FALS-YOLO: An Efficient and Lightweight Method for Automatic Brain Tumor Detection and Segmentation
Abstract
1. Introduction
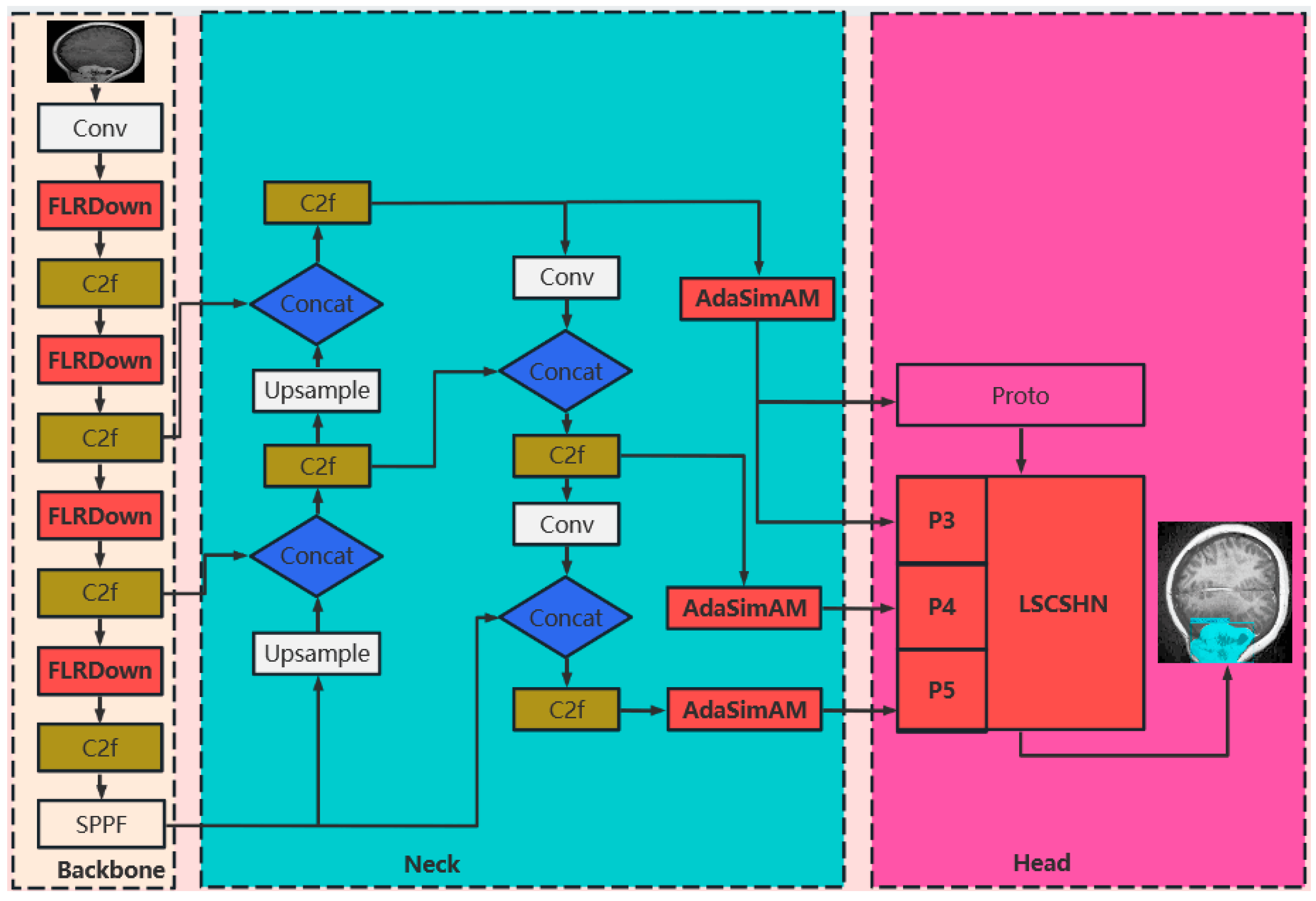
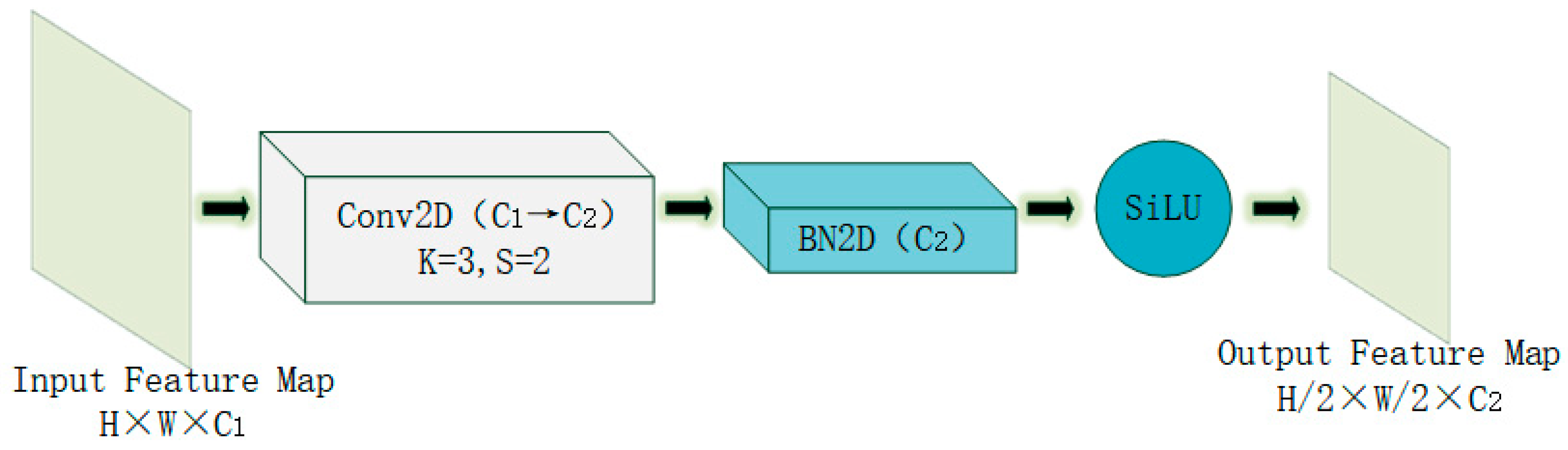
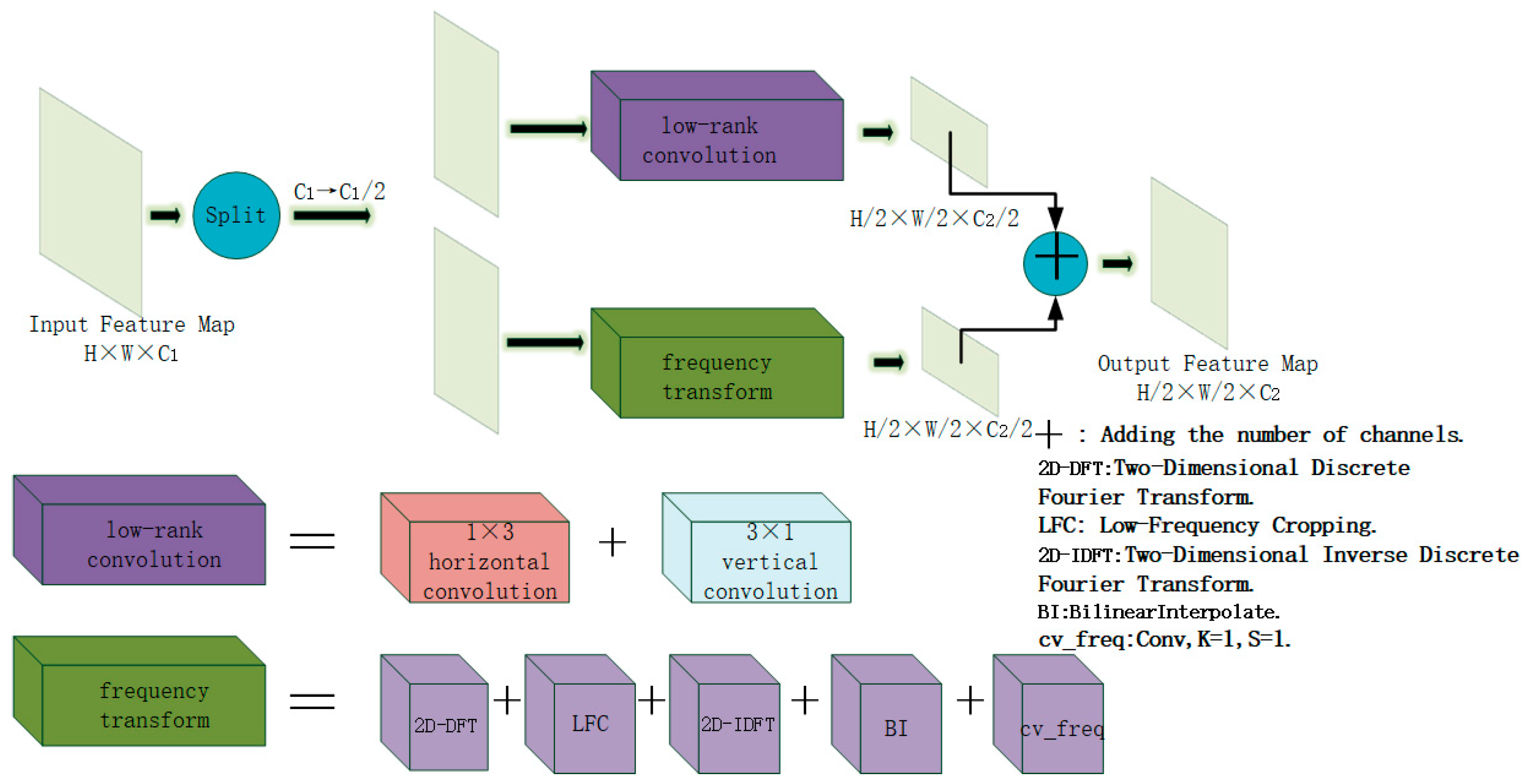
- Novel Downsampling Module FLRDown: This study designs a novel downsampling module FLRDown that integrates low-rank convolution and Fourier transform. While reducing spatial resolution, this module can more effectively preserve the low-frequency global information of images, enhance the model’s receptive field and feature expression ability, and at the same time lower computational complexity, thus providing stronger support for the recognition of multi-scale targets in medical images.
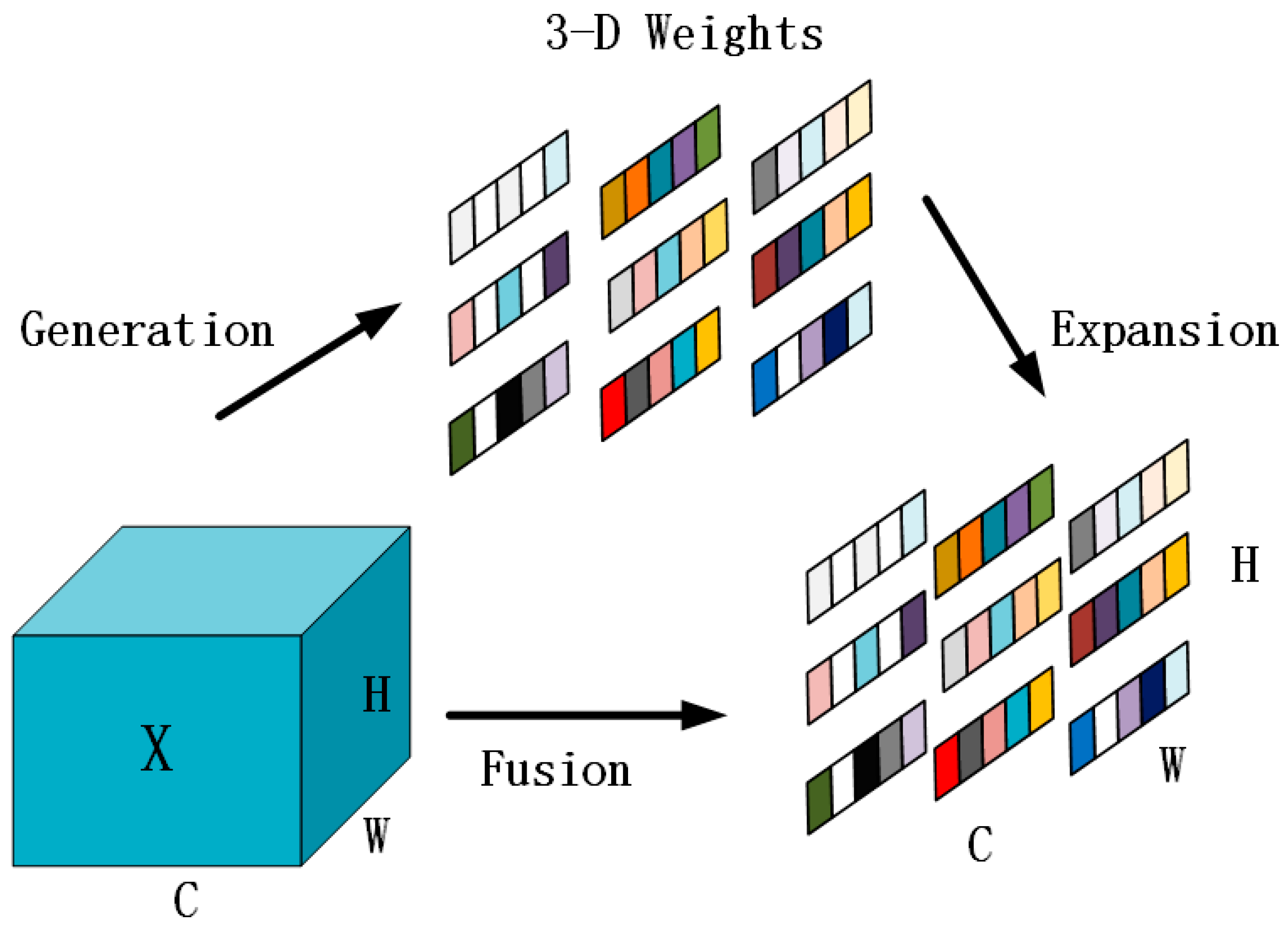
- Adaptive Simple Parameter-Free Attention Module AdaSimAM: Based on the SimAM attention mechanism, this study proposes a new attention module named AdaSimAM. AdaSimAM effectively reduces the interference of high-frequency noise on feature expression and enhances the feature response of local areas. Moreover, this module introduces an adaptive parameter-adjustment mechanism, enabling the model to more flexibly adapt to the needs of multi-scale features in visual tasks such as object detection and semantic segmentation.
- Lightweight Segmentation Convolutional Segmentation Head Network LSCSHN: In response to the issue of large parameter quantity and high resource consumption in the original YOLO series models, this invention designs the LSCSHN segmentation head network. It optimizes the head network structure and reduces redundant parameters. LSCSHN maintains high-precision output while significantly lowering the model’s computational load and enhancing its deployment capability in resource-constrained environments.
2. Related Work
3. Materials and Methods
3.1. FLRDown: An Efficient Downsampling Module Combining Low-Rank Convolution and Fourier Transform
3.2. AdaSimAM—An Adaptive Simple Parameter-Free Attention Module
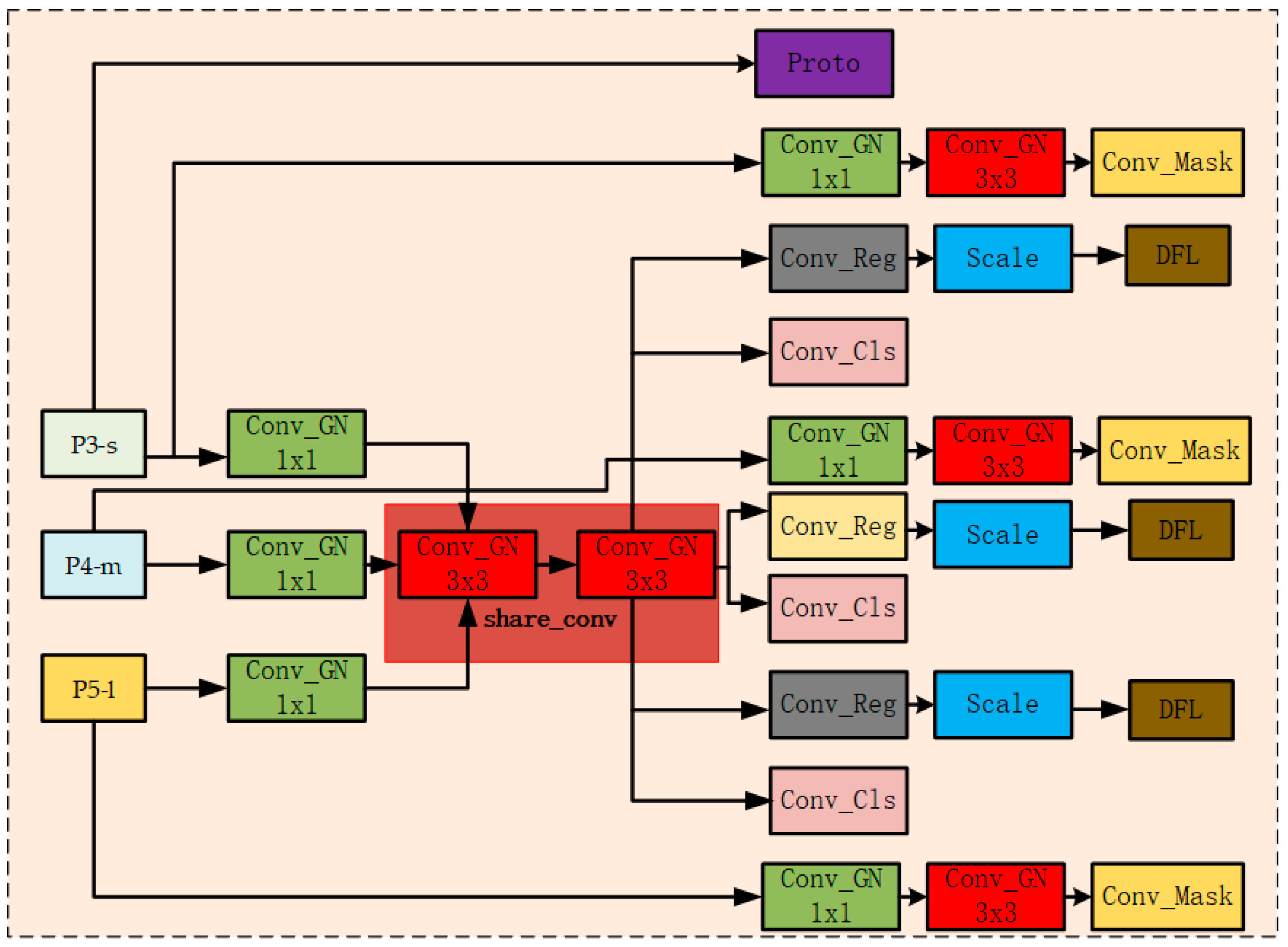
3.3. LSCSHN: A Lightweight Shared Convolutional Segmentation Head Network
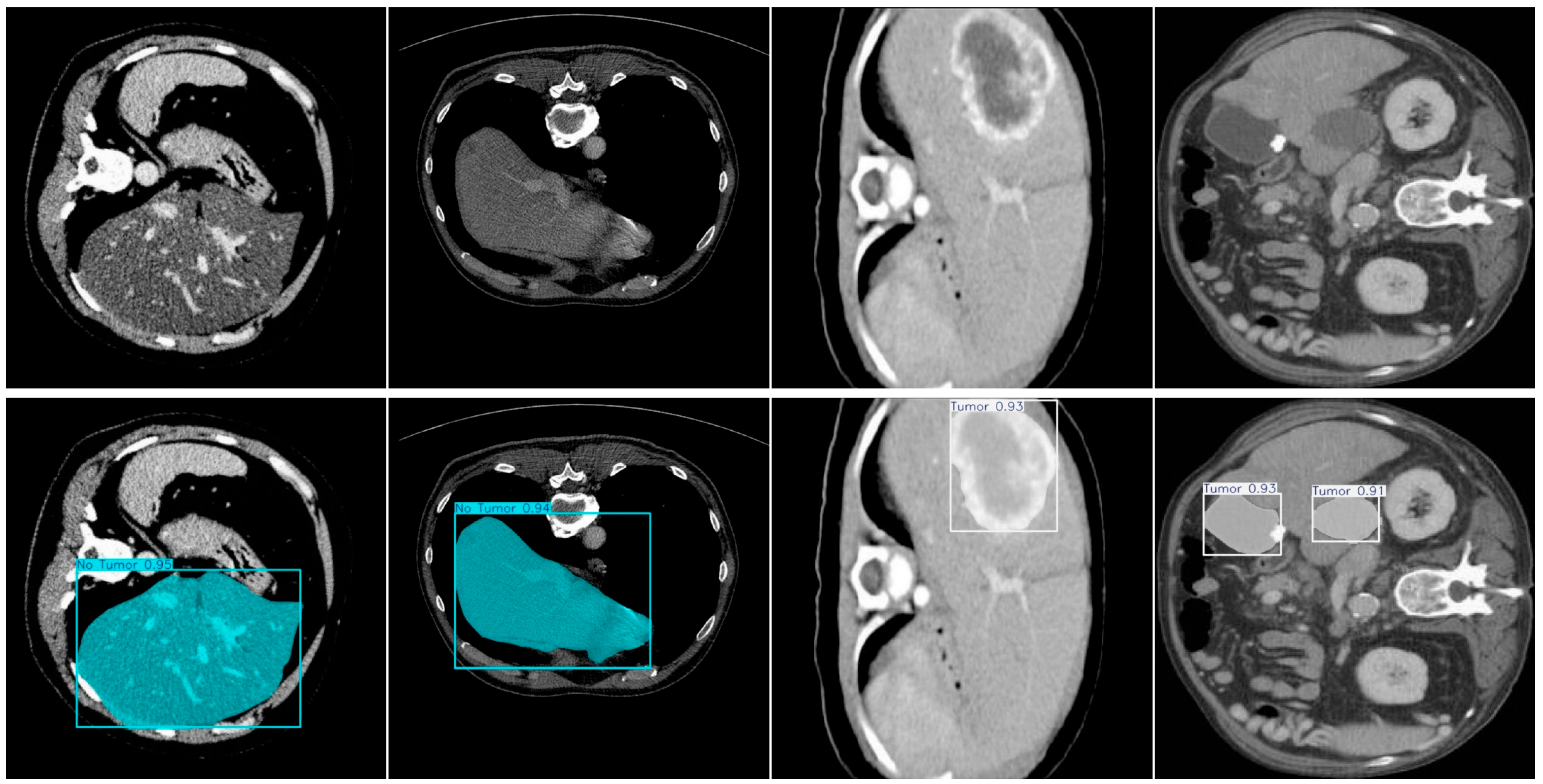
4. Experiments and Results
4.1. Dataset
4.2. Experimental Environment and Training Parameter Settings
4.3. Evaluation Metrics
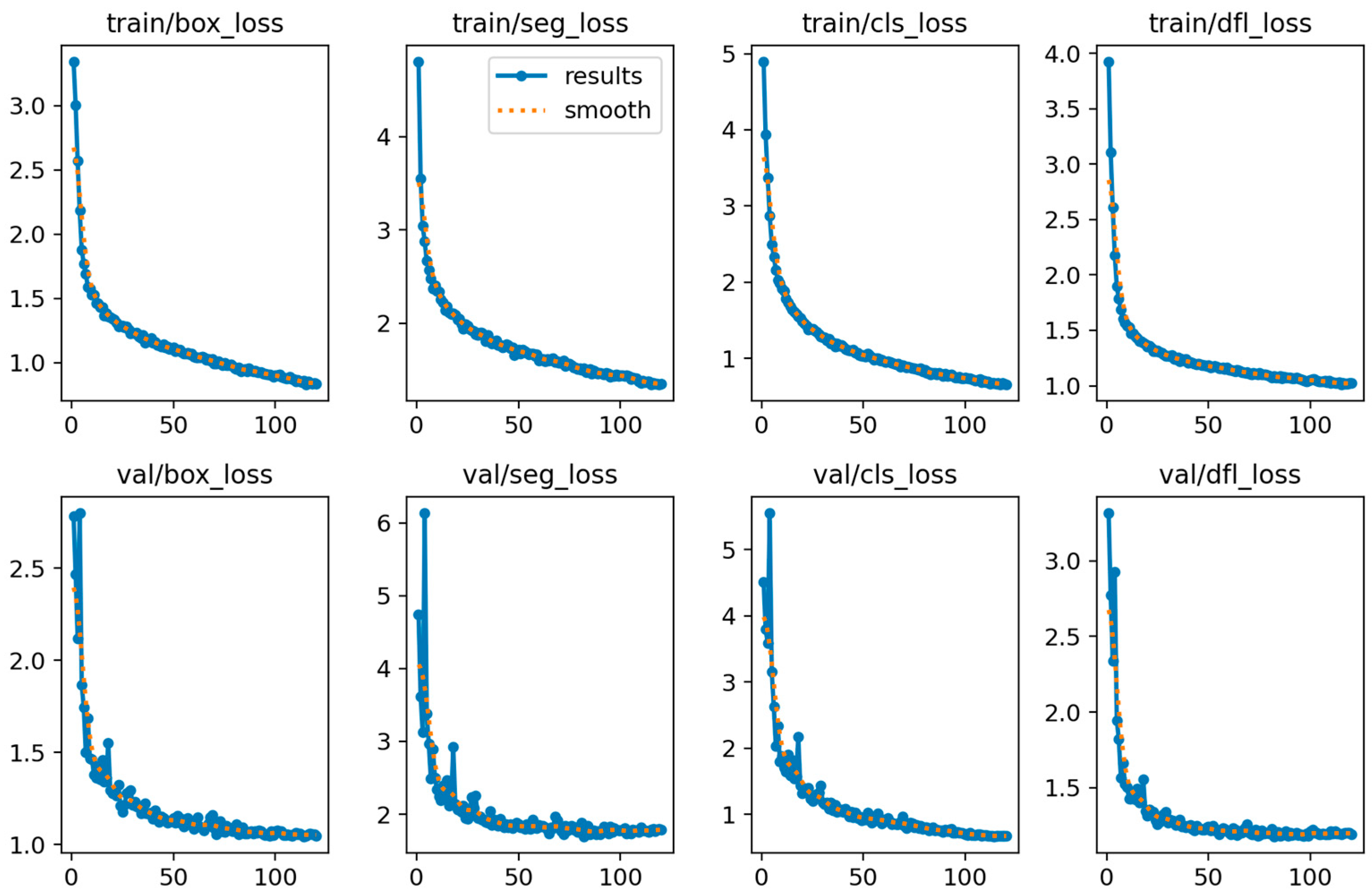
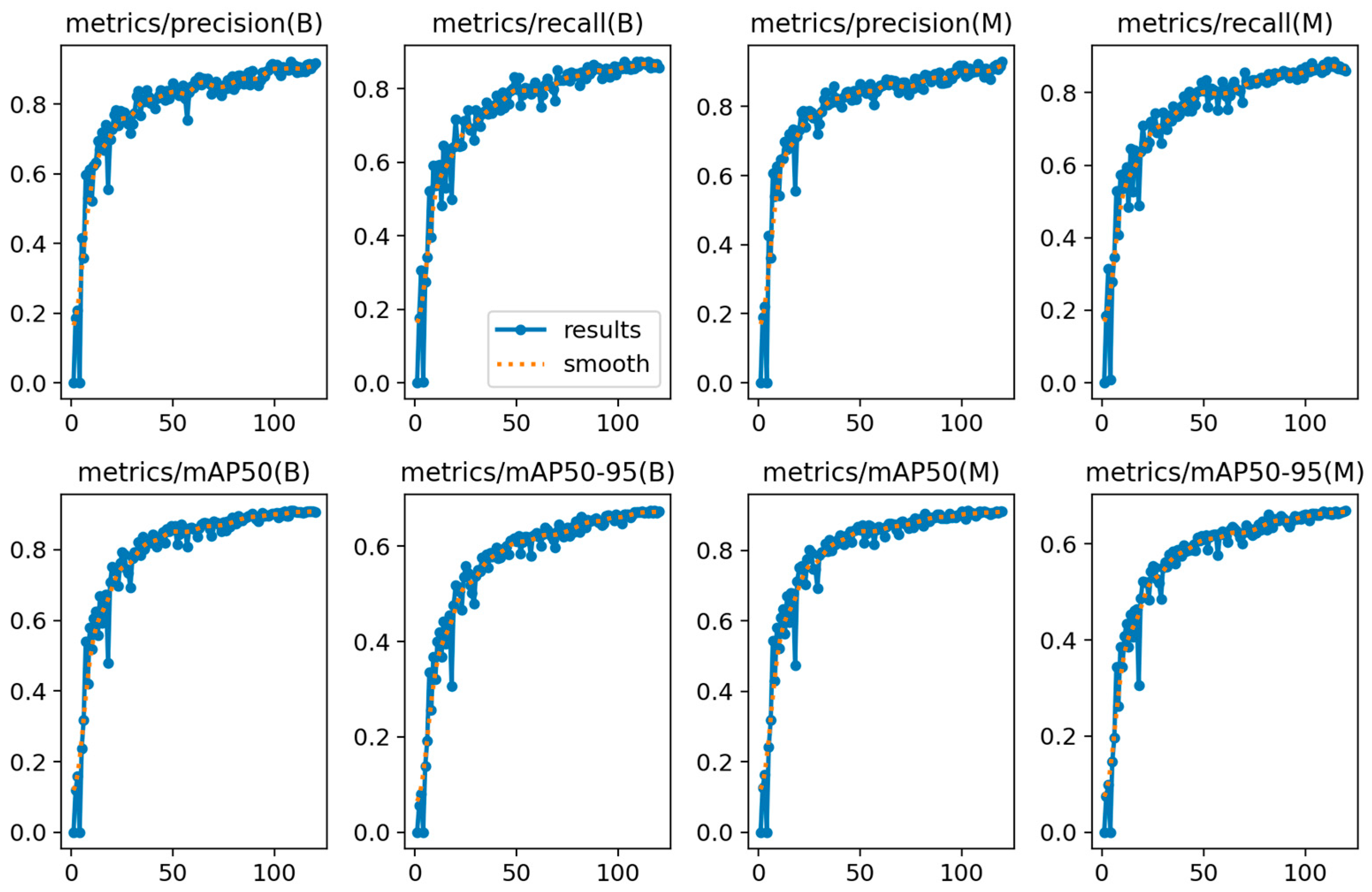
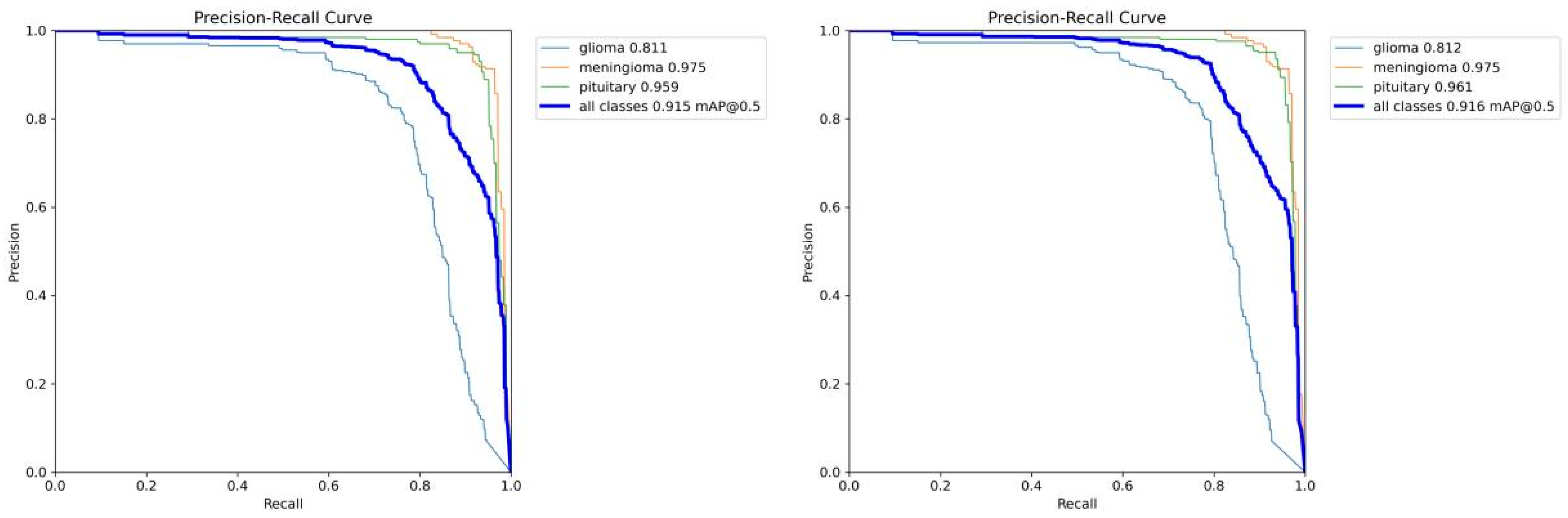
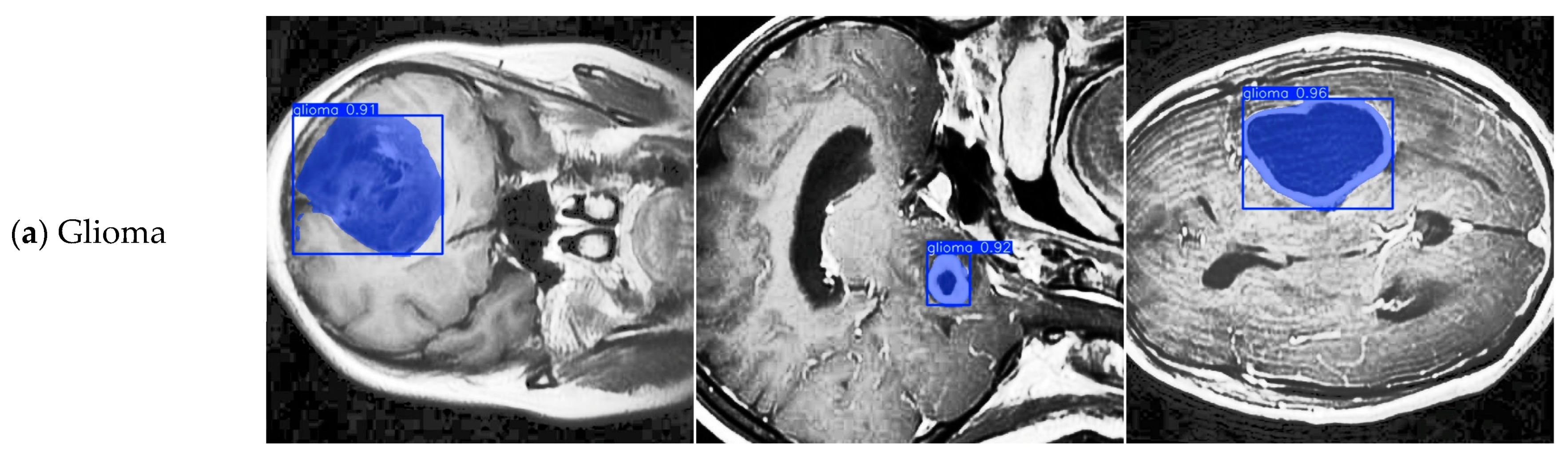
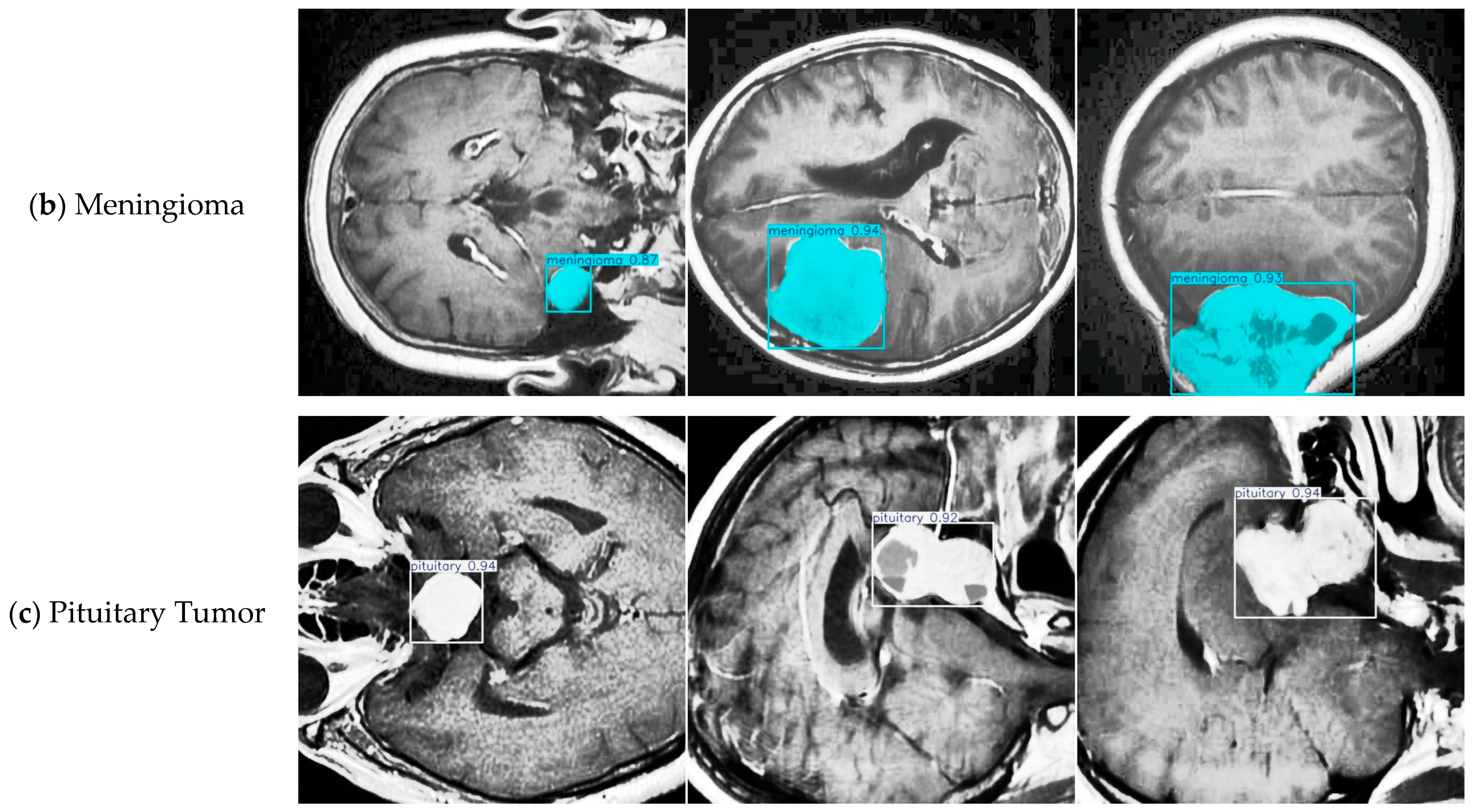
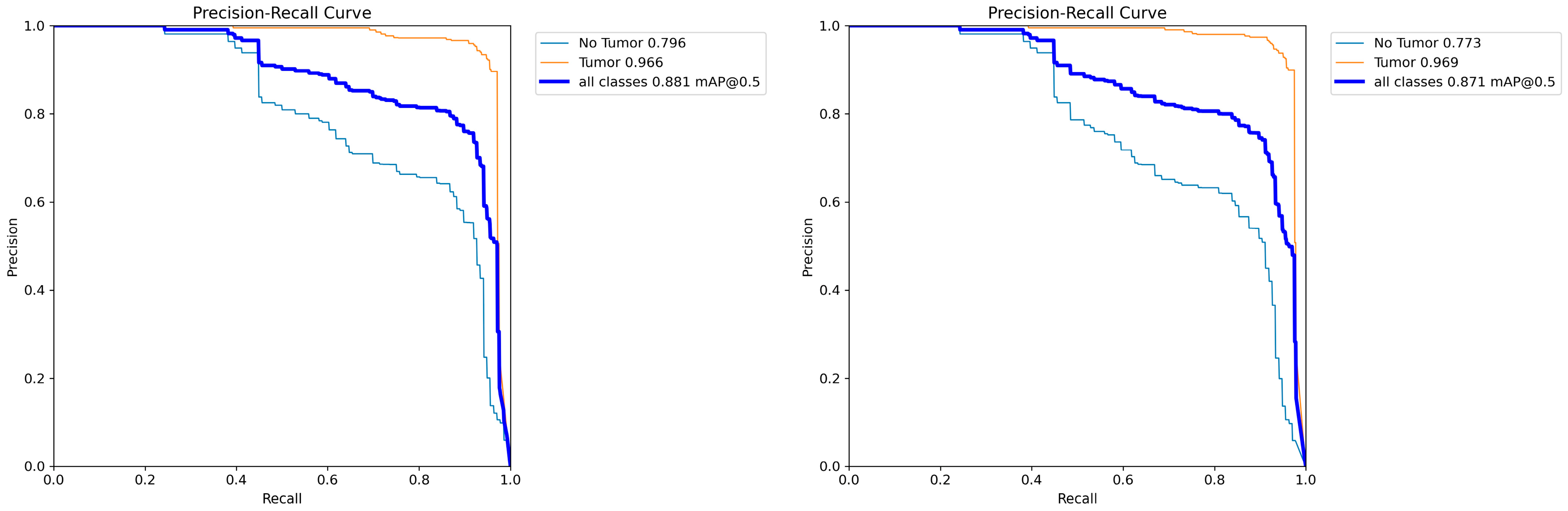
4.4. Model Training, Testing, and Results Analysis
4.5. Results and Analysis of Ablation Experiments
4.6. Results and Analysis of Comparative Experiment
4.7. Supplementary Experiment
4.8. Discussion on Brain Tumor Classification Ability
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- NC International Neuroscience Center. Understanding Brain Tumor Incidence and Mortality. International Neuroscience Information. 2025. Available online: https://www.incsg.cn/zixun/naozhongliukepu/59156.html (accessed on 13 August 2025).
- National Cancer Institute (NCI). SEER Cancer Statistics: Brain and Other Nervous System Cancer. SEER; 2024. Available online: https://seer.cancer.gov/statfacts/html/brain.html (accessed on 13 August 2025).
- Jiang, Z.; Lyu, X.; Zhang, J.; Zhang, Q.; Wei, X. Review of Deep Learning Methods for MRI Brain Tumor Image Segmentation. J. Image Graph. 2020, 25, 215–228. [Google Scholar] [CrossRef]
- Liang, F.; Yang, F.; Lu, L.; Yin, M. A Survey of Brain Tumor Segmentation Methods Based on Convolutional Neural Networks. J. Comput. Eng. Appl. 2021, 57, 7. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 26–30 June 2016; pp. 779–788. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. YOLO9000: Better, Faster, Stronger. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 6517–6525. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar] [CrossRef]
- Bochkovskiy, A.; Wang, C.-Y.; Liao, H.-Y.M. YOLOv4: Optimal Speed and Accuracy of Object Detection. arXiv 2020, arXiv:2004.10934. [Google Scholar] [CrossRef]
- Li, C.; Li, L.; Jiang, H.; Weng, K.; Geng, Y.; Li, L.; Ke, Z.; Li, Q.; Cheng, M.; Nie, W.; et al. YOLOv6: A Single-Stage Object Detection Framework for Industrial Applications. arXiv 2022, arXiv:2209.02976. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Bochkovskiy, A.; Liao, H.-Y.M. YOLOv7: Trainable Bag-of-Freebies Sets New State-of-the-Art for Real-Time Object Detectors. arXiv 2022, arXiv:2207.02696. [Google Scholar]
- Hussain, M. YOLOv1 to v8: Unveiling Each Variant–A Comprehensive Review of YOLO. IEEE Access 2024, 12, 42816–42833. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Yeh, I.-H.; Liao, H.-Y.M. YOLOv9: Learning What You Want to Learn Using Programmable Gradient Information. arXiv 2024, arXiv:2402.13616. [Google Scholar] [CrossRef]
- Wang, A.; Chen, H.; Liu, L.; Chen, K.; Lin, Z.; Han, J. YOLOv10: Real-Time End-to-End Object Detection. arXiv 2024, arXiv:2405.14458. [Google Scholar]
- Khanam, I.R.; Hussain, M. YOLOv11: An Overview of the Key Architectural Enhancements. arXiv 2024, arXiv:2410.17725. [Google Scholar] [CrossRef]
- Tian, Y.; Ye, Q.; Doermann, D. YOLOv12: Attention-Centric Real-Time Object Detectors. arXiv 2025, arXiv:2502.12524. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2015; Volume 9351, pp. 234–241. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar] [CrossRef]
- Terven, J.; Córdova-Esparza, D.-M.; Romero-González, J.-A. A Comprehensive Review of YOLO Architectures in Computer Vision: From YOLOv1 to YOLOv8 and YOLO-NAS. Mach. Learn. Knowl. Extr. 2023, 5, 1680–1716. [Google Scholar] [CrossRef]
- Ali, M.L.; Zhang, Z. The YOLO Framework: A Comprehensive Review of Evolution, Applications, and Benchmarks in Object Detection. Computers 2024, 13, 336. [Google Scholar] [CrossRef]
- Ranjbarzadeh, R.; Bagherian Kasgari, A.; Jafarzadeh Ghoushchi, S.; Anari, S.; Naseri, M.; Bendechache, M. Brain Tumor Segmentation Based on Deep Learning and an Attention Mechanism Using MRI Multi-Modalities Brain Images. Sci. Rep. 2021, 11, 10930. [Google Scholar] [CrossRef]
- Elazab, N.; Gab-Allah, W.A.; Elmogy, M. A Multi-Class Brain Tumor Grading System Based on Histopathological Images Using a Hybrid YOLO and RESNET Networks. Sci. Rep. 2024, 14, 4584. [Google Scholar] [CrossRef]
- Lin, Z.; Lin, J.; Jiang, Z. YOLOv8-DEC: An Enhanced YOLOv8 Model for Accurate Brain Tumor Detection. Prog. Electromagn. Res. M 2024, 129, 43–52. [Google Scholar] [CrossRef]
- Priyadharshini, S.; Bhoopalan, R.; Manikandan, D.; Ramaswamy, K. A Successive Framework for Brain Tumor Interpretation Using YOLO Variants. Sci. Rep. 2025, 15, 27973. [Google Scholar] [CrossRef]
- Mithun, M.S.; Jawhar, S.J. Detection and Classification on MRI Images of Brain Tumor Using YOLO NAS Deep Learning Model. J. Radiat. Res. Appl. Sci. 2024, 17, 101113. [Google Scholar] [CrossRef]
- Chen, A.; Lin, D.; Gao, Q. Enhancing Brain Tumor Detection in MRI Images Using YOLO-NeuroBoost Model. Front. Neurol. 2024, 15, 1445882. [Google Scholar] [CrossRef]
- Hashemi, S.M.H.; Safari, L.; Taromi, A.D. Realism in Action: Anomaly-Aware Diagnosis of Brain Tumors from Medical Images Using YOLOv8 and DeiT. arXiv 2024, arXiv:2401.03302. [Google Scholar] [CrossRef]
- Reddy, B.D.K.; Reddy, P.B.K.; Priya, L. Advanced Brain Tumor Segmentation and Detection Using YOLOv8. In Proceedings of the 2024 2nd International Conference on Sustainable Computing and Smart Systems (ICSCSS), IEEE, Coimbatore, India, 10–12 July 2024; pp. 1583–1588. [Google Scholar]
- Kang, M.; Ting, C.M.; Ting, F.F.; Phan, R.C.W. ASF-YOLO: A Novel YOLO Model with Attentional Scale Sequence Fusion for Cell Instance Segmentation. Image Vis. Comput. 2024, 147, 105057. [Google Scholar] [CrossRef]
- Almufareh, M.F.; Imran, M.; Khan, A.; Humayun, M.; Asim, M. Automated Brain Tumor Segmentation and Classification in MRI Using YOLO-Based Deep Learning. IEEE Access 2024, 12, 16189–16207. [Google Scholar] [CrossRef]
- Kasar, P.; Jadhav, S.; Kansal, V. Brain Tumor Segmentation Using U-Net and SegNet. In Proceedings of the International Conference on Signal Processing and Computer Vision (SIPCOV-2023); Atlantis Press: Paris, France, 2024; pp. 194–206. [Google Scholar] [CrossRef]
- Karthikeyan, S.; Lakshmanan, S. Image Segmentation for MRI Brain Tumor Detection Using Advanced AI Algorithm. In Proceedings of the 2024 OPJU International Technology Conference (OTCON) on Smart Computing for Innovation and Advancement in Industry 4.0, IEEE, Raigarh, India, 5–7 June 2024; pp. 1–8. [Google Scholar] [CrossRef]
- Iriawan, N.; Pravitasari, A.A.; Nuraini, U.S.; Nirmalasari, N.I.; Azmi, T.; Nasrudin, M.; Fandisyah, A.F.; Fithriasari, K.; Purnami, S.W.; Irhamah; et al. YOLO-UNet Architecture for Detecting and Segmenting the Localized MRI Brain Tumor Image. Appl. Comput. Intell. Soft Comput. 2024, 2024, 3819801. [Google Scholar] [CrossRef]
- Zafar, W.; Husnain, G.; Iqbal, A.; Alzahrani, A.S.; Irfan, M.A.; Ghadi, Y.Y.; Al-Zahrani, M.S.; Naidu, R.S. Enhanced TumorNet: Leveraging YOLOv8s and U-Net for Superior Brain Tumor Detection and Segmentation Utilizing MRI Scans. Results Eng. 2024, 18, 102994. [Google Scholar] [CrossRef]
- Karacı, A.; Akyol, K. YoDenBi-NET: YOLO + DenseNet + Bi-LSTM-Based Hybrid Deep Learning Model for Brain Tumor Classification. Neural Comput. Appl. 2023, 35, 12583–12598. [Google Scholar] [CrossRef]
- Lyu, Y.; Tian, X. MWG-UNet++: Hybrid Transformer U-Net Model for Brain Tumor Segmentation in MRI Scans. Bioengineering 2025, 12, 140. [Google Scholar] [CrossRef]
- Rastogi, D.; Johri, P.; Donelli, M.; Kadry, S.; Khan, A.A.; Espa, G.; Feraco, P.; Kim, J. Deep learning-integrated MRI brain tumor analysis: Feature extraction, segmentation, and survival prediction using replicator and volumetric networks. Sci. Rep. 2025, 15, 1437. [Google Scholar] [CrossRef]
- Shaikh, A.; Amin, S.; Zeb, M.A.; Sulaiman, A.; Al Reshan, M.S.; Alshahrani, H. Enhanced Brain Tumor Detection and Segmentation Using Densely Connected Convolutional Networks with Stacking Ensemble Learning. Comput. Biol. Med. 2025, 186, 109703. [Google Scholar] [CrossRef]
- Ganesh, A.; Kumar, P.; Singh, R.; Ray, M. Brain Tumor Segmentation and Detection in MRI Using Convolutional Neural Networks and VGG16. Cancer Biomark. 2025, 42, 18758592241311184. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, R.; Li, L.; Xie, X. SimAM: A Simple, Parameter-Free Attention Module for Convolutional Neural Networks. In Proceedings of the 38th International Conference on Machine Learning, Virtual, 18–24 July 2021; Proceedings of Machine Learning Research. Volume 139, pp. 11863–11874. [Google Scholar]
- Roboflow. Tumor Otak > YOLOv8 Crop 15-85. Roboflow Universe. Available online: https://universe.roboflow.com/sriwijaya-university-hivwu/tumor-otak-tbpou (accessed on 13 August 2025).
- Roboflow. BRAIN TUMOR > Roboflow Universe. Available online: https://universe.roboflow.com/college-piawa/brain-tumor-j0l2c (accessed on 11 September 2025).
- Roboflow. Project 5C Liver Tumor > Roboflow Universe. Available online: https://universe.roboflow.com/segmentasiliver/project-5c-liver-tumor-aikum (accessed on 11 September 2025).

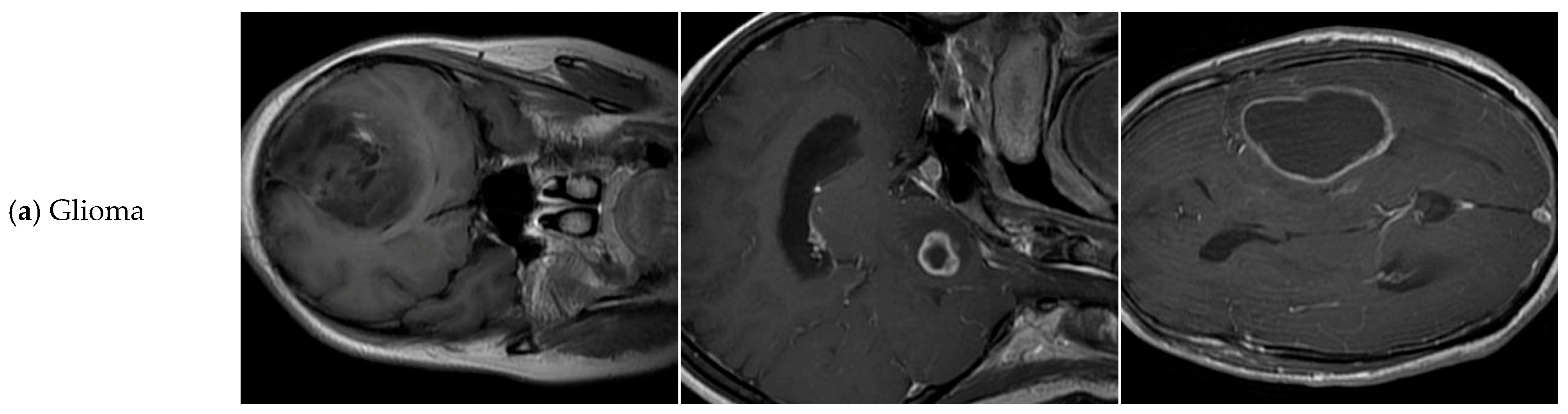
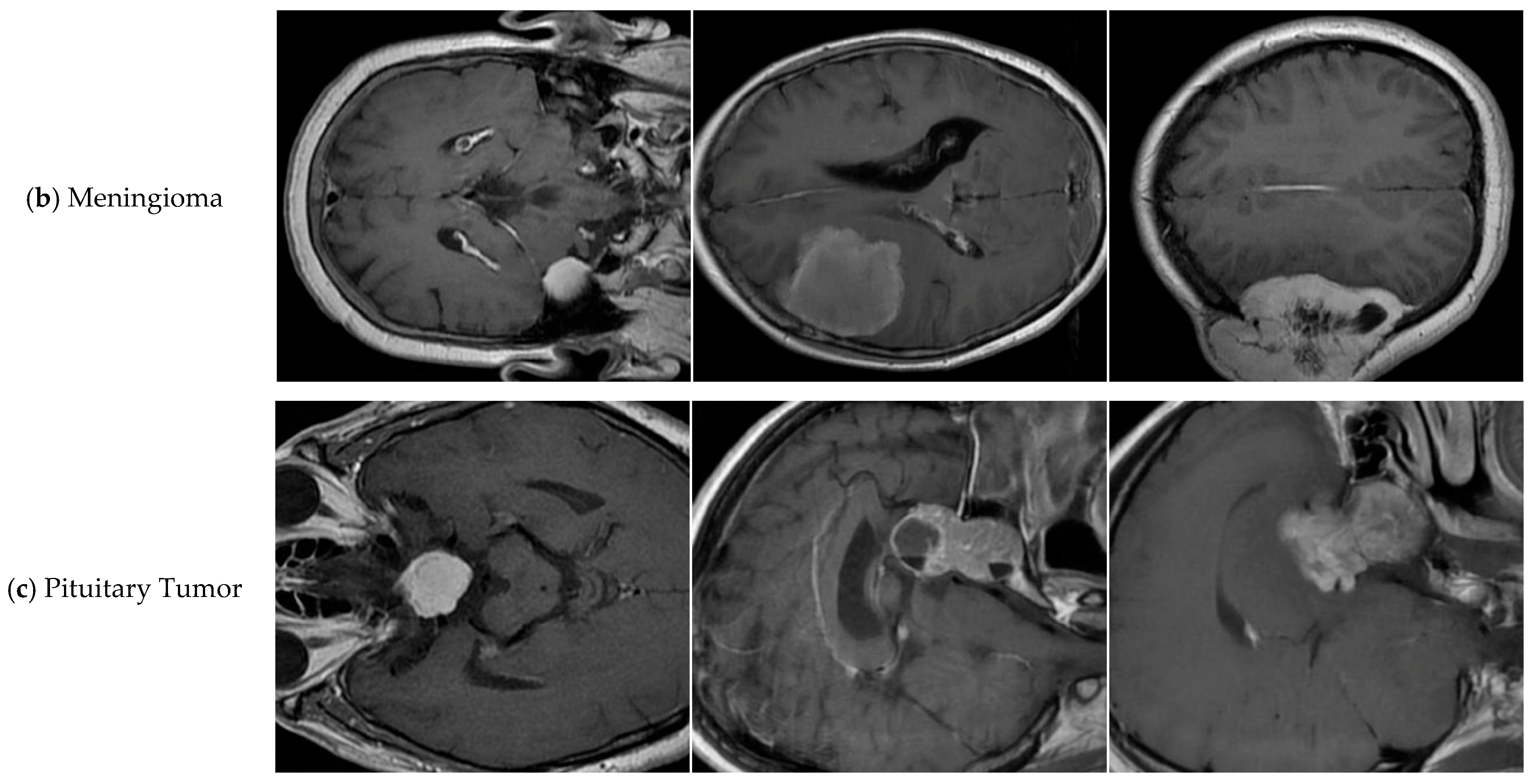
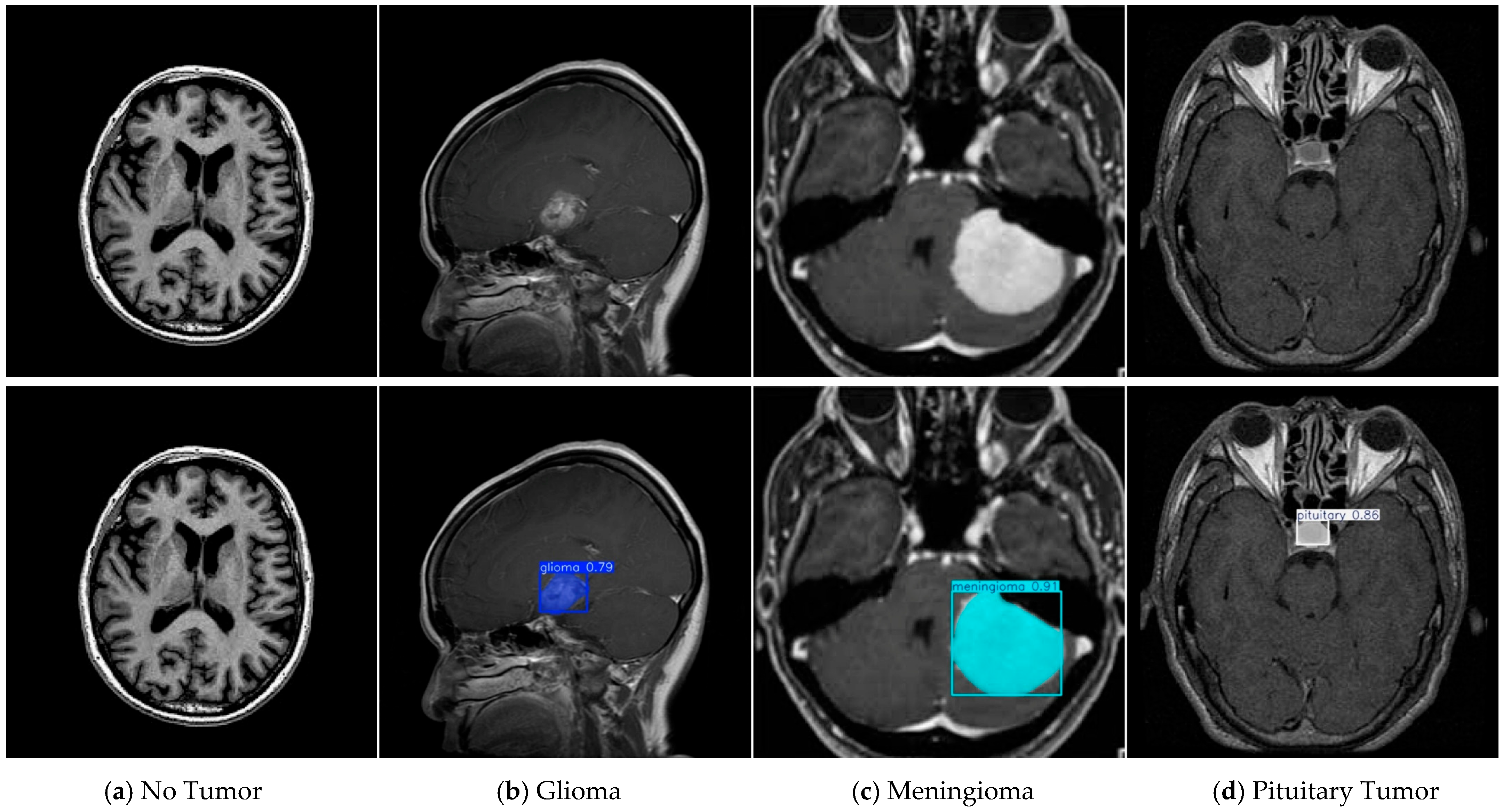

| Tumor-Otak | Total | Glioma | Meningioma | Pituitary Tumor |
|---|---|---|---|---|
| train | 2144 | 984 | 502 | 658 |
| val | 612 | 285 | 142 | 185 |
| test | 308 | 159 | 62 | 87 |
| Item | Version |
|---|---|
| Operating System | 64-bit Linux |
| Programming Language | Python 3.8.19 |
| PyTorch | 1.13.1 |
| CUDA | 11.7 |
| CPU | Intel(R) Xeon(R) Gold 5218 CPU @ 2.30 GHz |
| GPU | NVIDIA GeForce RTX 3090 × 1 (24 GB) |
| Item | Version |
|---|---|
| epoch | 120 |
| batch | 32 |
| workers | 4 |
| optimizer | SGD |
| learning rate | 0.001 |
| momentum | 0.973 |
| weight decay | 0.0005 |
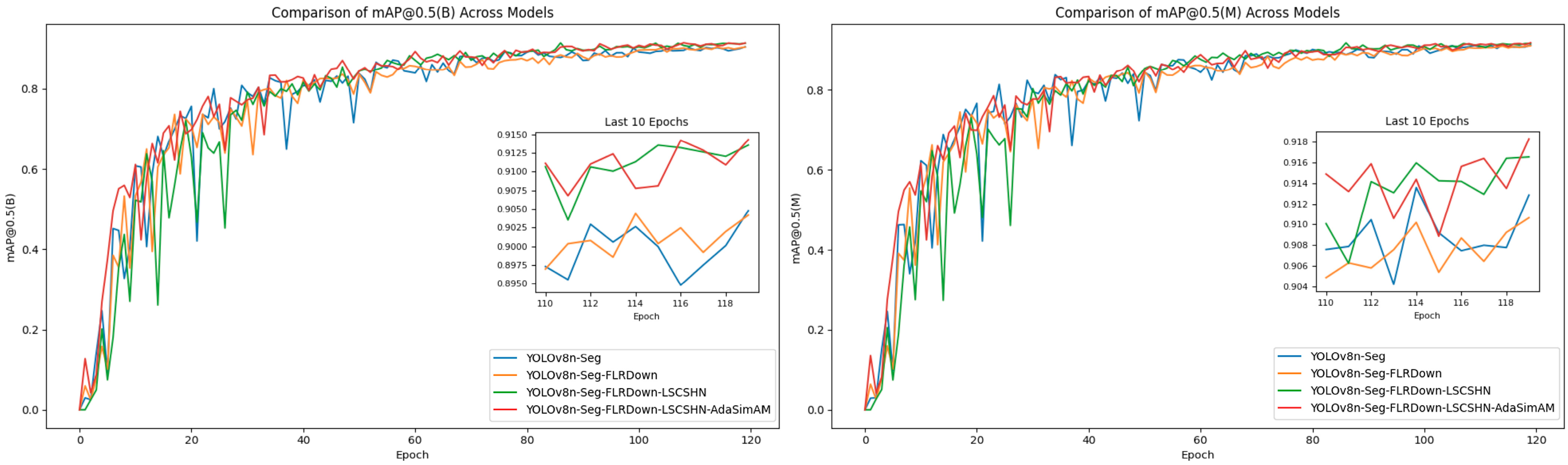
| FLRDown | LSCSHN | AdaSimAM | Precision (B) | Recall (B) | mAP@ 0.5(B) | Precision (M) | Recall (M) | mAP@ 0.5(M) | Param | GFLOPs | Model_Size |
|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | 0.870 | 0.843 | 0.897 | 0.881 | 0.858 | 0.909 | 3,258,649 | 12.0 | 6.5 M |
| √ | - | - | 0.894 | 0.823 | 0.900 | 0.896 | 0.84 | 0.905 | 2,975,529 | 11.5 | 5.9 M |
| √ | √ | - | 0.863 | 0.823 | 0.884 | 0.877 | 0.834 | 0.898 | 2,217,318 | 9.6 | 4.4 M |
| √ | √ | √ | 0.892 | 0.858 | 0.912 | 0.899 | 0.863 | 0.917 | 2,217,318 | 9.6 | 4.4 M |
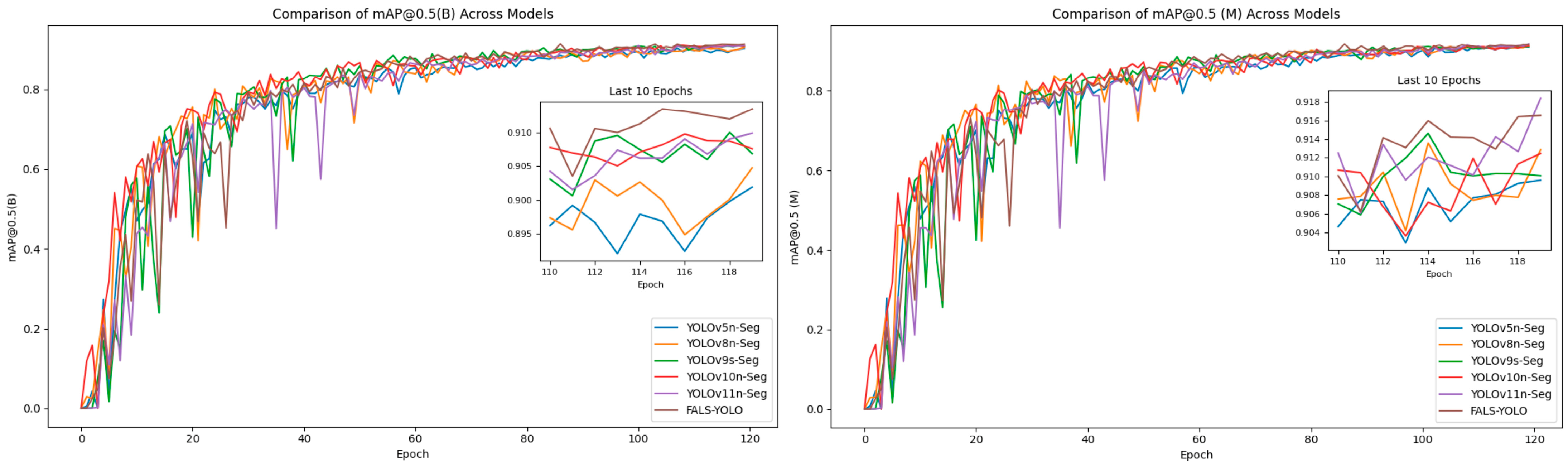
| Method | Precision (B) | Recall (B) | mAP@ 0.5 (B) | Precision (M) | Recall (M) | mAP@ 0.5 (M) | Param | GFLOPs | Model_Size |
|---|---|---|---|---|---|---|---|---|---|
| YOLOv5n-Seg | 0.873 | 0.834 | 0.900 | 0.883 | 0.842 | 0.913 | 2,755,945 | 11.0 | 5.5 M |
| YOLOv8n-Seg | 0.870 | 0.843 | 0.897 | 0.881 | 0.858 | 0.909 | 3,258,649 | 12.0 | 6.5 M |
| YOLOv9s-Seg | 0.863 | 0.852 | 0.895 | 0.862 | 0.881 | 0.906 | 8,521,017 | 75.4 | 17.1 M |
| YOLOv10n-Seg | 0.882 | 0.838 | 0.908 | 0.903 | 0.847 | 0.915 | 2,839,449 | 11.7 | 5.7 M |
| YOLOv11n-Seg | 0.876 | 0.875 | 0.907 | 0.883 | 0.883 | 0.913 | 2,835,153 | 10.2 | 5.7 M |
| FALS-YOLO (Ours) | 0.892 | 0.858 | 0.912 | 0.899 | 0.863 | 0.917 | 2,217,318 | 9.6 | 4.4 M |
| SimAM | AdaSimAM | Precision (B) | Recall (B) | mAP@ 0.5 (B) | Precision (M) | Recall (M) | mAP@ 0.5 (M) | Param | GFLOPs | Model_Size |
|---|---|---|---|---|---|---|---|---|---|---|
| - | - | 0.863 | 0.823 | 0.884 | 0.877 | 0.834 | 0.898 | 2,217,318 | 9.6 | 4.4 M |
| √ | - | 0.887 | 0.822 | 0.896 | 0.895 | 0.846 | 0.908 | 2,217,318 | 9.6 | 4.4 M |
| - | √ | 0.892 | 0.858 | 0.912 | 0.899 | 0.863 | 0.917 | 2,217,318 | 9.6 | 4.4 M |
| Class | FALS-YOLO Correct Classification | FALS-YOLO Misclassification | YOLOv8-Seg Correct Classification | YOLOv8-Seg Misclassification |
|---|---|---|---|---|
| Glioma | 132 | 40 | 124 | 37 |
| Meningioma | 58 | 10 | 56 | 6 |
| Pituitary | 81 | 10 | 78 | 5 |
| Background | - | 31 | - | 42 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, L.; Zheng, L.; Xin, Y. FALS-YOLO: An Efficient and Lightweight Method for Automatic Brain Tumor Detection and Segmentation. Sensors 2025, 25, 5993. https://doi.org/10.3390/s25195993
Sun L, Zheng L, Xin Y. FALS-YOLO: An Efficient and Lightweight Method for Automatic Brain Tumor Detection and Segmentation. Sensors. 2025; 25(19):5993. https://doi.org/10.3390/s25195993
Chicago/Turabian StyleSun, Liyan, Linxuan Zheng, and Yi Xin. 2025. "FALS-YOLO: An Efficient and Lightweight Method for Automatic Brain Tumor Detection and Segmentation" Sensors 25, no. 19: 5993. https://doi.org/10.3390/s25195993
APA StyleSun, L., Zheng, L., & Xin, Y. (2025). FALS-YOLO: An Efficient and Lightweight Method for Automatic Brain Tumor Detection and Segmentation. Sensors, 25(19), 5993. https://doi.org/10.3390/s25195993






