4.4.1. Quantitative Research
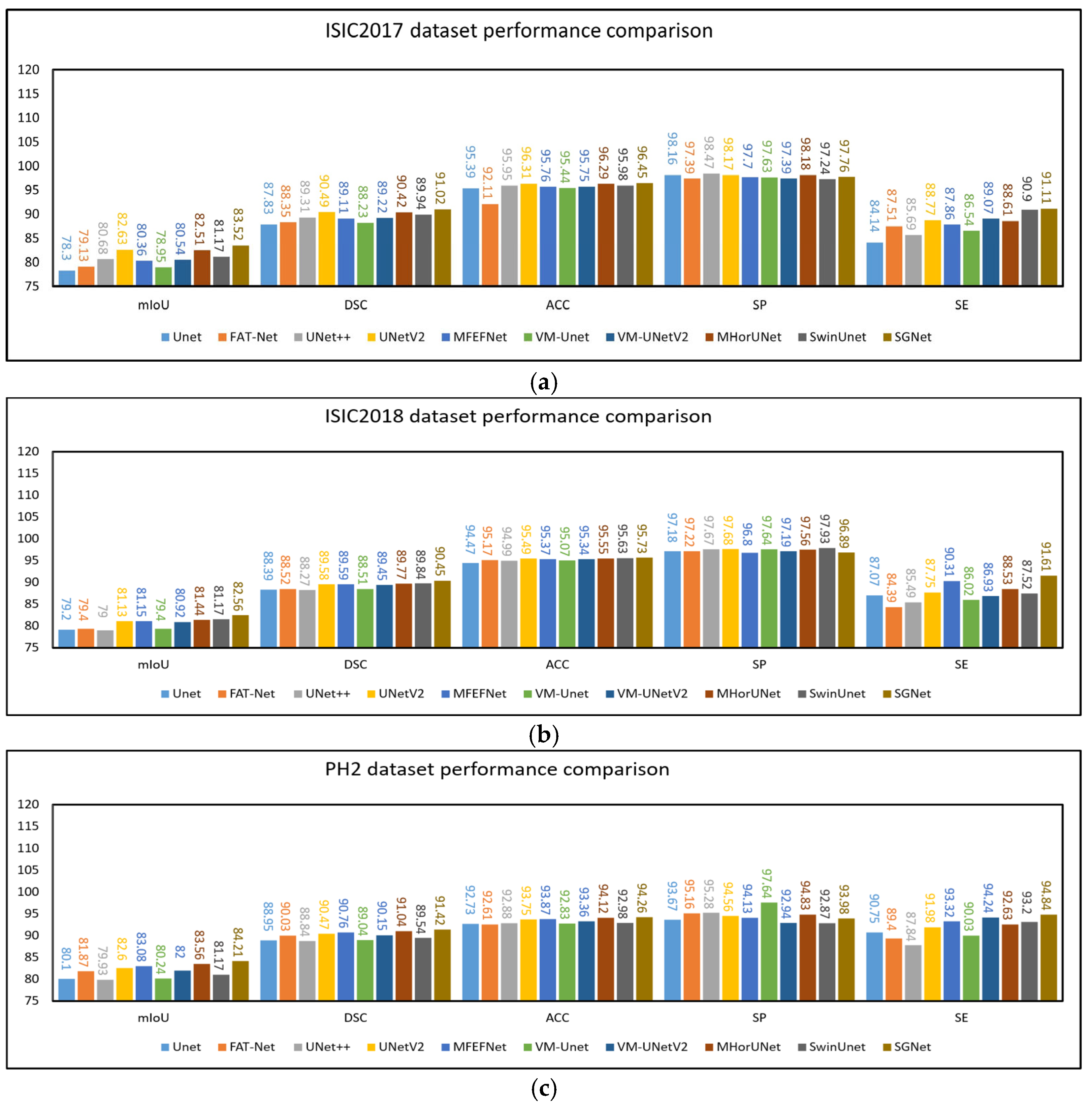
The experimental results based on the ISIC2017 dataset are presented in
Table 3. UNet++, UNetV2, MFEFNet, VM-UNetV2, MHorUNet, and SwinUnet all achieved mIoU values above 80% and DSC values exceeding 89%. Compared to UNet++, UNetV2, MFEFNet, VM-UNetV2, MHorUNet, and SwinUnet, SGNet improved the mIoU values by 3.48%, 1.07%, 3.95%, 3.68%, 1.46%, and 2.74%, respectively. The corresponding DSC improvements were 1.92%, 0.58%, 2.12%, 2.02%, 0.66%, and 1.21%. Compared to all other methods, SGNet improved the mIoU by an average of 4.01% and the DSC by 2.28%, demonstrating superior segmentation performance based on the ISIC2017 dataset.
These results underscore SGNet’s ability to deliver superior segmentation performance and robustness. By effectively addressing challenges such as complex lesion boundaries and image noise, SGNet demonstrates a significant advancement over existing methods.
The experimental results based on the ISIC2018 dataset are presented in
Table 4. UNetV2, MFEFNet, MHorUNet, SwinUnet, and VM-UNetV2 all achieved mIoU values above 80% and DSC values exceeding 89%. Compared to UNetV2, MFEFNet, MHorUNet, SwinUnet, and VM-UNetV2, SGNet improved the mIoU values by 1.76%, 1.74%, 1.38%, 1.23%, and 2.03%, respectively. The corresponding DSC improvements were 0.97%, 0.96%, 0.76%, 0.68%, and 1.12%. Compared to all of the other methods, SGNet improved the mIoU by an average of 2.87% and the DSC by 1.52%, demonstrating superior segmentation performance based on the ISIC2018 dataset.
Moreover, SGNet maintained competitive results in terms of accuracy, specificity, and sensitivity, demonstrating its ability to balance overall segmentation accuracy, specificity, and sensitivity. These results further validate the effectiveness of SGNet in addressing challenges such as complex lesion boundaries, variable contrast, and image noise.
To verify the generalization ability of the model, we used the PH2 dataset for external validation, where the model trained on the ISIC2018 dataset was directly applied to make predictions based on the PH2 dataset. The experimental results based on the PH2 dataset are presented in
Table 5. UNetV2, MFEFNet, MHorUNet, VM-UNetV2, and SGNet all achieved mIoU values above 82% and DSC values exceeding 90%. Compared to UNetV2, MFEFNet, MHorUNet, and VM-UNetV2, SGNet improved the mIoU values by 1.95%, 1.36%, 0.78%, and 2.70%, respectively. The corresponding DSC improvements were 1.05%, 0.73%, 0.42%, and 1.41%. Compared to all the other methods, SGNet improved the mIoU by an average of 3.22% and the DSC by 1.73%, further demonstrating its superior generalization ability and segmentation performance based on unseen data.
Although SGNet significantly outperforms other state-of-the-art methods in terms of the mIoU and DSC, its SP is slightly lower than that of some competing models based on the ISIC2017 and ISIC2018 datasets. This outcome may be attributed to the model’s design emphasis on enhancing sensitivity to lesion regions through the STFU and GMSR modules. The STFU module adaptively integrates shallow texture features with deep semantic representations, enabling the model to better highlight ambiguous or low-contrast lesion areas. Meanwhile, the GMSR progressively refines the segmentation output under semantic guidance and deep supervision, enhancing the model’s responsiveness to fine boundaries and subtle structural cues. However, this focus on sensitivity and detailed boundary delineation may inadvertently increase the risk of over-segmenting non-lesion areas with similar visual characteristics. As a result, SGNet may generate more false positive predictions, leading to a slight decline in specificity.
The experimental results from SGNet based on the PH2 dataset demonstrate that it not only achieves excellent performance based on internal datasets, but also exhibits strong generalization ability during external validation. The design concept of SGNet, particularly its GMSR and DDBE, allows it to showcase outstanding adaptability and robustness in the challenging task of skin lesion image segmentation. In order to illustrate the above comparative experiments more intuitively, as shown in
Figure 8a–c, this chapter presents the results of the comparative experiments based on the three datasets in the form of histograms.
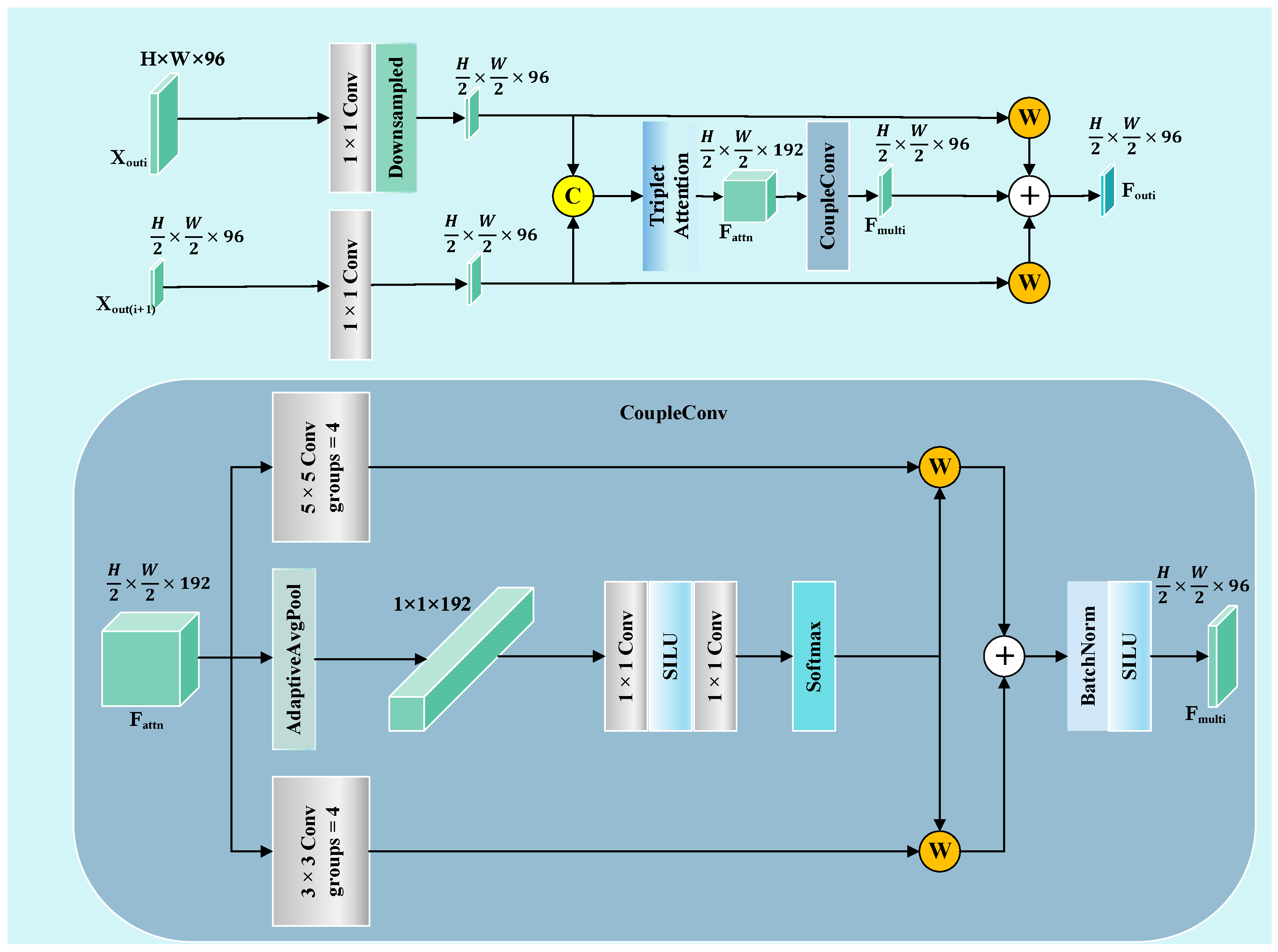
SGNet significantly reduces the computational complexity, while maintaining high segmentation accuracy through the DDBE module. As shown in
Table 6, SGNet has 30.26 M parameters, 7.92 G FLOPs, and an inference time of 0.012 s. Compared with other mainstream models, such as U-Net (32.08 M parameters, 16.59 G FLOPs, 0.008 s inference time), SwinUnet (41.39 M parameters, 11.37 G FLOPs, 0.214 s inference time), and UNet++ (47.20 M parameters, 65.58 G FLOPs, 0.112 s inference time), SGNet achieves an excellent balance between computational efficiency and segmentation performance. Compared to models with higher FLOPs, such as FAT-Net and UNet++, SGNet greatly reduces the computational cost, with only 7.92 G FLOPs required. Moreover, SGNet achieves a much shorter inference time of 0.012 s compared to SwinUnet, making it more suitable for real-time applications.
To further evaluate the performance of our proposed SGNet in regard to skin lesion segmentation tasks, we compared it with several representative models based on the ISIC2018 dataset. As shown in
Table 7, SGNet achieved the best performance in regard to both the DSC and HD95, with a DSC of 90.19 ± 0.17 and a HD95 of 13.10 ± 0.25, significantly outperforming other competing methods. The
p-values reported further highlight the superiority of SGNet in terms of DSC performance, indicating statistically significant improvements over all the baseline methods. These findings demonstrate that SGNet not only delivers superior overall segmentation accuracy, but also exhibits greater robustness and more precise boundary delineation.
4.4.2. Qualitative Research
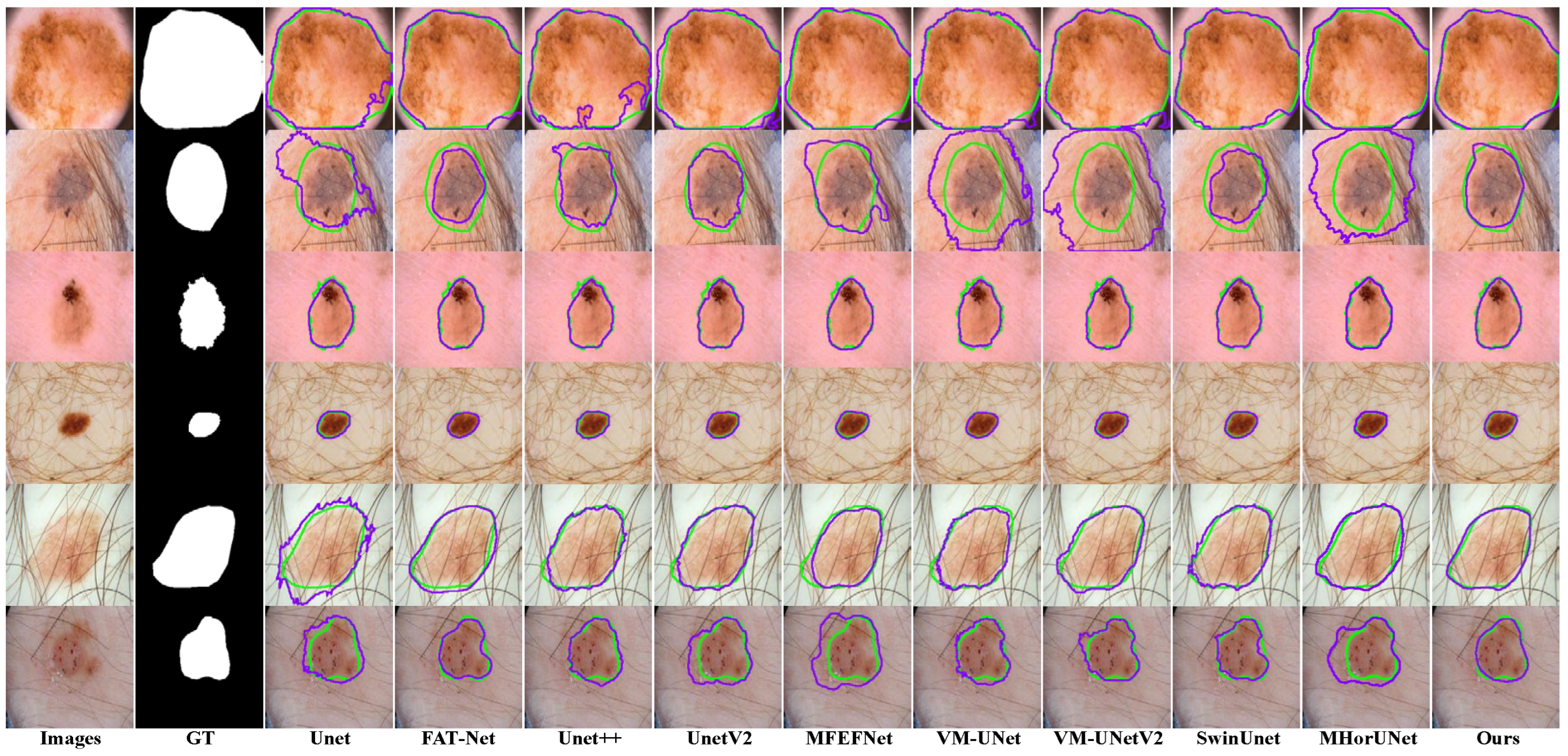
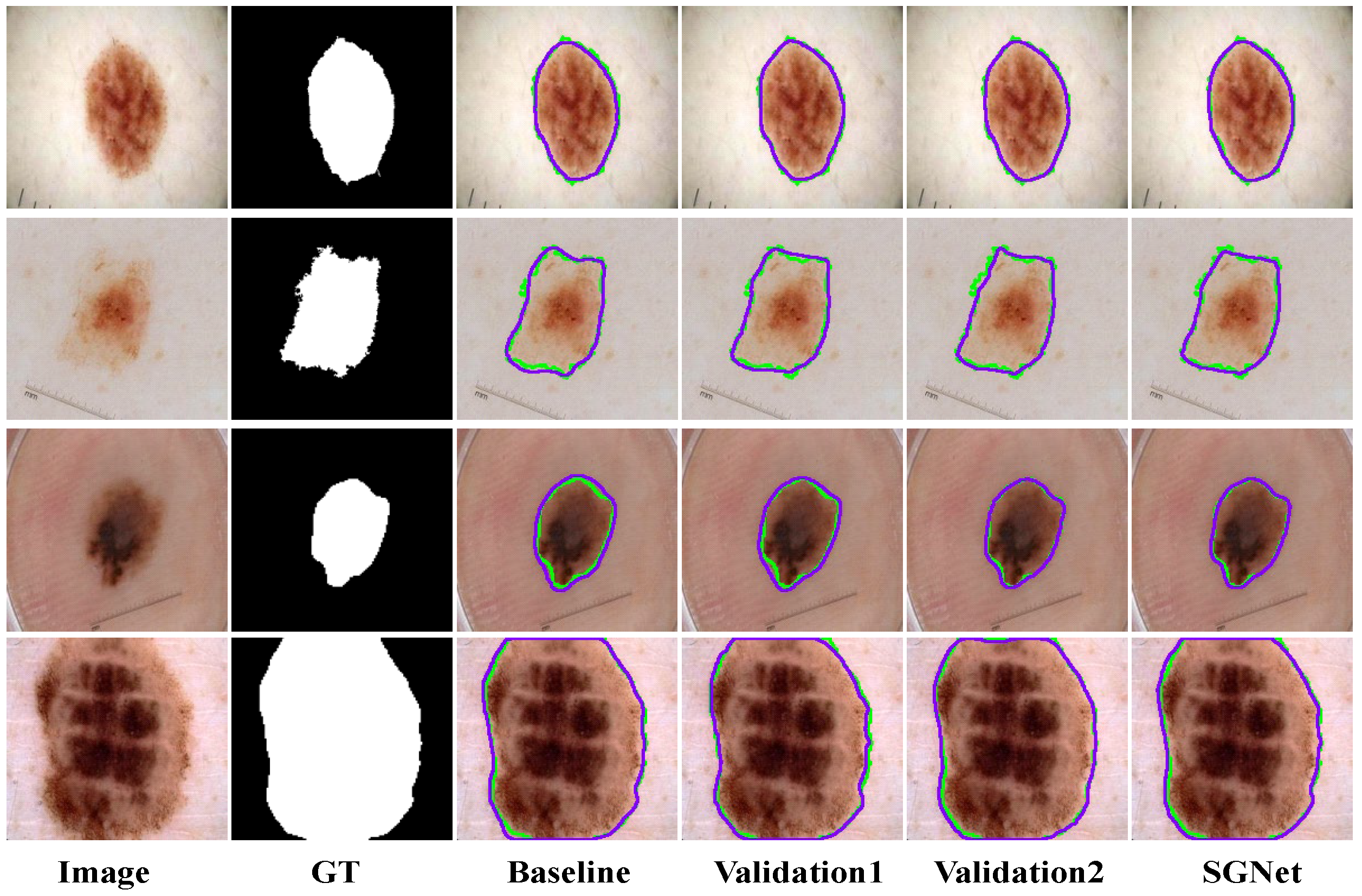
Existing models show clear limitations in segmenting the ISIC2017 dataset, as shown in
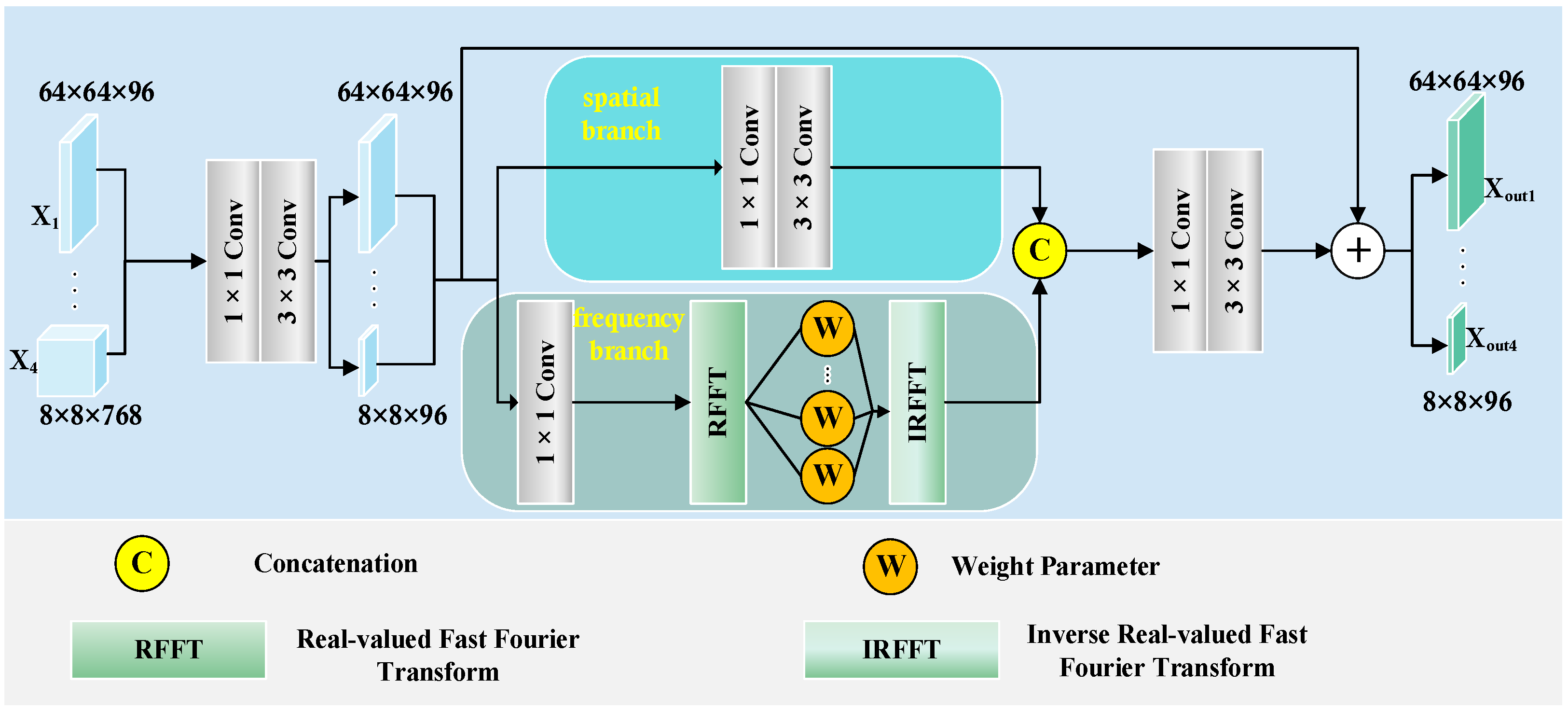
Figure 9. The UNet architecture struggles to capture complex lesions, due to its simple design. This results in missed target regions and lower accuracy for detailed areas. FAT-Net demonstrates an insufficient capability in capturing fine boundaries of small lesion targets, resulting in frequent mis-segmentation. MFEFNet and VM-UNet exhibit misclassification in regard to low-contrast skin images, where lesion-background differentiation becomes challenging. Furthermore, VM-UNetV2, MHorUNet, and SwinUnet models are susceptible to hair interference, causing incomplete lesion segmentation or erroneous classification of hair as lesions or background areas. SGNet leverages the DDBE and STFU for robust segmentation. The DDBE employs a dual-domain approach, wherein its frequency branch applies an RFFT to transform feature maps, emphasizing boundary-related frequency components, while attenuating high-frequency noise associated with hair artifacts. This noise suppression mechanism ensures accurate lesion segmentation without misclassifying hair. The STFU complements the DDBE by adaptively fusing these enhanced boundary features with semantic information through a triplet attention mechanism and multi-branch convolution blocks. This synergy enables SGNet to maintain morphological consistency in hair-occluded regions.
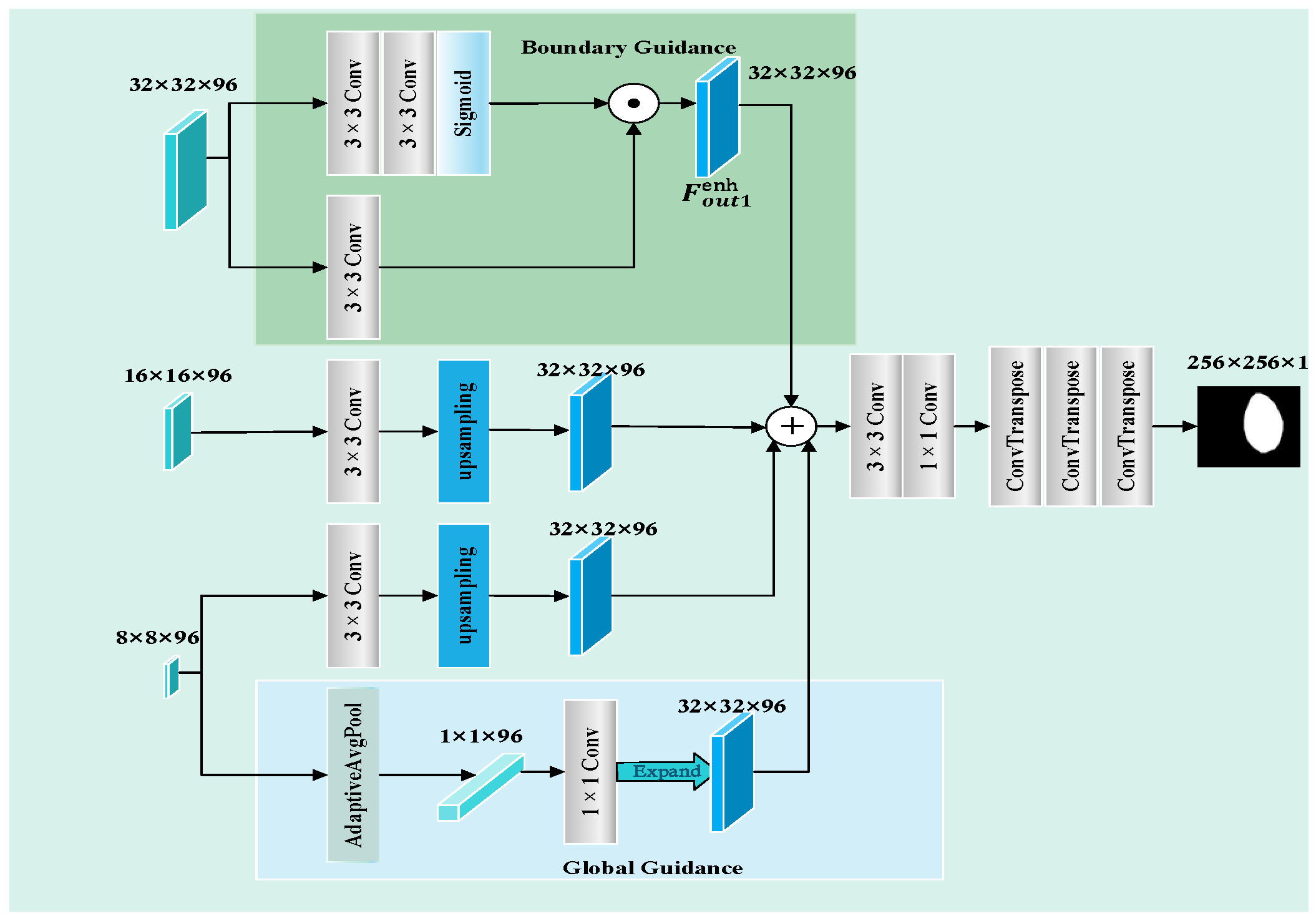
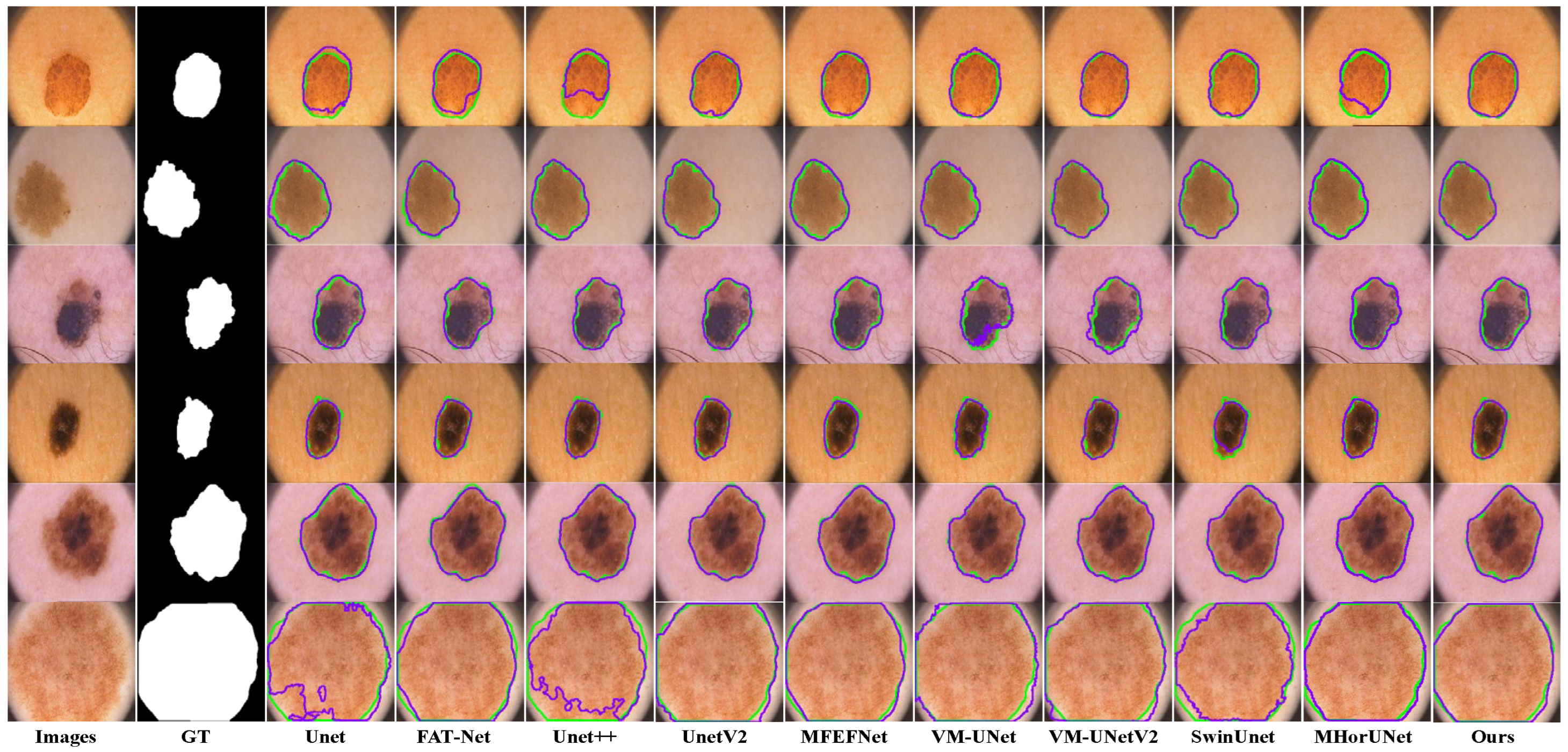
The segmentation results based on the ISIC2018 dataset, as shown in
Figure 10, reveal distinct limitations among the existing models. UNet++ and UNetV2 involve improved architectures compared to their predecessors, but remain inadequate in regard to detecting subtle lesions. FAT-Net and MFEFNet generate false positive predictions in scenarios with complex textural backgrounds, while VM-UNetV2 and MHorUNet fail to fully mitigate partial missed detections under hair occlusion. Notably, SwinUnet demonstrates enhanced robustness against hair interference; however, its performance degrades in low-contrast regions, producing blurred boundaries. SGNet introduces the SAGM and GMSR, following the boundary enhancement and semantic fusion provided by the DDBE and STFU, to further improve segmentation accuracy. The SAGM integrates multi-level fused features to produce a coarse lesion localization map, which provides structural guidance for more accurate lesion detection. Building on this, the GMSR utilizes the guidance to progressively refine the lesion boundaries through the use of a multi-scale semantic attention mechanism. This two-stage process effectively restores precise contours and ensures morphological consistency.
The segmentation results based on the PH2 dataset, as shown in
Figure 11, reveal distinct limitations among the existing models. UNet shows significant segmentation failures in the lesion regions, indicating weak generalization ability. FAT-Net and VM-UNet can roughly delineate the lesion regions, but with limited accuracy. MFEFNet, VM-UNetv2, MHorUNet, and SwinUnet can perform segmentation tasks successfully in most scenarios, with the segmentation results closely matching the ground truth, although their performance drops in low-contrast conditions. In contrast, SGNet accurately segments the lesion regions, unaffected by interference factors, and demonstrates strong generalization ability, outperforming the other networks.
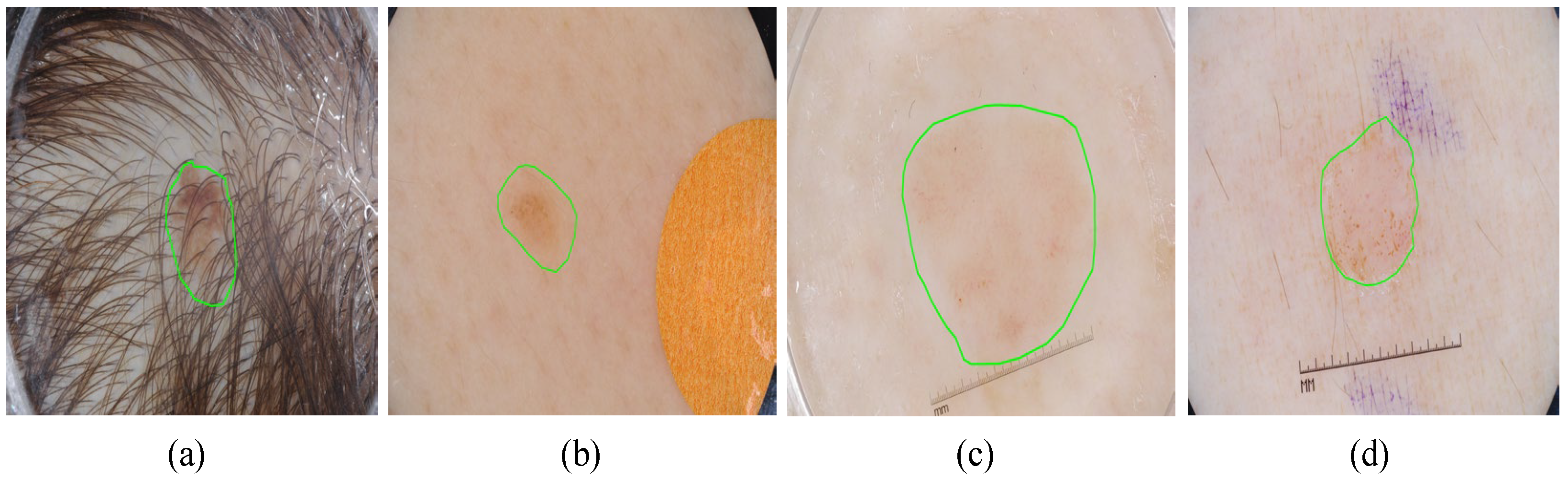
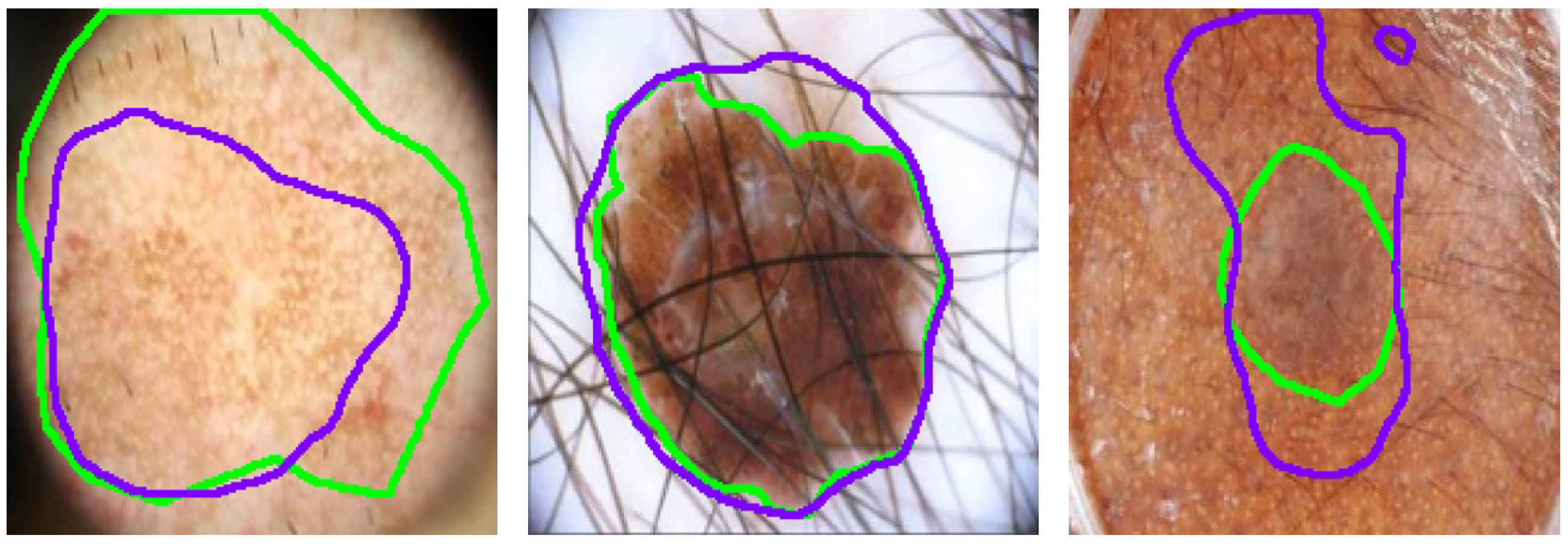
Despite SGNet’s robust performance, certain complex cases reveal limitations that require further investigation, as illustrated in
Figure 12. In scenarios with severe hair interference, frequency-domain modeling struggles to fully suppress high-frequency artifacts, leading to partial mis-segmentation or edge confusion around occluded regions. Although the dual-domain design enhances boundary perception, densely distributed hair or noise-like patterns can introduce spectral components that resemble lesion textures, thereby misleading the segmentation process. Additionally, in regard to low-contrast backgrounds, where the intensity difference between the lesion and healthy skin is minimal, the model often exhibits blurred or uncertain boundary predictions. This is particularly evident in regard to lesions with fuzzy perimeters or irregular morphological structures, where both spatial cues and semantic priors are weak. These challenges complicate accurate lesion delineation.