Generating Accurate Activity Patterns for Cattle Farm Management Using MCMC Simulation of Multiple-Sensor Data System
Abstract
1. Introduction
- Our work specifically addresses the analytical challenge of effectively integrating diverse sensor data streams for nuanced behavioral analysis.
- We build upon the foundational literature by demonstrating that the Markov Chain Monte Carlo (MCMC) simulation offers a robust, flexible, and interpretable framework for modeling the complex, dynamic nature of cattle behavior.
- This approach is becoming increasingly vital for providing nuanced, actionable insights that go beyond simple binary or multi-class classifications, representing a significant step forward in the application of advanced statistical and computational methods to the real-world problems of modern livestock farming.
2. Related Works
2.1. From Single-Sensor to Multi-Sensor Fusion
2.2. State-of-the-Art (SOTA) Computational Methods
- Deep Learning for Temporal Analysis: Studies like [8] have successfully employed Recurrent Neural Networks (RNNs) and Long Short-Term Memory (LSTM) models. These architectures are particularly effective at capturing temporal dependencies in time-series data, enabling accurate prediction of behavioral changes related to estrus detection and disease onset.
- Bayesian and Probabilistic Modeling: A notable advancement is the increasing adoption of Bayesian modeling techniques, with Markov Chain Monte Carlo (MCMC) as a core computational component. For example, ref. [9] developed a Bayesian hierarchical model to predict cow calving behavior from accelerometer data. This probabilistic approach is a significant improvement over traditional deterministic models because it provides not just a prediction, but also a quantifiable measure of uncertainty, which is crucial for high-stakes decision-making in farming.
- MCMC Applications and Theory: MCMC is a rapidly growing sampling method [10] that exploits the Markov property, where the next generated sample depends only on the current state [11]. This allows for the approximation of complex posterior distributions in Bayesian inference with a minimal number of samples, a necessity when analytical solutions are intractable [11]. The versatility of Bayesian and MCMC techniques—covering hierarchical, spatial, and nonparametric modeling—is explored in detail in the comprehensive literature reviews of [12,13,14,15], with practical introductions provided by [16] and advanced coverage in [10,17,18,19].
2.3. Focus on Animal Welfare and Research Gaps
3. Materials and Methods
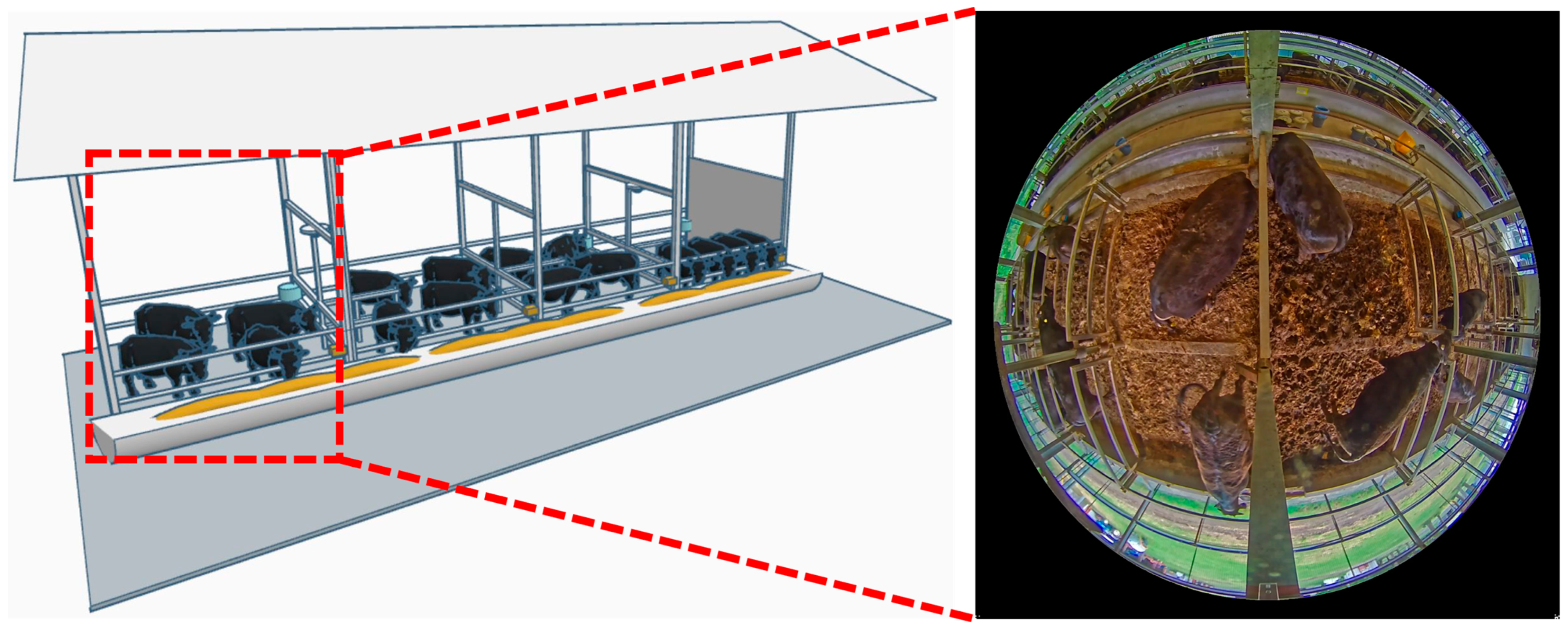
3.1. Study Subjects and Environmental Conditions
3.2. Data Collection and Sensor System
- Motion Data: The primary function is to capture movement data via its integrated accelerometer and gyroscope. This data is used to derive the activity and context features.
- Environmental Data (T and H): Crucially for this question, each U-Motion unit contains dedicated temperature and humidity sensors. These sensors provide real-time, localized readings of the immediate ambient environment surrounding the device.
Data Acquisition Synchronization
- 3D-Accelerometer Axes and Behavioral Monitoring
- The 3D accelerometer reports data along three axes relative to the cow’s movement and the device’s mounting position on the neck.
- X-Axis (Heave/Pitch): Measures motion along the length of the cow’s body (forward/backward movement and head nodding). This correlates highly with grazing posture changes and walking behavior.
- Y-Axis (Sway/Roll): Measures lateral motion (side-to-side, perpendicular to the neck). This correlates with head swinging during walking or searching movements.
- Z-Axis (Surge/Yaw): Measures vertical motion (up/down). This is critical for detecting head-up/head-down changes (e.g., transition from lying to standing, or lifting the head during feeding).
- 2.
- Mastication Detection (Acoustic/Vibration Sensor)
- The neck-mounted sensor monitors for mastication events. The movement/vibration associated with chewing, though originating in the jaw, transmits signals through the bone and soft tissue of the neck, where the sensor is mounted.
- Differentiation: As detailed in [27], the key to differentiating feeding from rumination lies in the mastication rate and pattern:
- ○
- Feeding: Characterized by rapid, high-intensity chewing bouts used for ingesting feed.
- ○
- Rumination: Characterized by slower, highly rhythmic, sustained chewing of the cud bolus. The U-Motion algorithm uses the frequency, duration, and amplitude signature of the detected micro-vibrations/acoustic pulses to classify the behavior with high specificity.
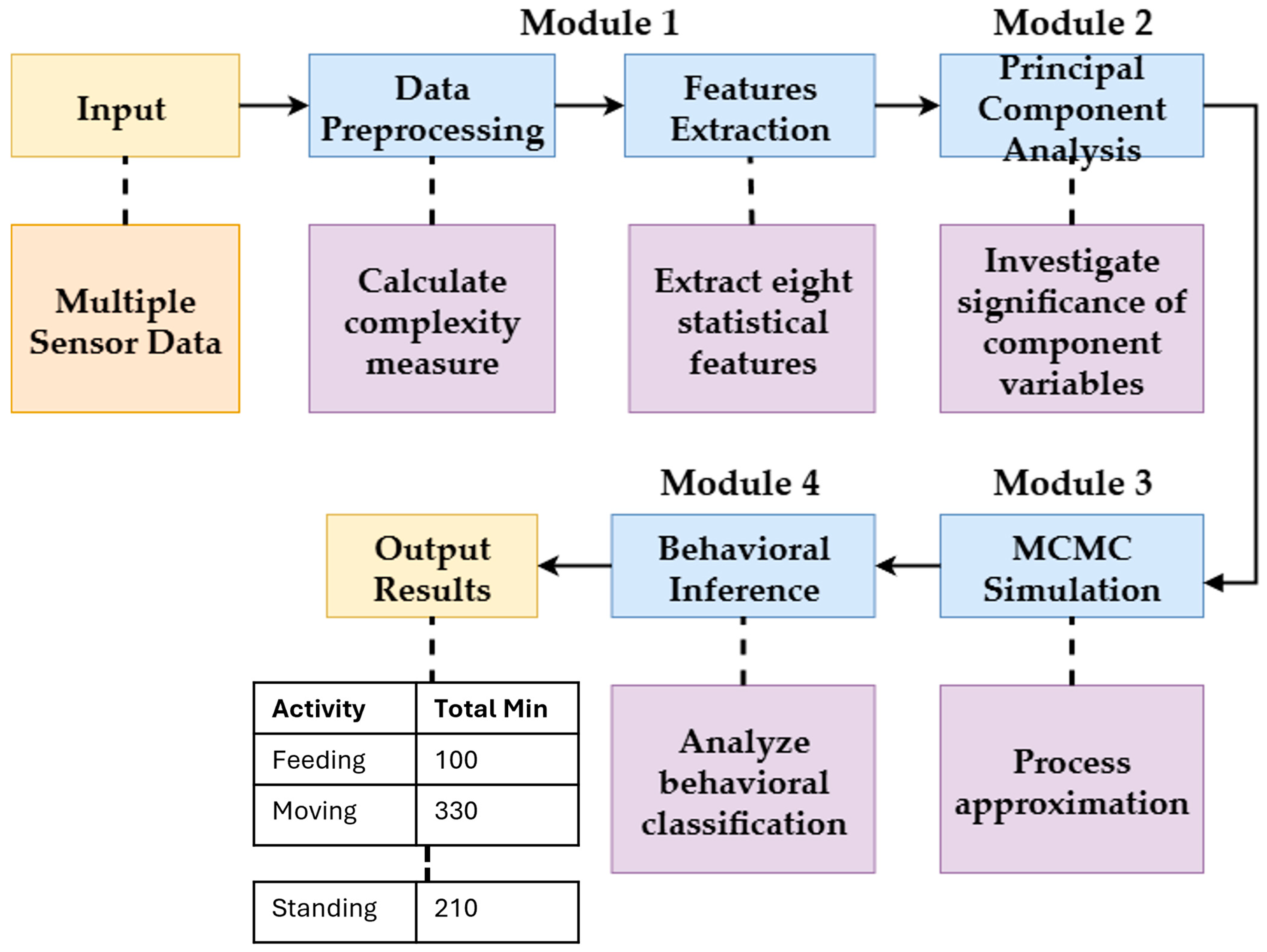
3.3. MCMC System Architecture
3.3.1. Module 1: Data Preprocessing and Feature Extraction
3.3.2. Module 2: Principal Component Analysis (PCA)
- Calculate the Covariance Matrix: An 8 × 8 covariance matrix was computed from the eight features of the MTS data.
- Calculate Eigenvalues and Eigenvectors: We determined the eigenvalues and eigenvectors of the covariance matrix. The principal components (eigenvectors) corresponding to the largest eigenvalues, which explained 90% of the total variance, were selected for further analysis.
- Data Transformation: The original data were transformed into a lower-dimensional space using the selected eigenvectors. This representation, which captured the most important information, was then used in the subsequent MCMC simulation.
3.3.3. Module 3: Markov Chain Monte Carlo (MCMC) Simulation
- Feeding (F): Time spent with the head down, detected by the acceleration sensor, combined with proximity to the feeding trough.
- Moving (M): Displacement and speed of the animal, determined by changes in the proximity sensor readings and acceleration data.
- Lying (L): Time spent in a recumbent position, identified by the acceleration sensor’s posture data.
- Standing (S): Time spent in an upright, non-moving posture.
- Rumination while standing (RS): The number of rumination boluses detected by the pneumatic sensor while the animal is in a standing posture.
- Rumination while lying (RL): The number of rumination boluses detected by the pneumatic sensor while the animal is in a lying posture.
3.3.4. Module 4: Behavioral Inference
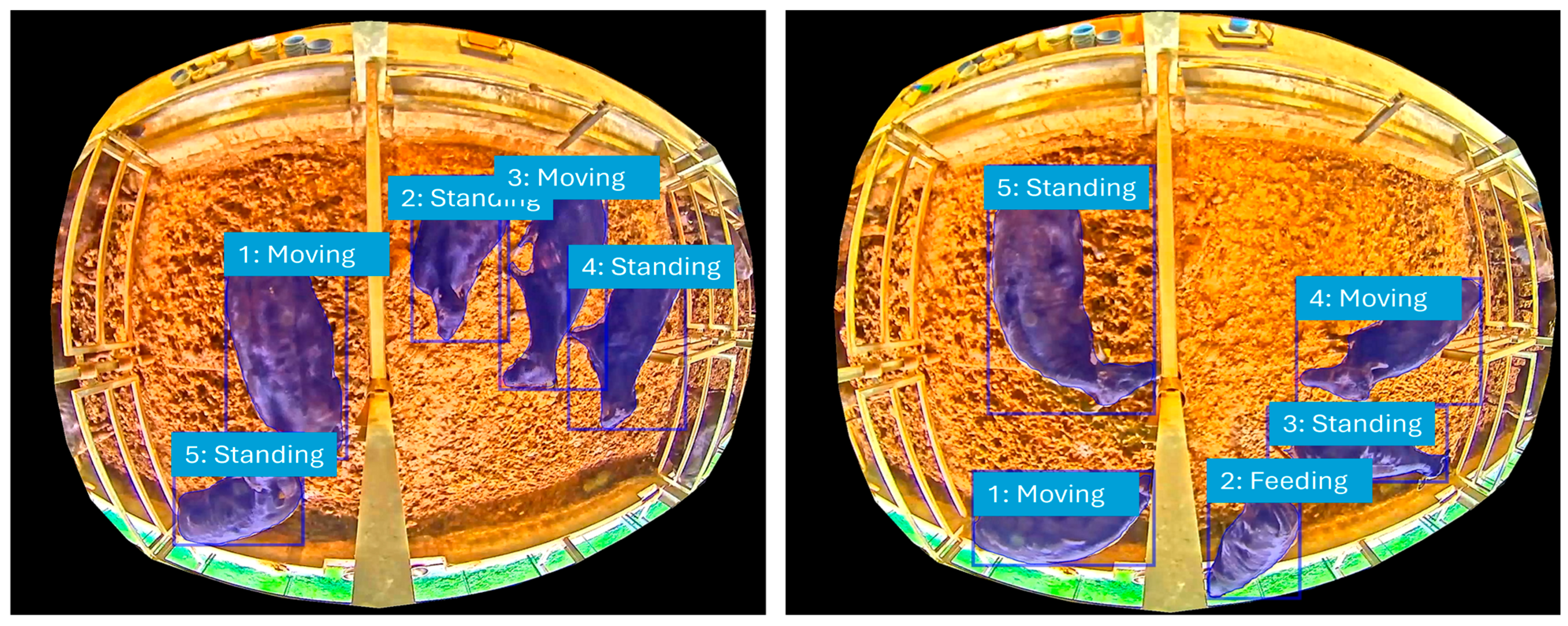
4. Experimental Design and Results
4.1. Experimental Design
4.1.1. Data Collection and Experimental Scale
- 3D-Acceleration Sensor: Captured movement data (e.g., activity counts, postural changes).
- Pneumatic Sensor: Recorded rumination and respiration patterns.
- Proximity Sensor: Logged social interactions and contact with feeding stations.
4.1.2. Data Pre-Processing and PCA for Variable Selection
- Standardization: All sensor data (e.g., acceleration values, respiration rates) were first standardized to have a mean of zero and a standard deviation of one. This ensures that variables with larger initial values do not disproportionately influence the analysis.
- Covariance Matrix Calculation: We computed the covariance matrix of the standardized data, which quantifies the relationships between all pairs of variables.
- Eigen-Decomposition: We performed eigen-decomposition on the covariance matrix to find its eigenvectors and eigenvalues. The eigenvectors represent the principal components (new axes), and the eigenvalues represent the amount of variance explained by each principal component.
- Variable Selection: We selected the top principal components that collectively explained over 95% of the total variance. The original variables that contributed most significantly to these selected principal components were identified. This process allowed us to confirm that the six selected behavioral activities (F, M, L, S, RS, RL) and the three environmental variables (T, H, THI) were indeed the most representative and influential variables for our model. This step ensures that our model is built on a robust and non-redundant set of features, enhancing its efficiency and interpretability.
4.1.3. Markov Chain Monte Carlo (MCMC) Simulation Details
- Burn-in Period: A burn-in period of 1000 iterations was used for all simulations. The first 1000 generated samples were discarded to allow the Markov chain to converge to its stationary distribution, ensuring that the subsequent samples are representative of the true underlying behavior patterns.
- Number of Iterations (n): We performed simulations with 3000, 4000, and 5000 iterations to assess the stability and accuracy of the model with increasing sample sizes. This range of iteration counts allowed us to observe the point at which the model’s performance plateaus.
- Prior Transition Matrix: The simulation was initialized using a prior transition matrix (P) derived from the collected sample data. This matrix represents the observed probabilities of transitioning from one state (e.g., Lying) to another (e.g., Standing). This prior knowledge provides a realistic starting point for the MCMC process.
- Proposal Distribution: We used a Metropolis–Hastings algorithm with a simple uniform proposal distribution. This ensures that the simulation explores the entire state space of possible transitions, preventing it from getting stuck in local optima.
4.2. Experimental Results
4.2.1. Dimensionality Reduction via PCA
4.2.2. MCMC Simulation and Model Accuracy
4.2.3. Performance Metrics
- Continuous Output: Our model’s objective is to predict the level of activity or environmental impact, which is represented as a continuous value (e.g., an activity intensity score, or a continuous measure of impact) that falls within a defined range, rather than a single, mutually exclusive category (like “Activity A” or “Activity B”). This output is conceptually treated as a regression target.
- Focus on Magnitude of Error: Average Absolute Error (AAE), Average Error in Squares (AES), and Root Mean Square Error (RMSE) are measures designed to quantify the magnitude of the prediction error. AAE provides the average size of the error in the original units. RMSE (and relatedly, AES) places a higher penalty on large errors (outliers) due to the squaring of the difference.
- Inapplicability of Classification Metrics: Metrics like Precision, Recall, and F1 Score are designed for discrete classification problems where the prediction is either entirely correct or incorrect (e.g., predicting “Cat” vs. “Dog”). These metrics are calculated based on a confusion matrix (True Positives, False Positives, etc.), which is not applicable when the model output is a continuous number. Forcing a continuous score into a discrete class for these metrics would discard valuable information about the predicted intensity or degree.
5. Discussion
5.1. Interpretation of Key Findings
- Multivariate Data Integration: The use of a multivariate time series was crucial, allowing the model to capture the complex, interconnected nature of animal behavior (e.g., the relationship between locomotion, feed intake, and resting).
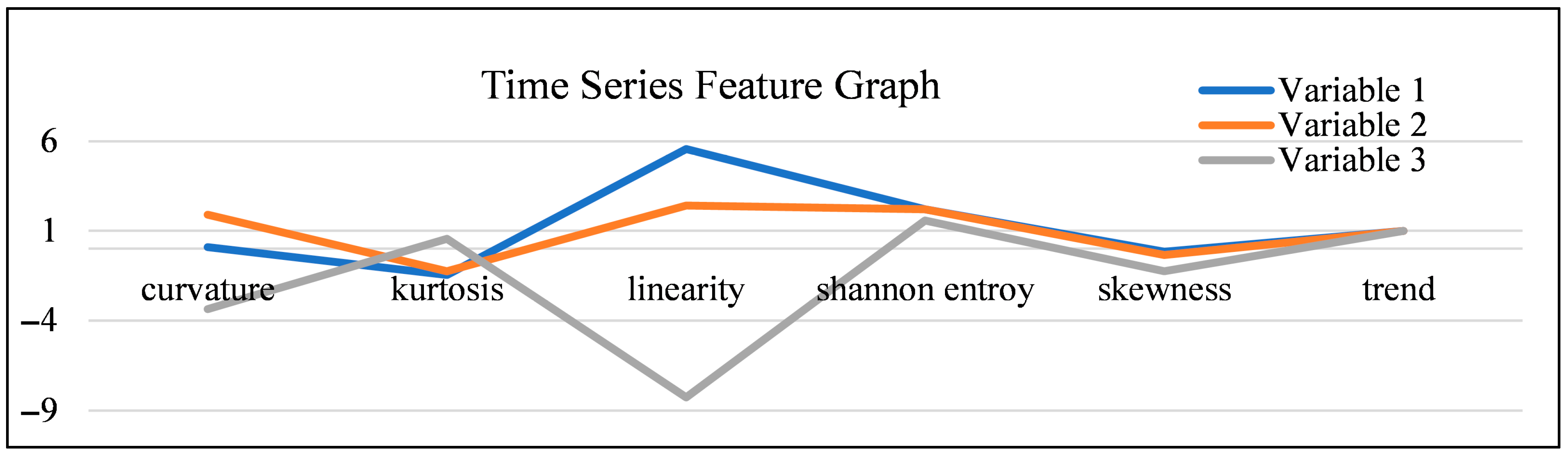
- Dimensionality Reduction and Feature Selection: Our data pre-processing steps, specifically Principal Component Analysis (PCA), ensured that the model focused on the most influential and non-redundant behavioral and environmental variables, enhancing computational efficiency and model clarity. The selection of these critical variables was guided by their time series features, as illustrated in Figure 5.
- MCMC for Temporal Dynamics: The MCMC simulation’s core strength lies in its ability to explicitly model temporal dependencies and transition probabilities. This allowed the model to accurately learn and reproduce realistic sequences of activities, such as the natural transition from feeding to standing, and then to lying down for rumination. The observation that the model’s accuracy improved with more iterations suggests that, even starting with a limited initial dataset, a sufficiently long simulation can generate a highly representative and statistically reliable synthetic dataset for further, deeper analysis.
5.1.1. Analysis of Time Series Features
5.1.2. Comparison of Variables
- Variables 1 (Blue) and 2 (Orange): These variables exhibit similar feature profiles, especially in their measures of distribution: their values for kurtosis (peakedness) and skewness (asymmetry) are close to zero and near each other, suggesting that their probability distributions are also statistically similar. Furthermore, their high, positive, and similar values for curvature and linearity reflect an underlying seasonal or cyclic pattern that, while present, does not significantly affect the overall time series trend. These characteristics suggest they are strong candidates for inclusion in the core behavioral model.
- Variable 3 (Grey): In contrast, Variable 3 exhibits significant differences in all features, reflecting its unique characteristics and lower utility for this specific behavioral analysis. Its feature profile is highly volatile, particularly its deep negative values for linearity and curvature. This indicates a lack of consistent temporal structure or linear relationship, suggesting that this variable may not be essential for modeling the specific behavioral sequences we are targeting.
5.1.3. Justification for Variable Selection
5.2. Comparison to Existing Literature
- Understand Transition Probabilities: It models the likelihood of a cow moving from one state to another (e.g., Standing→Lying).
- Determine Stationary Distributions: It identifies the typical proportion of time an animal spends in each behavioral state over a long period.
5.3. Practical Implications and Future Directions
5.3.1. Practical Implications
- Reduced Data Collection Burden: It minimizes the need for extensive, long-term, and costly data collection, lowering the barrier to entry for smaller or less technologically advanced farms.
- Proactive Management and Welfare: The model’s predictive power enables proactive herd management. An early deviation in an animal’s predicted behavior from the established, healthy baseline can serve as an early disease or stress indicator, allowing farmers to intervene rapidly. This not only improves animal welfare but also enhances overall farm productivity and economic returns.
- Enhanced Calving Management: The demonstrated success in calving event prediction offers a direct method for optimizing resource allocation and ensuring a safe, supervised birth, which is critical for the health of both the cow and the calf.
5.3.2. Future Directions
- Multi-Modal Data Fusion: Incorporating additional, complementary data streams, such as physiological measurements (e.g., heart rate, body temperature, respiration rate), to build even more comprehensive and biologically informed models.
- Generalizability and Robustness: Applying and validating this MCMC methodology to other livestock species (e.g., swine, sheep, poultry) and across diverse environmental and farming conditions to rigorously test the model’s generalizability and practical utility across the industry.
- Real-Time Deployment: Developing a more efficient, computationally lightweight implementation of the MCMC inference process suitable for real-time edge computing on the farm, accelerating the delivery of actionable insights to the farmer.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aquilani, C.; Confessore, A.; Bozzi, R.; Sirtori, F.; Pugliese, C. Review: Precision Livestock Farming technologies in pasture-based livestock systems. Animal 2021, 16, 100429. [Google Scholar] [CrossRef]
- Džermeikaitė, K.; Bačėninaitė, D.; Antanaitis, R. Innovations in Cattle Farming: Application of Innovative Technologies and Sensors in the Diagnosis of Diseases. Animals 2023, 13, 780. [Google Scholar] [CrossRef]
- Sadiq, M.B.; Ramanoon, S.Z.; Shaik Mossadeq, W.M.; Mansor, R.; Syed-Hussain, S.S. Association between Lameness and Indicators of Dairy Cow Welfare Based on Locomotion Scoring, Body and Hock Condition, Leg Hygiene and Lying Behavior. Animals 2017, 7, 79. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Antanaitis, R.; Džermeikaitė, K.; Bespalovaitė, A.; Ribelytė, I.; Rutkauskas, A.; Japertas, S.; Baumgartner, W. Assessment of Ruminating, Eating, and Locomotion Behavior during Heat Stress in Dairy Cattle by Using Advanced Technological Monitoring. Animals 2023, 13, 2825. [Google Scholar] [CrossRef]
- Chen, G.; Li, C.; Guo, Y.; Shu, H.; Cao, Z.; Xu, B. Recognition of Cattle’s Feeding Behaviors Using Noseband Pressure Sensor with Machine Learning. Front. Vet. Sci. 2022, 9, 822621. [Google Scholar] [CrossRef] [PubMed]
- Bishop-Hurley, G.; Henry, D.; Smith, D.; Dutta, R.; Hills, J.; Rawnsley, R.; Hellicar, A.; Timms, G.; Morshed, A.; Rahman, A.; et al. An investigation of cow feeding behavior using motion sensors. In Proceedings of the 2014 IEEE International Instrumentation and Measurement Technology Conference (I2MTC), Montevideo, Uruguay, 12–15 May 2014; pp. 1285–1290. [Google Scholar]
- Aguilar-Lazcano, C.A.; Espinosa-Curiel, I.E.; Ríos-Martínez, J.A.; Madera-Ramírez, F.A.; Pérez-Espinosa, H. Machine Learning-Based Sensor Data Fusion for Animal Monitoring: Scoping Review. Sensors 2023, 23, 5732. [Google Scholar] [CrossRef]
- Tsay, R.S. Multivariate Time Series Analysis: With R and Financial Applications; Wiley: Hoboken, NJ, USA, 2014. [Google Scholar]
- Brooks, S.; Gelman, A.; Jones, G.L.; Meng, X.-L. (Eds.) Handbook of Markov Chain Monte Carlo; Chapman and Hall/CRC: Boca Raton, FL, USA, 2011. [Google Scholar]
- Zakaria, A.; Ismail, F.B.; Lipu, M.H.; Hannan, M.A. Uncertainty models for stochastic optimization in renewable energy applications. Renew. Energy 2020, 145, 1543–1571. [Google Scholar] [CrossRef]
- Li, J. An application of MCMC simulation in mortality projection for populations with limited data. Demogr. Res. 2014, 30, 1–14. [Google Scholar] [CrossRef]
- Neethirajan, S. Recent advances in wearable sensors for animal health management. Sens. Bio-Sens. Res. 2017, 12, 15–29. [Google Scholar] [CrossRef]
- Alipio, M.; Villena, M.L. Intelligent wearable devices and biosensors for monitoring cattle health conditions: A review and classification. Smart Health 2023, 27, 100369. [Google Scholar] [CrossRef]
- Yan, Y.; Sosa, J.; Martínez, C.A. Advances in Bayesian Modeling: Applications and Methods. arXiv 2025, arXiv:2502.11321. [Google Scholar] [CrossRef]
- Gamerman, D.; Lopes, H.F. Markov Chain Monte Carlo: Stochastic Simulation for Bayesian Inference, 2nd ed.; Chapman and Hall/CRC: Boca Raton, FL, USA, 2006. [Google Scholar] [CrossRef]
- Jackman, S. Estimation and Inference via Bayesian Simulation: An Introduction to Markov Chain Monte Carlo. Am. J. Political Sci. 2000, 44, 375–404. [Google Scholar] [CrossRef]
- Hasan, K.N.; Preece, R.; Milanović, J.V. Existing approaches and trends in uncertainty modelling and probabilistic stability analysis of power systems with renewable generation. Renew. Sustain. Energy Rev. 2019, 101, 168–180. [Google Scholar] [CrossRef]
- Chen, F. Missing No More: Using the MCMC Procedure to Model Missing Data. In Proceedings of the SAS Global Forum 2013 Conference, San Francisco, CA, USA, 28 April–1 May 2013; SAS Institute Inc.: Cary, NC, USA, 2013. Available online: https://support.sas.com/resources/papers/proceedings13/436-2013.pdf (accessed on 6 September 2025).
- Niloofar, P.; Lazarova-Molnar, S.; Francis, D.P.; Vulpe, A.; Suciu, G.; Balanescu, M. Modeling and Simulation for Decision Support in Precision Livestock Farming. In Proceedings of the 2020 Winter Simulation Conference, WSC 2020, Orlando, FL, USA, 14–18 December 2020. [Google Scholar] [CrossRef]
- Omar, S. Internet of Things (IoT) for Smart Farming: A Systematic Review. Int. J. Comput. Appl. 2021, 174, 47–54. [Google Scholar] [CrossRef]
- Aydin, A.; Bahr, C.; Berckmans, D. A real-time monitoring tool to automatically measure the feed intakes of multiple broiler chickens by sound analysis. J. Comput. Electron. Agric. 2015, 114, 1–6. [Google Scholar] [CrossRef]
- Mg, W.H.E.; Tin, P.; Aikawa, M.; Kobayashi, I.; Horii, Y.; Honkawa, K.; Zin, T.T. Customized tracking algorithm for robust cattle detection and tracking in occlusion environments. Sensors 2024, 24, 1181. [Google Scholar] [CrossRef]
- Sumi, K.; Maw, S.Z.; Zin, T.T.; Tin, P.; Kobayashi, I.; Horii, Y. Activity-Integrated Hidden Markov Model to Predict Calving Time. Animals 2021, 11, 385. [Google Scholar] [CrossRef]
- Dhanaraju, M.; Chenniappan, P.; Ramalingam, K.; Pazhanivelan, S.; Kaliaperumal, R. Smart Farming: Internet of Things (IoT)-Based Sustainable Agriculture. Agriculture 2022, 12, 1745. [Google Scholar] [CrossRef]
- Akhigbe, B.I.; Munir, K.; Akinade, O.; Akanbi, L.; Oyedele, L.O. IoT Technologies for Livestock Management: A Review of Present Status, Opportunities, and Future Trends. Big Data Cogn. Comput. 2021, 5, 10. [Google Scholar] [CrossRef]
- Brown, D.D.; Kays, R.; Wikelski, M.; Wilson, R.; Klimley, A.P. Observing the unwatchable through acceleration logging of animal behavior. Anim. Biotelemetry 2013, 1, 20. [Google Scholar] [CrossRef]
- Ruuska, S.; Kajava, S.; Mughal, M.; Zehner, N.; Mononen, J. Validation of a pressure sensor-based system for measuring eating, rumination and drinking behaviour of dairy cattle. Appl. Anim. Behav. Sci. 2016, 174, 19–23. [Google Scholar] [CrossRef]
- Chelotti, J.O.; Martinez-Rau, L.S.; Ferrero, M.; Vignolo, L.D.; Galli, J.R.; Planisich, A.M.; Rufiner, H.L.; Giovanini, L.L. Livestock feeding behaviour: A review on automated systems for ruminant monitoring. Biosyst. Eng. 2024, 246, 150–177. [Google Scholar] [CrossRef]

| Var. | Curvature | Kurtosis | Linearity | Shannon Entropy | Skewness | Trend |
|---|---|---|---|---|---|---|
| 1 | 0.086 | −1.471 | 5.550 | 2.190 | −0.165 | 0.995 |
| 2 | 1.903 | −1.272 | 2.415 | 2.189 | −0.359 | 0.995 |
| 3 | −3.381 | 0.552 | −8.262 | 1.564 | −1.269 | 0.981 |
| No | T | H | THI | F | M | L | S | RS | RL | TOTAL_MIN |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 31.2 | 88.7 | 86.1 | 100 | 330 | 470 | 210 | 70 | 260 | 1440 |
| 2 | 31.3 | 85.4 | 85.7 | 100 | 300 | 450 | 140 | 170 | 280 | 1440 |
| 3 | 31.9 | 80.8 | 86 | 70 | 250 | 450 | 220 | 180 | 270 | 1440 |
| 4 | 32 | 76.1 | 85.3 | 70 | 240 | 480 | 200 | 170 | 280 | 1440 |
| 5 | 31.9 | 76.8 | 85.2 | 70 | 330 | 490 | 150 | 130 | 270 | 1440 |
| 6 | 32 | 77.1 | 85.5 | 130 | 190 | 600 | 150 | 160 | 210 | 1440 |
| 7 | 31.6 | 78.7 | 85.2 | 170 | 220 | 520 | 170 | 120 | 230 | 1430 |
| 8 | 31.6 | 78.6 | 85 | 130 | 320 | 380 | 210 | 170 | 230 | 1440 |
| 9 | 32 | 75.8 | 85.3 | 130 | 290 | 410 | 130 | 140 | 340 | 1440 |
| 10 | 31.9 | 79.2 | 85.7 | 170 | 310 | 390 | 150 | 70 | 350 | 1440 |
| 11 | 32.1 | 76.3 | 85.36 | 80 | 370 | 450 | 150 | 80 | 310 | 1440 |
| 12 | 32.2 | 73.9 | 85.2 | 160 | 370 | 480 | 50 | 90 | 290 | 1440 |
| 13 | 33.2 | 70.5 | 86 | 100 | 440 | 450 | 180 | 60 | 210 | 1440 |
| 14 | 32.3 | 73.7 | 85.4 | 140 | 440 | 380 | 210 | 100 | 170 | 1440 |
| 15 | 32.5 | 72.9 | 85.5 | 170 | 360 | 440 | 70 | 140 | 260 | 1440 |
| 16 | 31.7 | 74.9 | 84.7 | 210 | 510 | 340 | 160 | 70 | 150 | 1440 |
| 17 | 30.9 | 79.7 | 84.2 | 120 | 390 | 420 | 120 | 130 | 260 | 1440 |
| 18 | 32 | 77 | 85.4 | 160 | 430 | 470 | 90 | 100 | 190 | 1440 |
| 19 | 32.9 | 73.3 | 86.2 | 240 | 360 | 490 | 80 | 80 | 190 | 1440 |
| 20 | 31.1 | 80.9 | 84.8 | 260 | 360 | 430 | 110 | 20 | 260 | 1440 |
| 21 | 31.5 | 81 | 85.3 | 170 | 330 | 510 | 150 | 120 | 160 | 1440 |
| 22 | 31.4 | 80.6 | 85.1 | 180 | 380 | 490 | 90 | 50 | 250 | 1440 |
| 23 | 31.5 | 77.3 | 84.7 | 80 | 450 | 530 | 120 | 80 | 180 | 1440 |
| 24 | 30.4 | 81.6 | 83.6 | 70 | 400 | 550 | 130 | 70 | 220 | 1440 |
| 25 | 29.1 | 77.8 | 81 | 110 | 350 | 430 | 170 | 70 | 310 | 1440 |
| 26 | 29.6 | 73.2 | 81.2 | 70 | 290 | 560 | 160 | 110 | 250 | 1440 |
| 27 | 29.9 | 71.4 | 81.3 | 100 | 350 | 540 | 150 | 100 | 200 | 1440 |
| 28 | 30.1 | 74.7 | 82.1 | 120 | 370 | 510 | 130 | 80 | 230 | 1440 |
| 29 | 30.8 | 80.1 | 84.1 | 120 | 340 | 520 | 90 | 60 | 310 | 1440 |
| 30 | 31.2 | 82.2 | 85.1 | 70 | 360 | 490 | 160 | 130 | 230 | 1440 |
| Cow ID | Performance | Simulation (n) Number of Iterations | ||
|---|---|---|---|---|
| Measures | n = 3000 | n = 4000 | n = 5000 | |
| ID 1 | AAE | 0.046923 | 0.029635 | 0.021482 |
| AES | 0.003663 | 0.002304 | 0.000958 | |
| RMSE | 0.060527 | 0.048000 | 0.030953 | |
| ID 2 | AAE | 0.047203 | 0.035593 | 0.028498 |
| AES | 0.003804 | 0.002005 | 0.002093 | |
| RMSE | 0.061676 | 0.044773 | 0.045748 | |
| ID 3 | AAE | 0.045708 | 0.038125 | 0.028295 |
| AES | 0.003886 | 0.002914 | 0.001550 | |
| RMSE | 0.062339 | 0.053983 | 0.039373 | |
| ID 4 | AAE | 0.038484 | 0.038125 | 0.025685 |
| AES | 0.002362 | 0.002137 | 0.001056 | |
| RMSE | 0.048605 | 0.046233 | 0.032495 | |
| ID 5 | AAE | 0.041612 | 0.035671 | 0.027747 |
| AES | 0.002782 | 0.002010 | 0.001993 | |
| RMSE | 0.052740 | 0.044836 | 0.044639 | |
| Average | 0.034821 | 0.028423 | 0.022171 | |
| Accuracy | 0.96 | 0.97 | 0.98 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hashimoto, Y.; Zin, T.T.; Tin, P.; Kobayashi, I.; Hama, H. Generating Accurate Activity Patterns for Cattle Farm Management Using MCMC Simulation of Multiple-Sensor Data System. Sensors 2025, 25, 6781. https://doi.org/10.3390/s25216781
Hashimoto Y, Zin TT, Tin P, Kobayashi I, Hama H. Generating Accurate Activity Patterns for Cattle Farm Management Using MCMC Simulation of Multiple-Sensor Data System. Sensors. 2025; 25(21):6781. https://doi.org/10.3390/s25216781
Chicago/Turabian StyleHashimoto, Yukie, Thi Thi Zin, Pyke Tin, Ikuo Kobayashi, and Hiromitsu Hama. 2025. "Generating Accurate Activity Patterns for Cattle Farm Management Using MCMC Simulation of Multiple-Sensor Data System" Sensors 25, no. 21: 6781. https://doi.org/10.3390/s25216781
APA StyleHashimoto, Y., Zin, T. T., Tin, P., Kobayashi, I., & Hama, H. (2025). Generating Accurate Activity Patterns for Cattle Farm Management Using MCMC Simulation of Multiple-Sensor Data System. Sensors, 25(21), 6781. https://doi.org/10.3390/s25216781







