R103 and R115 Affinity Mutants of ATeam ATP Biosensors
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
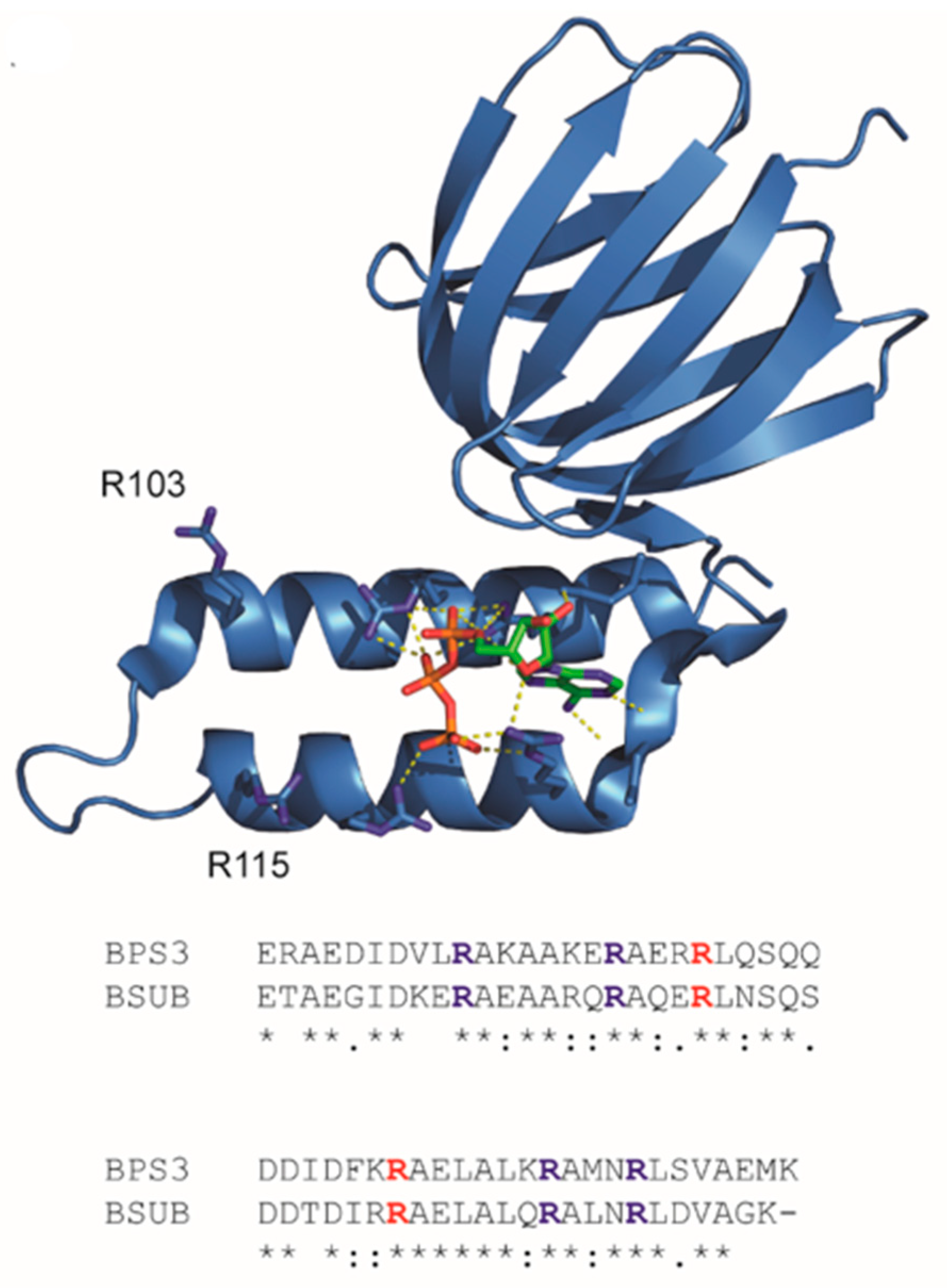
3.1. Tuning a High-Affinity Family of ATeam3.10 Sensors
3.2. Tuning Affinities of ATeam1.03 and ATeam1.03YEMK
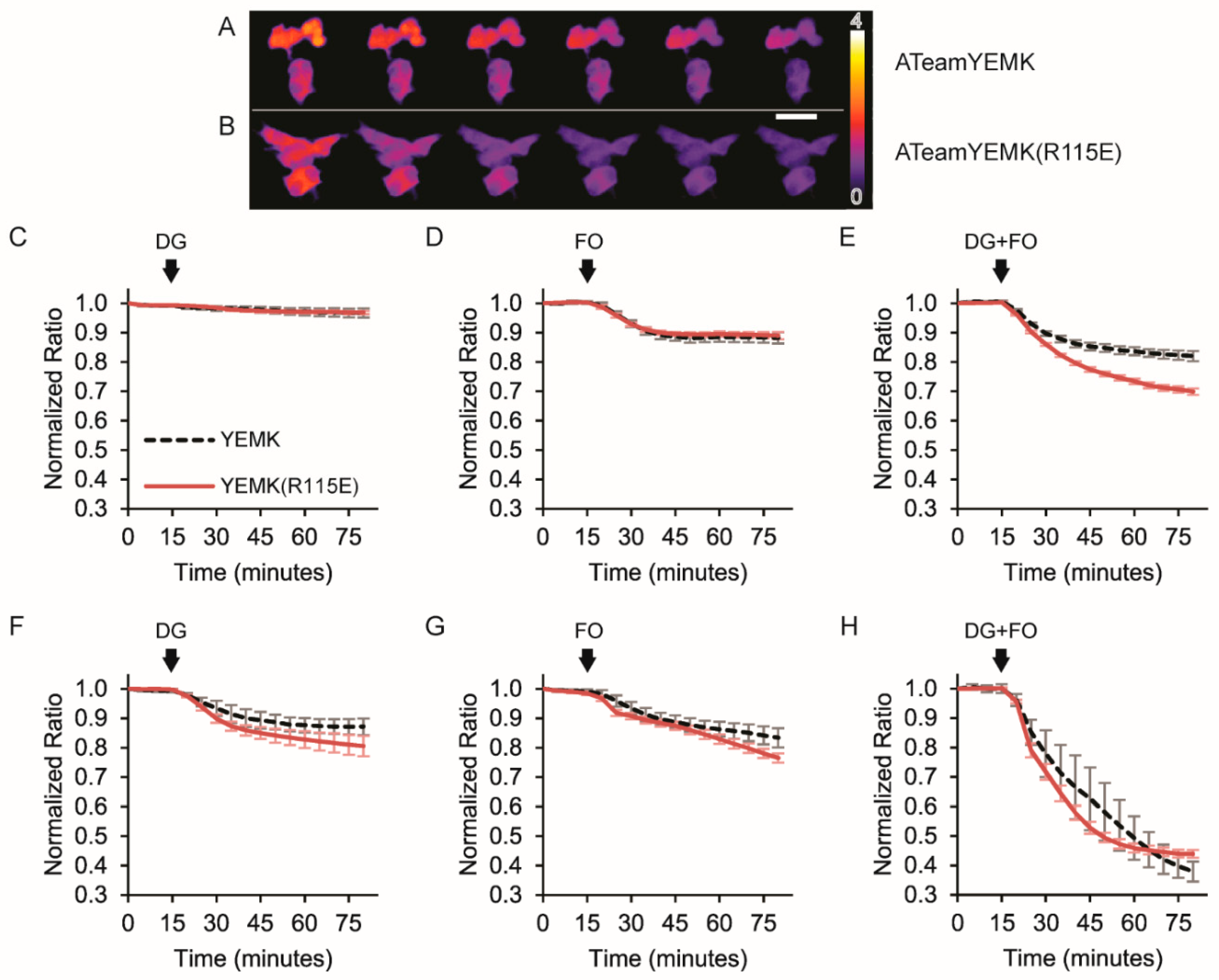
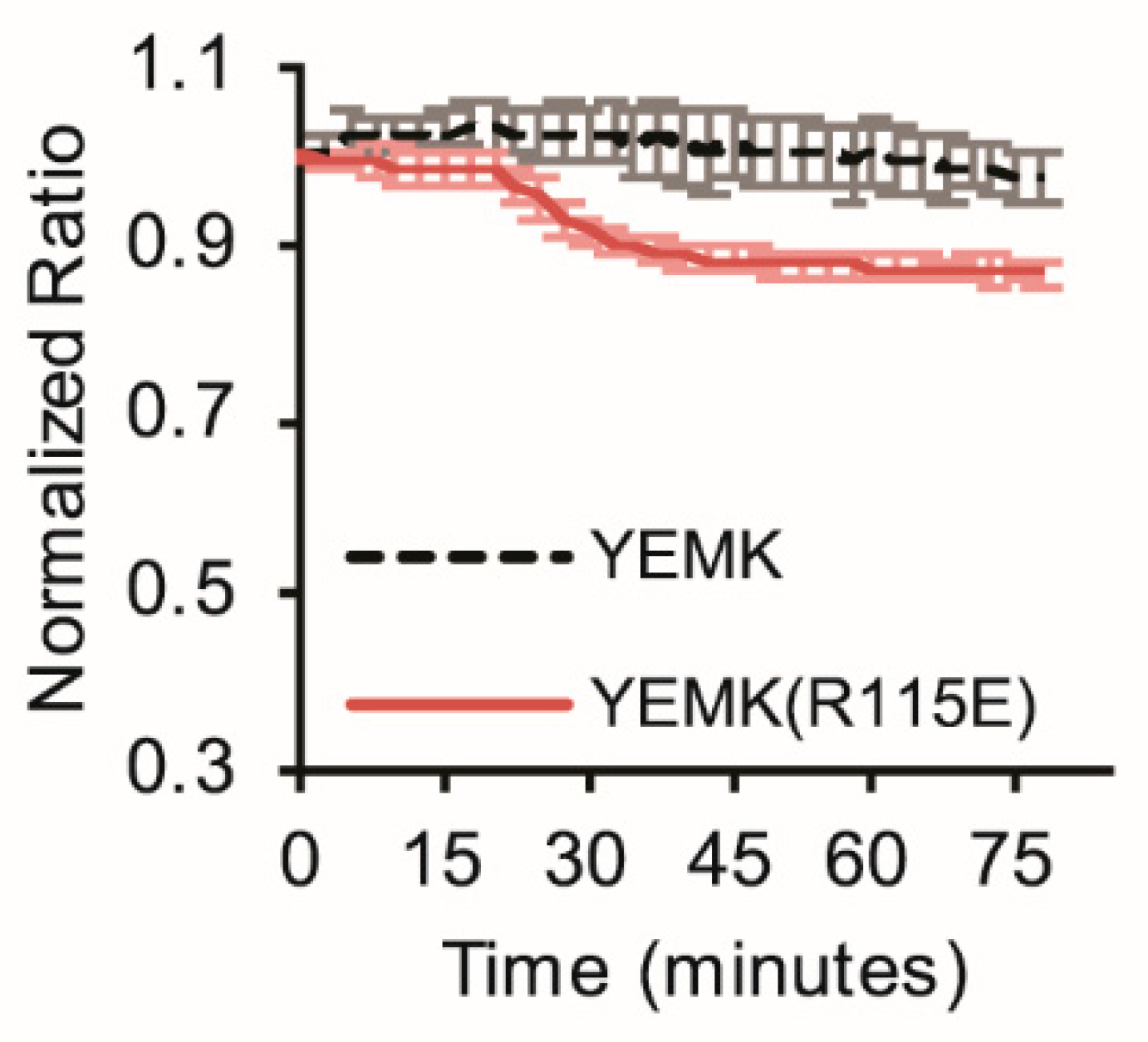
3.3. Imaging Metabolic Dynamics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Imamura, H.; Nhat, K.P.H.; Togawa, H.; Saito, K.; Iino, R.; Kato-Yamada, Y.; Nagai, T.; Noji, H. Visualization of ATP Levels inside Single Living Cells with Fluorescence Resonance Energy Transfer-Based Genetically Encoded Indicators. Proc. Natl. Acad. Sci. USA 2009, 106, 15651–15656. [Google Scholar] [CrossRef]
- Pankratov, Y.; Lalo, U.; Verkhratsky, A.; North, R.A. Vesicular Release of ATP at Central Synapses. Pflugers Arch. 2006, 452, 589–597. [Google Scholar] [CrossRef]
- Burnstock, G. An Introduction to the Roles of Purinergic Signalling in Neurodegeneration, Neuroprotection and Neuroregeneration. Neuropharmacology 2016, 104, 4–17. [Google Scholar] [CrossRef] [PubMed]
- Kato-Yamada, Y. Isolated Epsilon Subunit of Bacillus Subtilis F1-ATPase Binds ATP. FEBS Lett. 2005, 579, 6875–6878. [Google Scholar] [CrossRef] [PubMed]
- Kato-Yamada, Y.; Yoshida, M. Isolated Epsilon Subunit of Thermophilic F1-ATPase Binds ATP. J. Biol. Chem. 2003, 278, 36013–36016. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, M.; Kato-Yamada, Y. ATP-Binding Affinity of the ε Subunit of Thermophilic F1-ATPase under Label-Free Conditions. Biochem. Biophys. Rep. 2020, 21, 100725. [Google Scholar] [CrossRef]
- Nakano, M.; Imamura, H.; Nagai, T.; Noji, H. Ca2+ Regulation of Mitochondrial ATP Synthesis Visualized at the Single Cell Level. ACS Chem. Biol. 2011, 6, 709–715. [Google Scholar] [CrossRef]
- Tsuyama, T.; Kishikawa, J.; Han, Y.-W.; Harada, Y.; Tsubouchi, A.; Noji, H.; Kakizuka, A.; Yokoyama, K.; Uemura, T.; Imamura, H. In Vivo Fluorescent Adenosine 5′-Triphosphate (ATP) Imaging of Drosophila Melanogaster and Caenorhabditis Elegans by Using a Genetically Encoded Fluorescent ATP Biosensor Optimized for Low Temperatures. Anal. Chem. 2013, 85, 7889–7896. [Google Scholar] [CrossRef]
- Yaginuma, H.; Kawai, S.; Tabata, K.V.; Tomiyama, K.; Kakizuka, A.; Komatsuzaki, T.; Noji, H.; Imamura, H. Diversity in ATP Concentrations in a Single Bacterial Cell Population Revealed by Quantitative Single-Cell Imaging. Sci. Rep. 2014, 4, 6522. [Google Scholar] [CrossRef]
- Yoshida, T.; Kakizuka, A.; Imamura, H. BTeam, a Novel BRET-Based Biosensor for the Accurate Quantification of ATP Concentration within Living Cells. Sci. Rep. 2016, 6, 39618. [Google Scholar] [CrossRef]
- Conley, J.M.; Radhakrishnan, S.; Valentino, S.A.; Tantama, M. Imaging Extracellular ATP with a Genetically-Encoded, Ratiometric Fluorescent Sensor. PLoS ONE 2017, 12, e0187481. [Google Scholar] [CrossRef] [PubMed]
- Arai, S.; Kriszt, R.; Harada, K.; Looi, L.-S.; Matsuda, S.; Wongso, D.; Suo, S.; Ishiura, S.; Tseng, Y.-H.; Raghunath, M.; et al. RGB-Color Intensiometric Indicators to Visualize Spatiotemporal Dynamics of ATP in Single Cells. Angew. Chem. Int. Ed. Engl. 2018, 57, 10873–10878. [Google Scholar] [CrossRef]
- Lobas, M.A.; Tao, R.; Nagai, J.; Kronschläger, M.T.; Borden, P.M.; Marvin, J.S.; Looger, L.L.; Khakh, B.S. A Genetically Encoded Single-Wavelength Sensor for Imaging Cytosolic and Cell Surface ATP. Nat. Commun. 2019, 10, 711. [Google Scholar] [CrossRef]
- Marvin, J.S.; Kokotos, A.C.; Kumar, M.; Pulido, C.; Tkachuk, A.N.; Yao, J.S.; Brown, T.A.; Ryan, T.A. iATPSnFR2: A High-Dynamic-Range Fluorescent Sensor for Monitoring Intracellular ATP. Proc. Natl. Acad. Sci. USA 2024, 121, e2314604121. [Google Scholar] [CrossRef]
- Zhong, C.; Arai, S.; Okada, Y. Development of Fluorescence Lifetime Biosensors for ATP, cAMP, Citrate, and Glucose Using the mTurquoise2-Based Platform. Cell Rep. Methods 2024, 4, 100902. [Google Scholar] [CrossRef]
- Kato, S.; Yoshida, M.; Kato-Yamada, Y. Role of the Epsilon Subunit of Thermophilic F1-ATPase as a Sensor for ATP. J. Biol. Chem. 2007, 282, 37618–37623. [Google Scholar] [CrossRef]
- Kato-Yamada, Y. High Affinity Nucleotide-Binding Mutant of the ε Subunit of Thermophilic F1-ATPase. Biochem. Biophys. Res. Commun. 2016, 469, 1129–1132. [Google Scholar] [CrossRef]
- Krah, A.; Kato-Yamada, Y.; Takada, S. The Structural Basis of a High Affinity ATP Binding ε Subunit from a Bacterial ATP Synthase. PLoS ONE 2017, 12, e0177907. [Google Scholar] [CrossRef] [PubMed]
- Krah, A.; Bond, P.J. Single Mutations in the ε Subunit from Thermophilic Bacillus PS3 Generate a High Binding Affinity Site for ATP. PeerJ 2018, 6, e5505. [Google Scholar] [CrossRef]
- Yagi, H.; Kajiwara, N.; Tanaka, H.; Tsukihara, T.; Kato-Yamada, Y.; Yoshida, M.; Akutsu, H. Structures of the Thermophilic F1-ATPase Epsilon Subunit Suggesting ATP-Regulated Arm Motion of Its C-Terminal Domain in F1. Proc. Natl. Acad. Sci. USA 2007, 104, 11233–11238. [Google Scholar] [CrossRef] [PubMed]
- Tantama, M.; Martínez-François, J.R.; Mongeon, R.; Yellen, G. Imaging Energy Status in Live Cells with a Fluorescent Biosensor of the Intracellular ATP-to-ADP Ratio. Nat. Commun. 2013, 4, 2550. [Google Scholar] [CrossRef]
- Steinbach, P.J.; Ionescu, R.; Matthews, C.R. Analysis of Kinetics Using a Hybrid Maximum-Entropy/Nonlinear-Least-Squares Method: Application to Protein Folding. Biophys. J. 2002, 82, 2244–2255. [Google Scholar] [CrossRef] [PubMed]
- Steinbach, P.J. Filtering Artifacts from Lifetime Distributions When Maximizing Entropy Using a Bootstrapped Model. Anal. Biochem. 2012, 427, 102–105. [Google Scholar] [CrossRef]
- Chakraborty, S.; Steinbach, P.J.; Paul, D.; Mu, H.; Broyde, S.; Min, J.-H.; Ansari, A. Enhanced Spontaneous DNA Twisting/Bending Fluctuations Unveiled by Fluorescence Lifetime Distributions Promote Mismatch Recognition by the Rad4 Nucleotide Excision Repair Complex. Nucleic. Acids. Res. 2018, 46, 1240–1255. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; He, K.; Chen, Y.; Li, H.; Pan, S.; Li, B.; Liu, T.; Xi, F.; Deng, F.; Wang, H.; et al. A Sensitive GRAB Sensor for Detecting Extracellular ATP in Vitro and in Vivo. Neuron 2022, 110, 770–782.e5. [Google Scholar] [CrossRef] [PubMed]
- Horovitz, A. Double-Mutant Cycles: A Powerful Tool for Analyzing Protein Structure and Function. Fold. Des. 1996, 1, R121–R126. [Google Scholar] [CrossRef]
- Thestrup, T.; Litzlbauer, J.; Bartholomäus, I.; Mues, M.; Russo, L.; Dana, H.; Kovalchuk, Y.; Liang, Y.; Kalamakis, G.; Laukat, Y.; et al. Optimized Ratiometric Calcium Sensors for Functional in Vivo Imaging of Neurons and T Lymphocytes. Nat. Methods 2014, 11, 175–182. [Google Scholar] [CrossRef]
- Ferguson, S.A.; Cook, G.M.; Montgomery, M.G.; Leslie, A.G.W.; Walker, J.E. Regulation of the Thermoalkaliphilic F1-ATPase from Caldalkalibacillus Thermarum. Proc. Natl. Acad. Sci. USA 2016, 113, 10860–10865. [Google Scholar] [CrossRef]
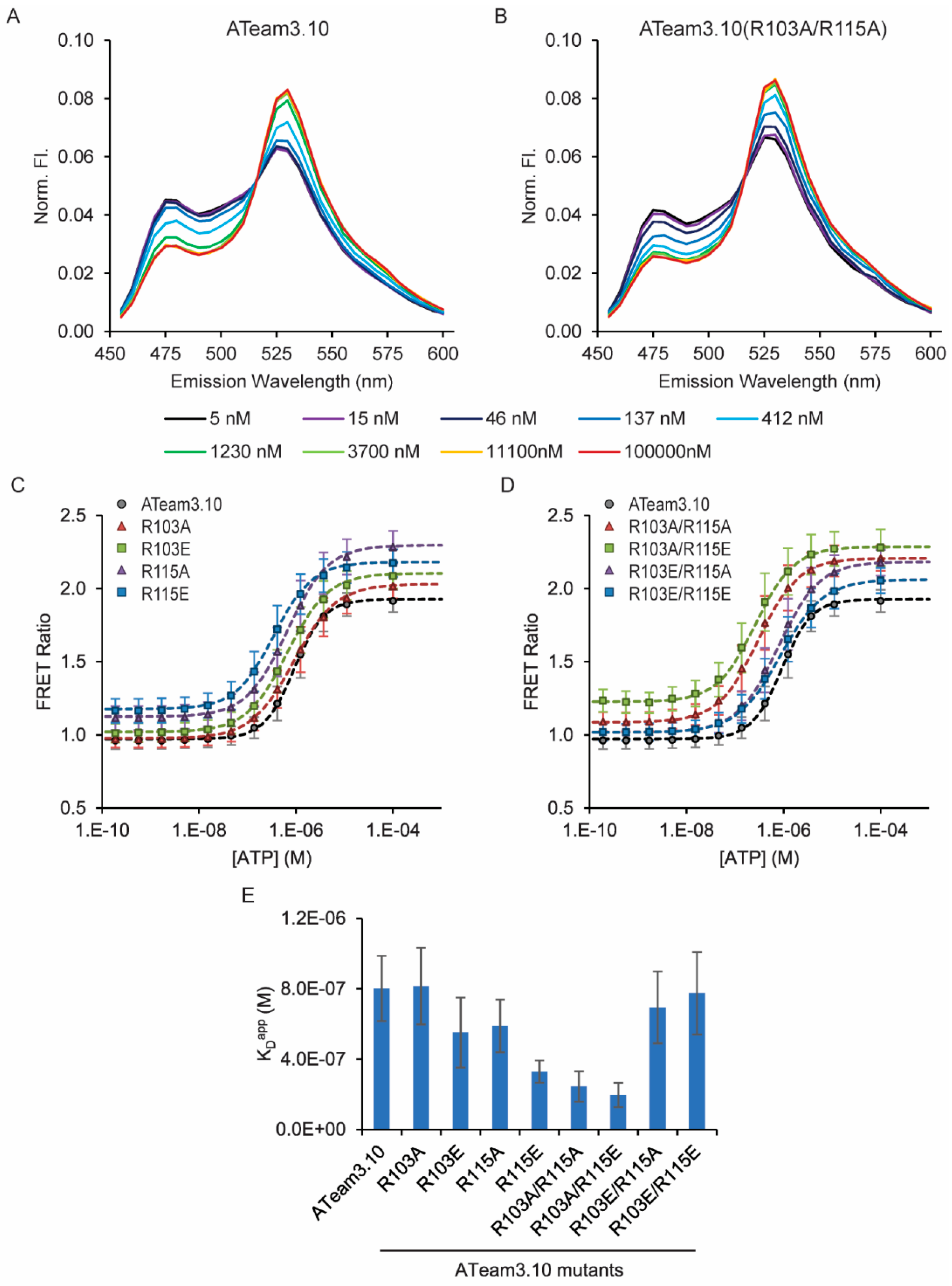
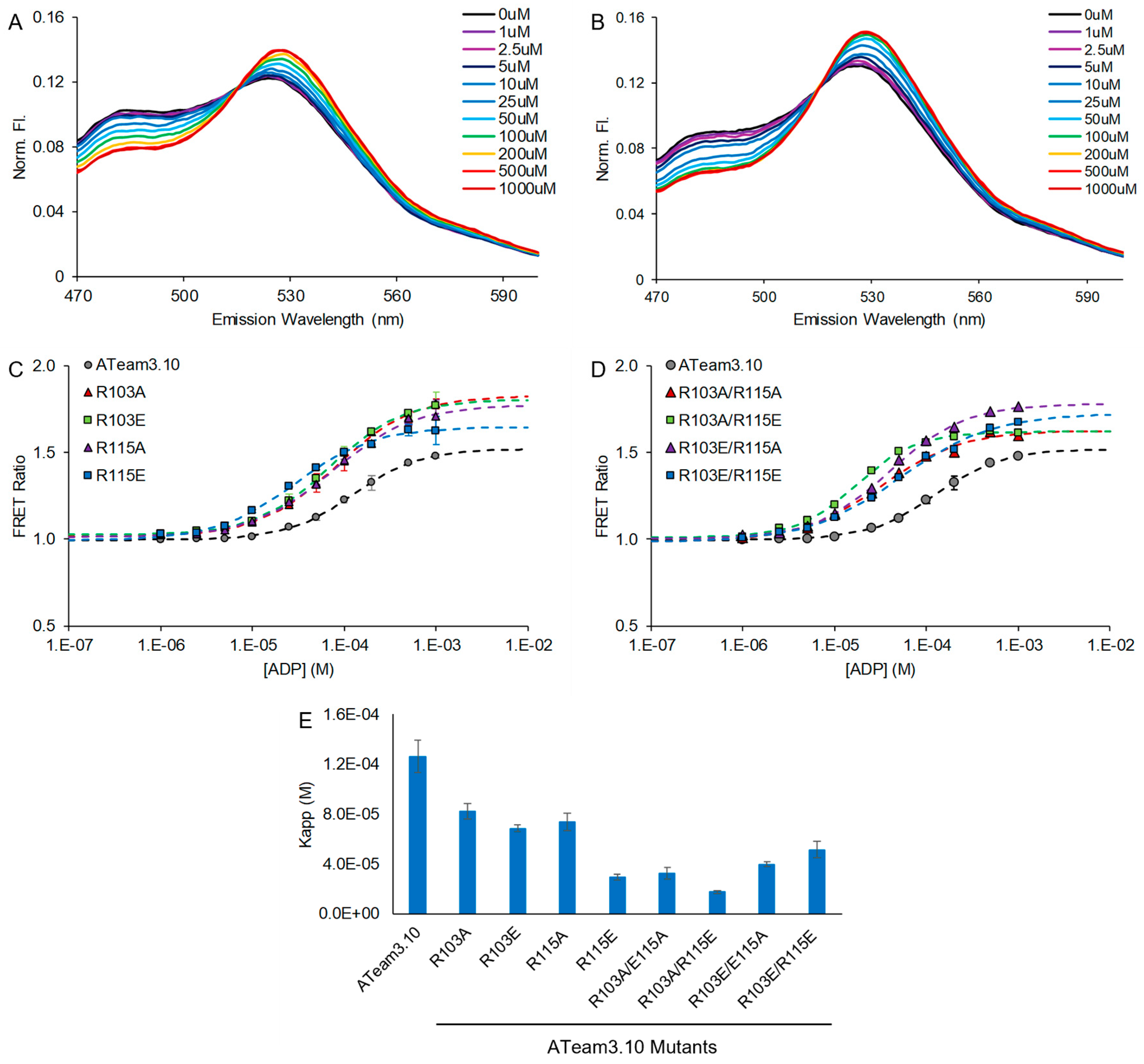

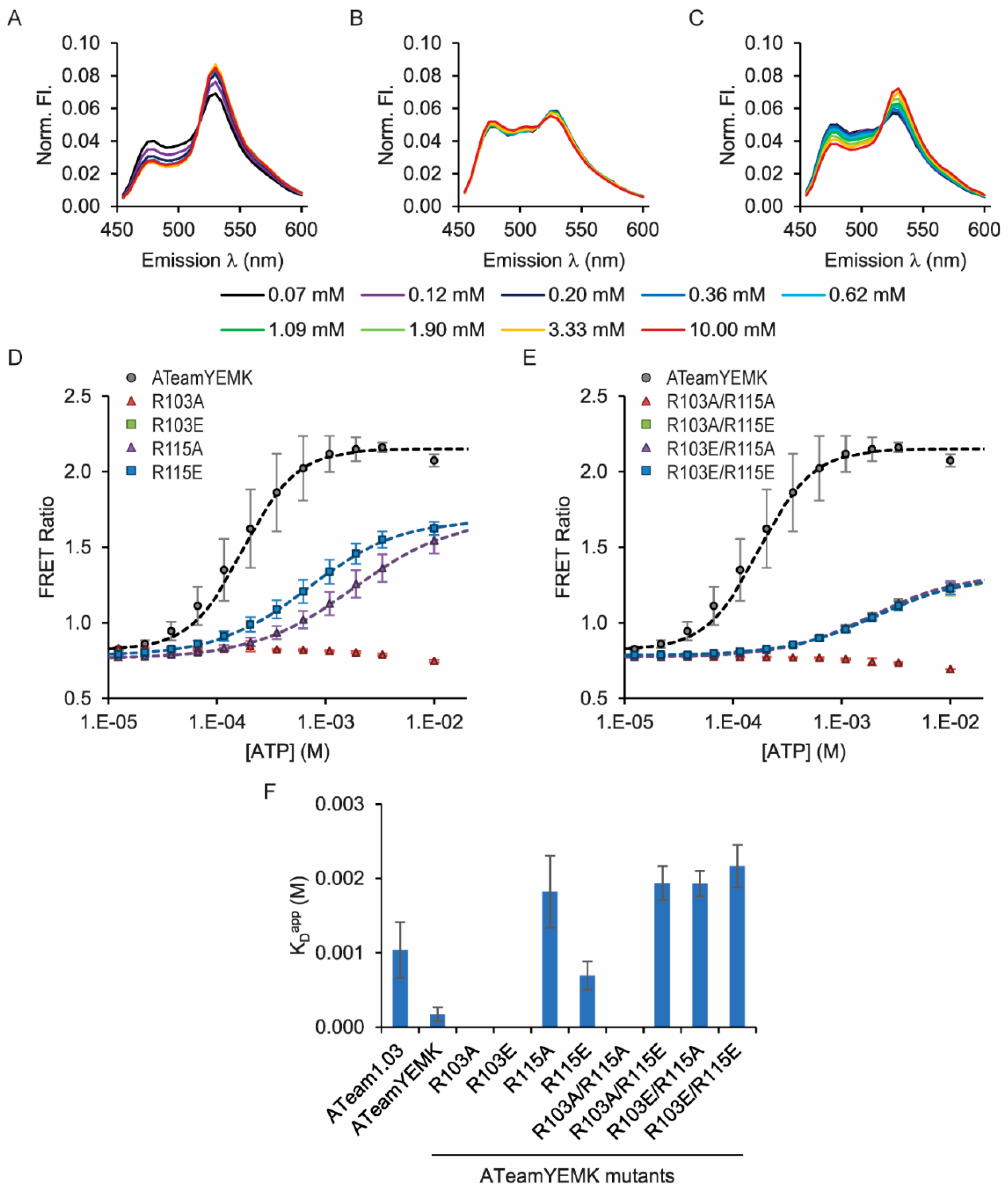
| ATeam3.10 Mutation | ATP Affinity Kapp 1 |
|---|---|
| WT | 800 ± 300 (200) nM |
| R103A | 800 ± 300 (200) nM |
| R103E | 600 ± 300 (200) nM |
| R115A | 600 ± 200 (200) nM |
| R115E | 300 ± 100 (60) nM * |
| R103A/R115A | 200 ± 100 (90) nM * |
| R103A/R115E | 200 ± 100 (70) nM * |
| R103E/R115A | 700 ± 300 (200) nM |
| R103E/R115E | 800 ± 400 (200) nM |
| ATeam3.10 Mutation | ADP Affinity Kapp |
|---|---|
| WT | 130 ± 10 µM |
| R103A | 82 ± 6 µM |
| R103E | 68 ± 3 µM |
| R115A | 74 ± 7 µM |
| R115E | 29 ± 2 µM |
| R103A/R115A | 32 ± 5 µM |
| R103A/R115E | 17 ± 1 µM |
| R103E/R115A | 40 ± 2 µM |
| R103E/R115E | 51 ± 6 µM |
| ATeam1.03 Mutation | ATP Affinity Kapp 1 | ATeam1.03YEMK Mutation | ATP Affinity Kapp 1 |
|---|---|---|---|
| WT | 1.0 ± 0.5 (0.3) mM | WT | 0.2 ± 0.1 (0.09) mM |
| R103A | Non-Binding | R103A | Non-Binding |
| R103E | Non-Binding | R103E | ND 2 |
| R115A | Non-Binding | R115A | 1.8 ± 0.5 (0.5) mM |
| R115E | Non-Binding | R115E | 0.7 ± 0.2 (0.2) mM |
| R103A/R115A | Non-Binding | R103A/R115A | Non-Binding |
| R103A/R115E | Non-Binding | R103A/R115E | 1.9 ± 0.2 (0.2) mM |
| R103E/R115A | Non-Binding | R103E/R115A | 1.9 ± 0.2 (0.2) mM |
| R103E/R115E | Non-Binding | R103E/R115E | 2.2 ± 0.3 (0.3) mM |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cholger, A.; Conley, J.M.; Valentino, S.A.; Colomb, E.; de Cuba, O.; Kress, J.; Tantama, M. R103 and R115 Affinity Mutants of ATeam ATP Biosensors. Sensors 2025, 25, 6180. https://doi.org/10.3390/s25196180
Cholger A, Conley JM, Valentino SA, Colomb E, de Cuba O, Kress J, Tantama M. R103 and R115 Affinity Mutants of ATeam ATP Biosensors. Sensors. 2025; 25(19):6180. https://doi.org/10.3390/s25196180
Chicago/Turabian StyleCholger, Autumn, Jason M. Conley, Stephen A. Valentino, Elaine Colomb, Olivia de Cuba, Jacob Kress, and Mathew Tantama. 2025. "R103 and R115 Affinity Mutants of ATeam ATP Biosensors" Sensors 25, no. 19: 6180. https://doi.org/10.3390/s25196180
APA StyleCholger, A., Conley, J. M., Valentino, S. A., Colomb, E., de Cuba, O., Kress, J., & Tantama, M. (2025). R103 and R115 Affinity Mutants of ATeam ATP Biosensors. Sensors, 25(19), 6180. https://doi.org/10.3390/s25196180






