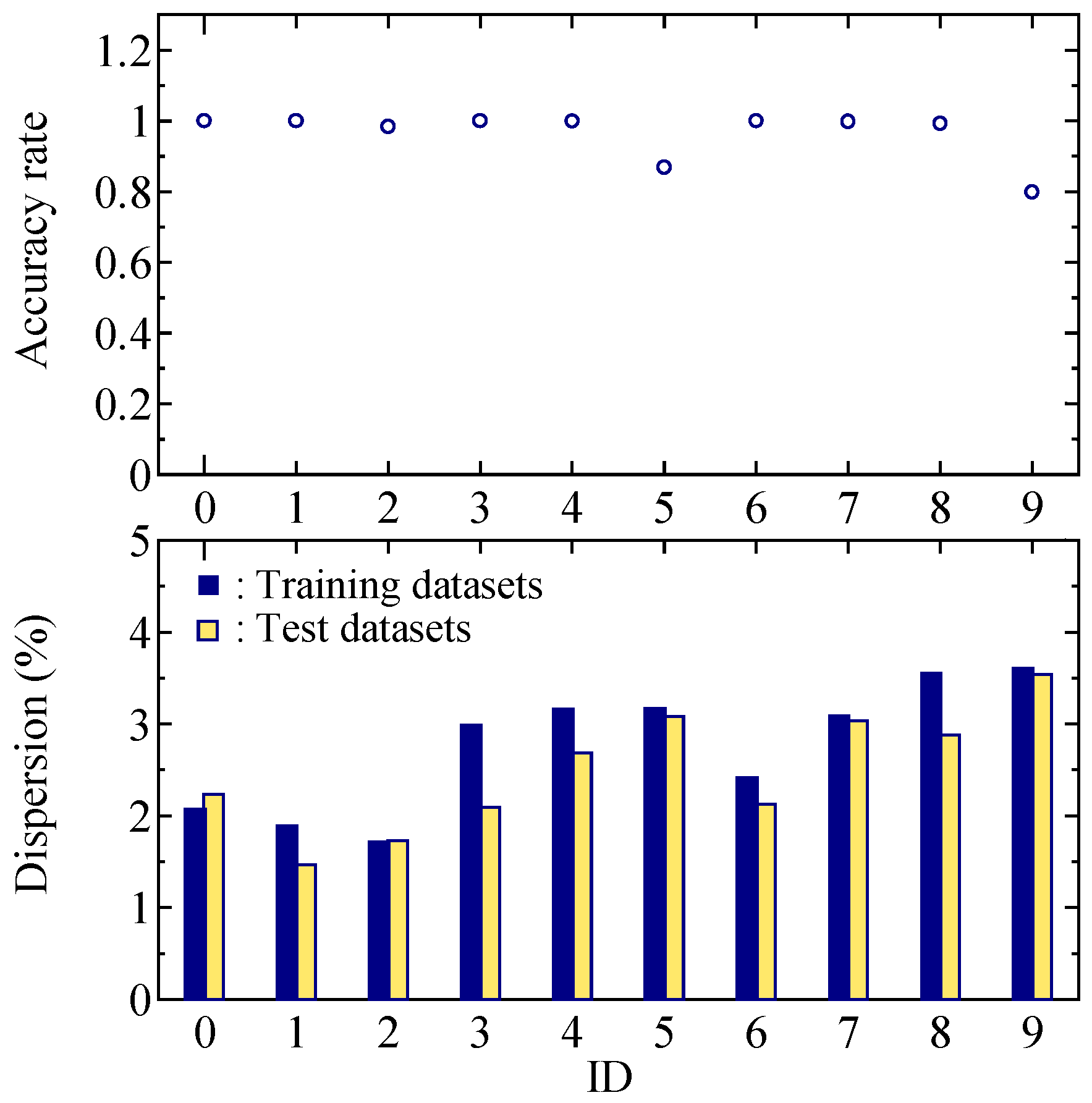
3.2. Five-Class Classification Results
4. Discussion
4.1. Five-Class Classification
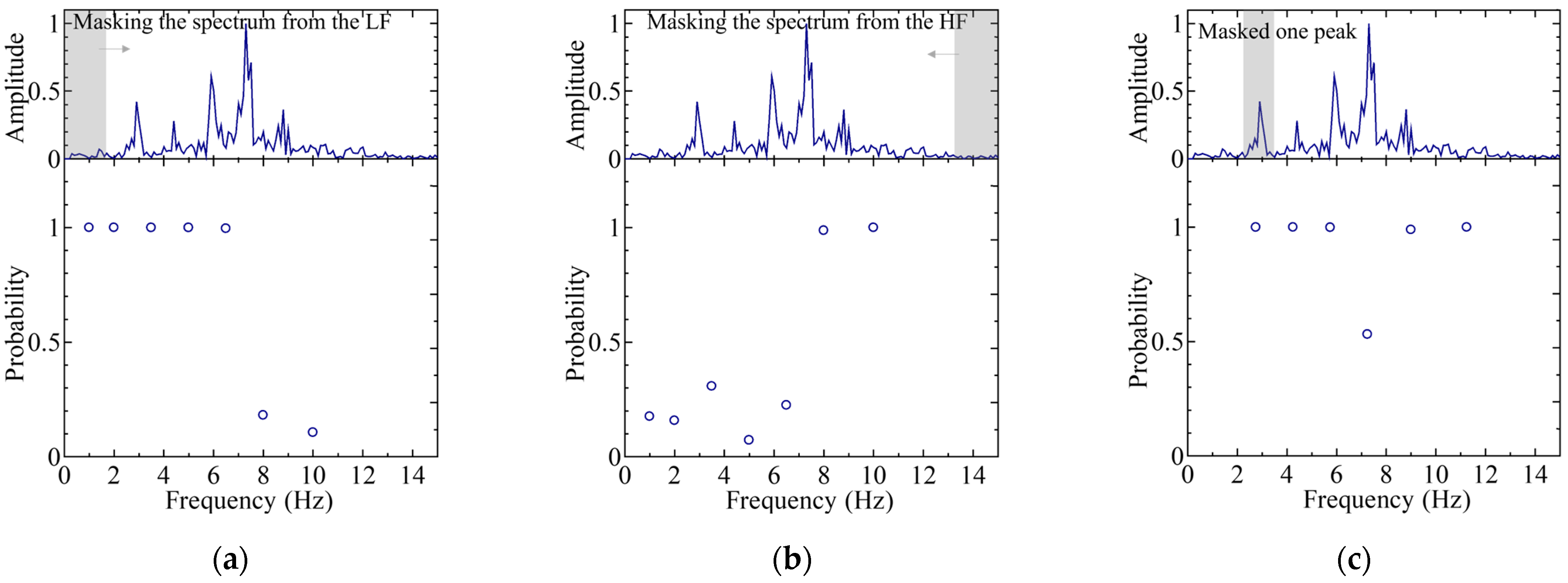
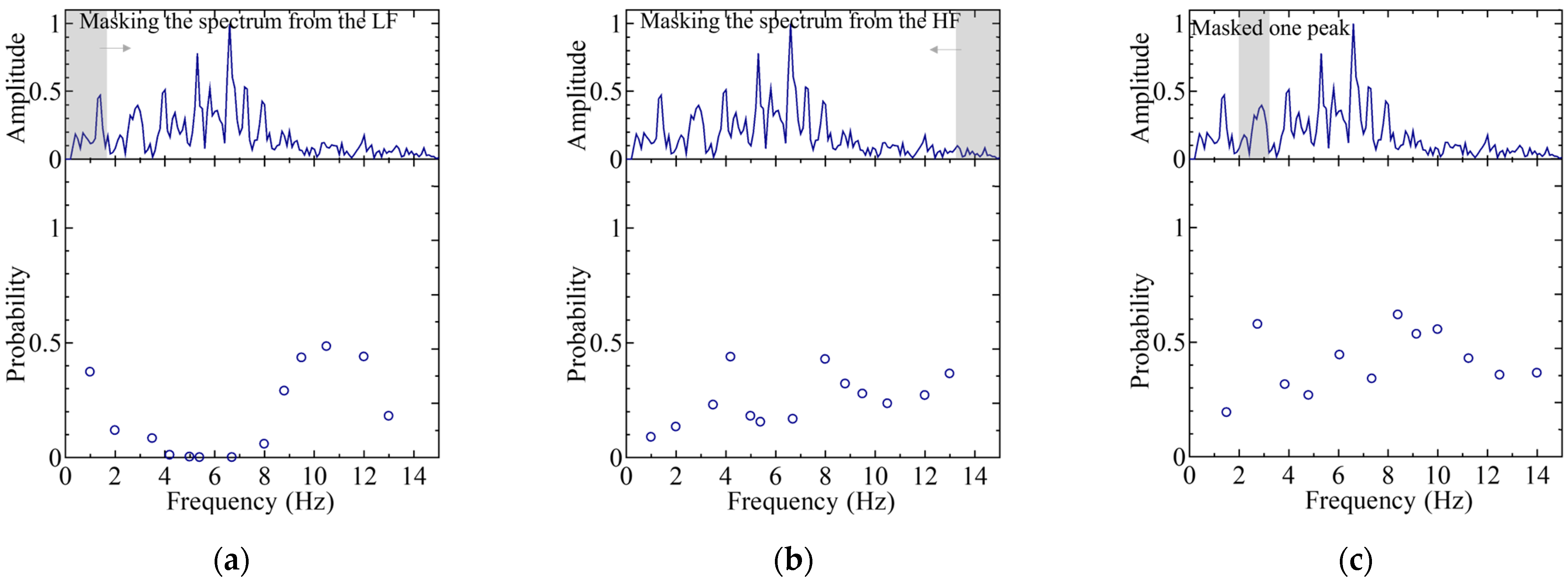
4.2. Features in Regard to the Worst Case
4.3. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Mshali, H.; Lemlouma, T.; Moloney, M.; Magoni, D. A survey on health monitoring system for health smart homes. Int. J. Ind. Ergon. 2018, 66, 26–56. [Google Scholar] [CrossRef]
- Almarashdeh, I.; Alsmadi, M.K.; Farag, T.; Albahussain, A.S.; Badawi, U.A.; Altuwaijri, N.; Almaimoni, H.; Asiry, F.; Alowaid, S.; Alshabanah, M.; et al. Real-Time Elderly Healthcare Monitoring Expert System Using Wireless Sensor Network. Int. J. Appl. Eng. Res. 2018, 13, 3517–3523. [Google Scholar] [CrossRef]
- Majumder, S.; Roy, A.K.; Jiang, W.; Modal, T.; Deen, M.J. Smart Textiles to Enable In-Home Health Care: State of the Art, Challenges, and Future Perspectives. IEEE J. Flex. Electron. 2023, 3, 120–150. [Google Scholar] [CrossRef]
- Shaik, T.; Tao, X.; Higgins, N.; Li, L.; Gururajan, R.; Zhou, X.; Acharya, U.R. Remote patient monitoring using artificial intelligence: Current state, applications, and challenges. WIREs Data Min. Knowl Discov. 2023, 13, e1485. [Google Scholar] [CrossRef]
- Malasinghe, L.P.; Ramzan, N.; Dahal, K. Remote patient monitoring: A comprehensive study. J. Ambient. Intell. Humaniz. Comput. 2019, 10, 57–76. [Google Scholar] [CrossRef]
- Boikanyo, K.; Zungeru, A.M.; Sigweni, B.; Yahya, A.; Lebekwe, C. Remote patient monitoring system: Applications, architecture, and challenges. Sci. Afr. 2023, 20, e01638. [Google Scholar] [CrossRef]
- Rajala, S.; Lekkala, J. Film-Type Sensor Materials PVDF and EMFi in Measurement of Cardiorespiratory Signals—A Review. IEEE Sens. J. 2012, 12, 3. [Google Scholar] [CrossRef]
- Lei, K.-F.; Hsieh, Y.-Z.; Chiu, Y.-Y.; Wu, M.-H. The Structure Design of Piezoelectric Poly (vinylidene Fluoride) (PVDF) Polymer-Based Sensor Patch for the Respiration Monitoring under Dynamic Walking Conditions. Sensors 2015, 15, 18801–18812. [Google Scholar] [CrossRef] [PubMed]
- Morokuma, S.; Hayashi, T.; Kanegae, M.; Mizukami, Y.; Asano, S.; Kimura, I.; Tatezumi, Y.; Ueno, H.; Ikeda, S.; Niizeki, K. Deep learning-based sleep stage classification with cardiorespiratory and body movement activities in individuals with suspected sleep disorders. Sci. Rep. 2023, 13, 17730. [Google Scholar] [CrossRef] [PubMed]
- Ueno, H. A Piezoelectric Sensor Signal Analysis Method for Identifying Persons Groups. Sensors 2019, 19, 733. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zheng, Q.; Liu, B.; Gao, X. EEG Conformer: Convolutional Transformer for EEG Decoding and Visualization. IEEE Trans. Neural Syst. Rehabil. Eng. 2023, 31, 710–719. [Google Scholar] [CrossRef] [PubMed]
- Tangkraingkij, P.; Lursinsap, C.; Sanguansintukul, S.; Desudchit, T. Personal Identification by EEG Using ICA and Neural Network. In Proceedings of the 2010 international conference on Computational Science and Its Applications, Fukuoka, Japan, 23–26 March 2010; Volume 6018, pp. 419–430. [Google Scholar] [CrossRef]
- Akbarnia, Y.; Daliri, M.R. EEG-based identification system using deep neural networks with frequency features. Heliyon 2024, 10, e25999. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Elwarfalli, I. Deep Learning-Powered ECG-Based Biometric Authentication. In Proceedings of the 2023 International Conference on Next-Generation Computing, IoT and Machine Learning (NCIM), Gazipur, Bangladesh, 16–17 June 2023; pp. 1–6. [Google Scholar] [CrossRef]
- Al-Jibreen, A.; Al-Ahmadi, S.; Islam, S.; Artoli, A.M. Person identification with arrhythmic ECG signals using deep convolution neural network. Sci. Rep. 2024, 14, 4431. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Ding, G.; Wang, H.; Xu, F. Person Identification With Millimeter-Wave Radar in Realistic Smart Home Scenarios. IEEE Geosci. Remote Sens. Lett. 2022, 19, 1–5. [Google Scholar] [CrossRef]
- Zhao, P.; Lu, C.X.; Wang, J.; Chen, C.; Wang, W.; Trigoni, N.; Markham, A. Human tracking and identification through a millimeter wave radar. Ad Hoc Netw. 2021, 116, 102475. [Google Scholar] [CrossRef]
- Zhuang, Y.; Chuah, J.H.; Chow, C.O.; Lim, M.G. Iris Recognition using Convolutional Neural Network. In Proceedings of the 2020 IEEE 10th International Conference on System Engineering and Technology (ICSET), Shah Alam, Malaysia, 9 November 2020; pp. 134–138. [Google Scholar] [CrossRef]
- Tang, Y.; Gao, F.; Feng, J.; Liu, Y. FingerNet: An unified deep network for fingerprint minutiae extraction. In Proceedings of the 2017 IEEE International Joint Conference on Biometrics (IJCB), Denver, CO, USA, 1–4 October 2017; pp. 108–116. [Google Scholar] [CrossRef]
- Takahashi, K.; Ueno, H. Ballistocardial Signal-Based Personal Identification Using Deep Learning for the Non-Invasive and Non-Restrictive Monitoring of Vital Signs. Sensors 2024, 24, 2527. [Google Scholar] [CrossRef] [PubMed]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, K.; Tanno, Y.; Ueno, H. Identification of People in a Household Using Ballistocardiography Signals Through Deep Learning. Sensors 2025, 25, 1732. https://doi.org/10.3390/s25061732
Takahashi K, Tanno Y, Ueno H. Identification of People in a Household Using Ballistocardiography Signals Through Deep Learning. Sensors. 2025; 25(6):1732. https://doi.org/10.3390/s25061732
Chicago/Turabian StyleTakahashi, Karin, Yoshinobu Tanno, and Hitoshi Ueno. 2025. "Identification of People in a Household Using Ballistocardiography Signals Through Deep Learning" Sensors 25, no. 6: 1732. https://doi.org/10.3390/s25061732
APA StyleTakahashi, K., Tanno, Y., & Ueno, H. (2025). Identification of People in a Household Using Ballistocardiography Signals Through Deep Learning. Sensors, 25(6), 1732. https://doi.org/10.3390/s25061732





