Cross-Modal Data Fusion via Vision-Language Model for Crop Disease Recognition
Abstract
1. Introduction
- (1)
- To solve the multi-modal data fusion of the image and text description features of crop diseases, we propose a cross-modal data fusion framework through a visual-language model. Extensive experiments on multiple crop datasets show that our proposed method can achieve better performance than state-of-the-art methods that rely only on image data to recognize crop diseases.
- (2)
- Compared with other image-only models, our proposed model has smaller parameters and is more suitable for deployment in edge devices to achieve high-precision crop disease recognition.
2. Related Works
2.1. Convolutional Neural Network for Crop Disease Recognition
2.1.1. Traditional CNNs
2.1.2. Lightweight CNNs
2.2. Multimodel Data Fusion for Crop Disease Recognition
2.3. Vision-Language Model
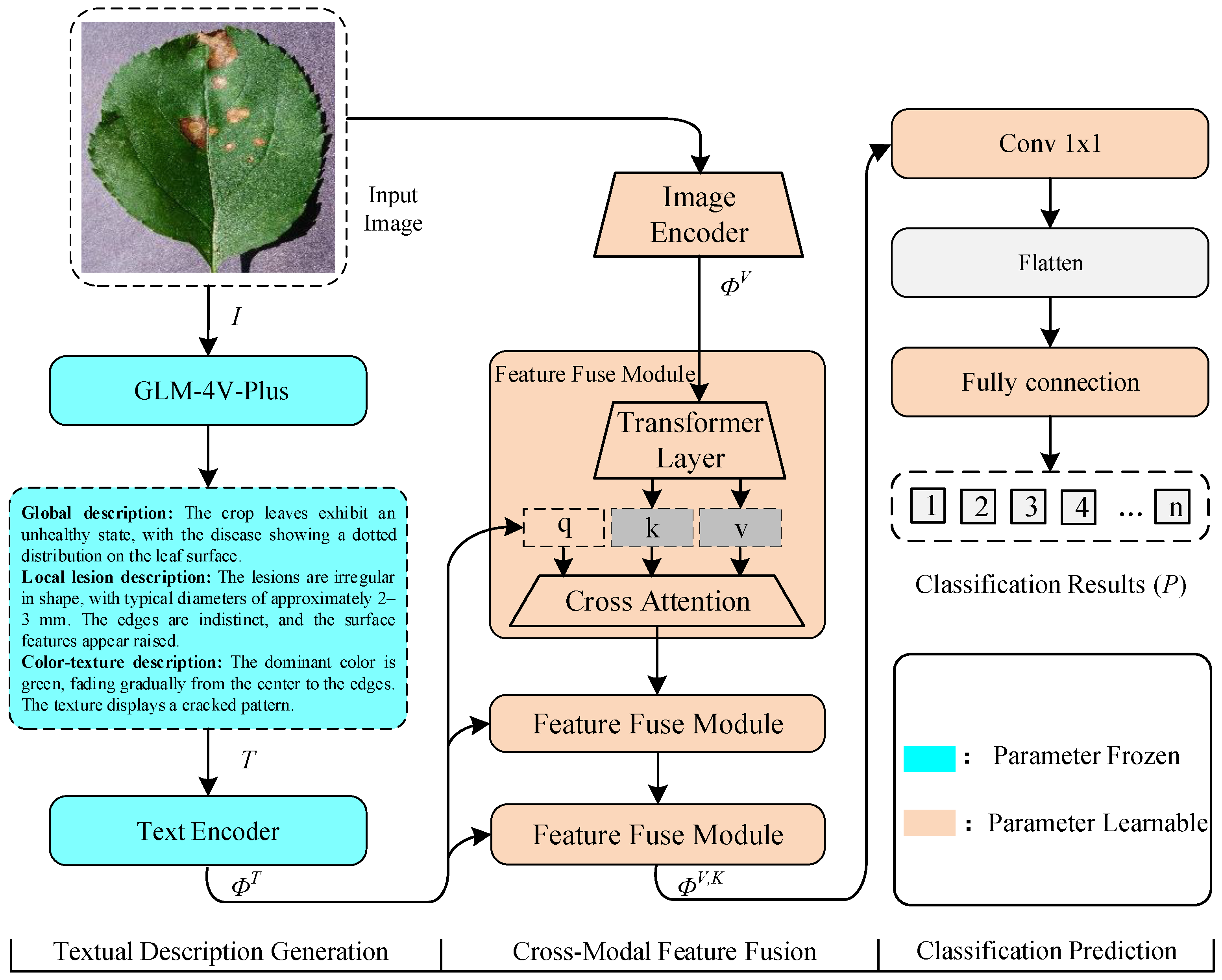
3. Methodology
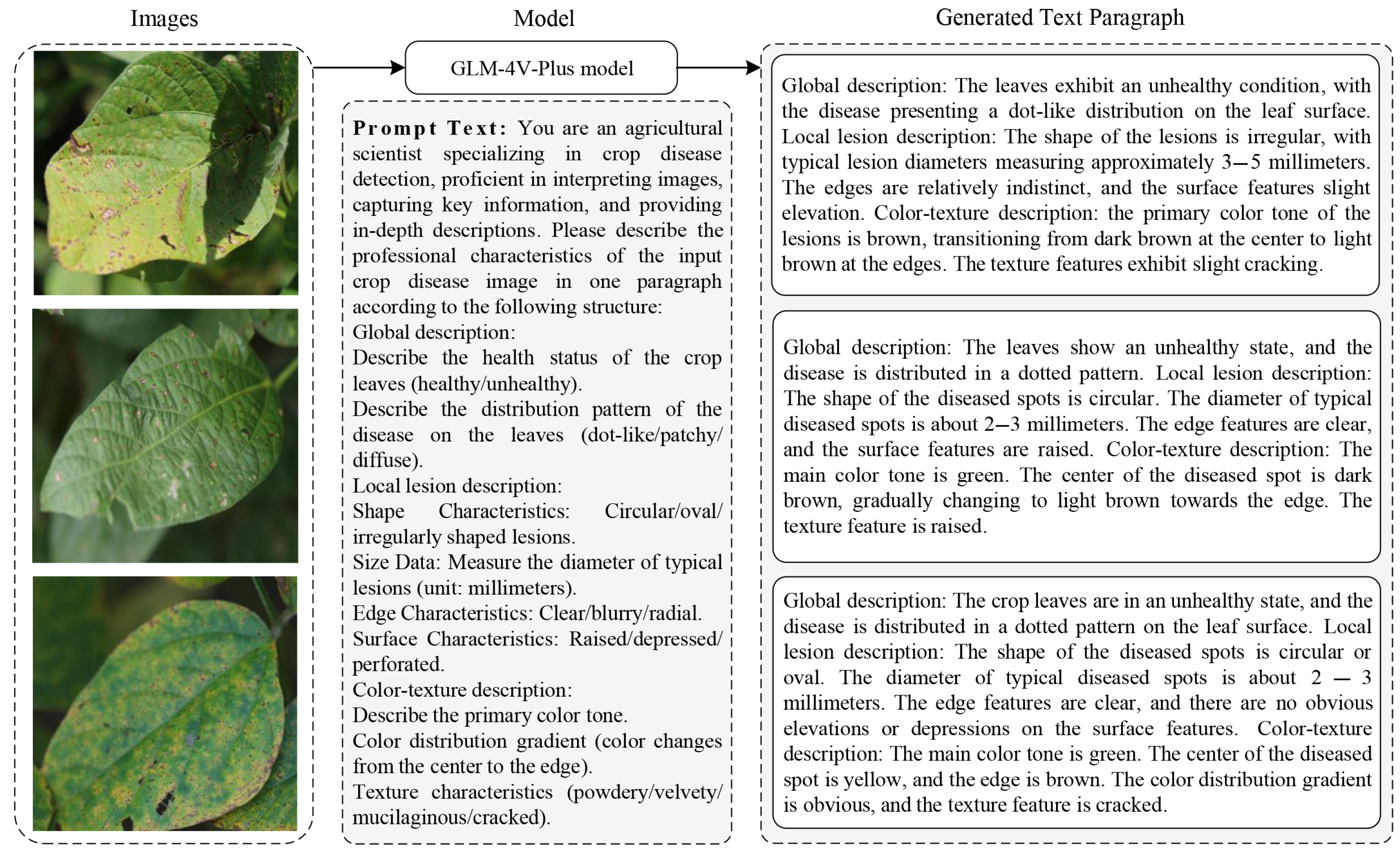
3.1. Textual Description Generation
3.2. Cross-Model Feature Fusion
3.3. Classification Prediction
3.4. Crop Disease Multimodal Dataset Construction
4. Experiment and Analysis
4.1. Implementation Details
4.2. Ablation Studies
4.3. Image Classification on the Crop Disease Datasets
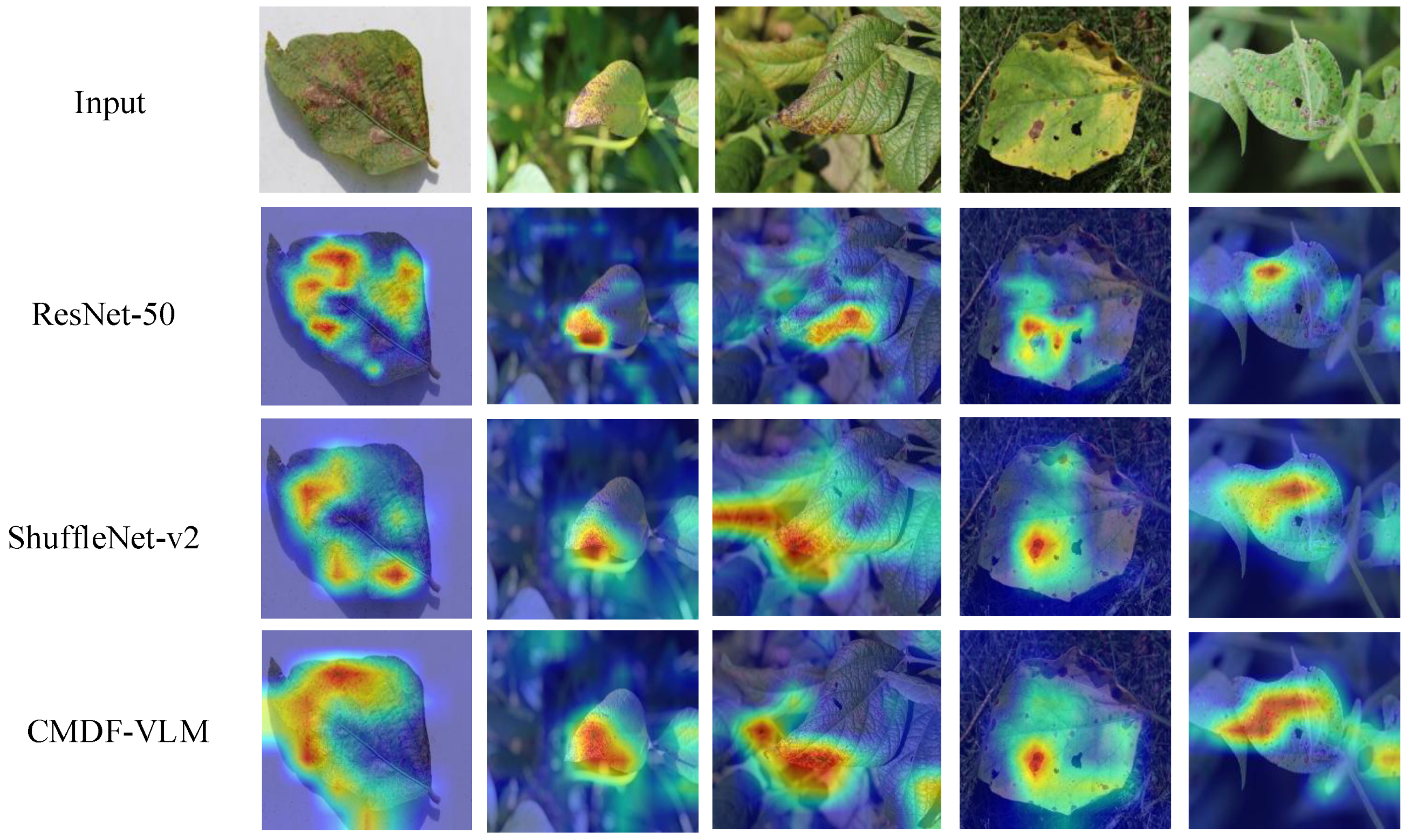
4.4. Network Visualization with Grad-CAM
5. Discussion and Analysis
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef]
- Elmer, W.; White, J.C. The future of nanotechnology in plant pathology. Annu. Rev. Phytopathol. 2018, 56, 111–133. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Li, G.; Qu, P.; Xie, X.; Pan, X.; Zhang, W. Research on plant disease identification based on CNN. Cogn. Robot. 2022, 2, 155–163. [Google Scholar] [CrossRef]
- Benti, N.E.; Chaka, M.D.; Semie, A.G.; Warkineh, B.; Soromessa, T. Transforming agriculture with Machine Learning, Deep Learning, and IoT: Perspectives from Ethiopia—challenges and opportunities. Discov. Agric. 2024, 2, 63. [Google Scholar] [CrossRef]
- Lei, L.; Yang, Q.; Yang, L.; Shen, T.; Wang, R.; Fu, C. Deep learning implementation of image segmentation in agricultural applications: A comprehensive review. Artif. Intell. Rev. 2024, 57, 149. [Google Scholar] [CrossRef]
- Elashmawy, R.; Uysal, I. Precision agriculture using soil sensor driven machine learning for smart strawberry production. Sensors 2023, 23, 2247. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Huang, Z.; Zhang, Z. Feature fusion transferability aware transformer for unsupervised domain adaptation. In Proceedings of the 2025 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Tucson, AZ, USA, 26 February–6 March 2025; IEEE: Piscataway, NJ, USA, 2025; pp. 6752–6761. [Google Scholar]
- Terentev, A.; Dolzhenko, V.; Fedotov, A.; Eremenko, D. Current state of hyperspectral remote sensing for early plant disease detection: A review. Sensors 2022, 22, 757. [Google Scholar] [CrossRef]
- Geetharamani, G.; Pandian, A. Identification of plant leaf diseases using a nine-layer deep convolutional neural network. Comput. Electr. Eng. 2019, 76, 323–338. [Google Scholar]
- Barbedo, J.G.A. Plant disease identification from individual lesions and spots using deep learning. Biosyst. Eng. 2019, 180, 96–107. [Google Scholar] [CrossRef]
- Liu, W.; Wu, C.; Ren, F. Stochastic channel reuse residual networks for plant disease severity detection. In Proceedings of the 2019 IEEE 6th International Conference on Cloud Computing and Intelligence Systems (CCIS), Singapore, 19–21 December 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 57–61. [Google Scholar]
- Chen, J.; Jia, J. Automatic Recognition of Tea Diseases Based on Deep Learning. In Advances in Forest Management Under Global Change; Zhang, L., Ed.; IntechOpen: Rijeka, Croatia, 2020; Chapter 8. [Google Scholar] [CrossRef]
- Karthik, R.; Hariharan, M.; Anand, S.; Mathikshara, P.; Johnson, A.; Menaka, R. Attention embedded residual CNN for disease detection in tomato leaves. Appl. Soft Comput. 2020, 86, 105933. [Google Scholar]
- Zhong, Y.; Zhao, M. Research on deep learning in apple leaf disease recognition. Comput. Electron. Agric. 2020, 168, 105146. [Google Scholar] [CrossRef]
- Wagle, S.A.; R, H. Comparison of Plant Leaf Classification Using Modified AlexNet and Support Vector Machine. Trait. Signal 2021, 38, 79–87. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. Shufflenet: An extremely efficient convolutional neural network for mobile devices. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 6848–6856. [Google Scholar]
- Ma, N.; Zhang, X.; Zheng, H.T.; Sun, J. Shufflenet v2: Practical guidelines for efficient cnn architecture design. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 116–131. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar]
- Han, K.; Wang, Y.; Tian, Q.; Guo, J.; Xu, C.; Xu, C. Ghostnet: More features from cheap operations. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 14–19 June 2020; pp. 1580–1589. [Google Scholar]
- Thakur, P.S.; Sheorey, T.; Ojha, A. VGG-ICNN: A Lightweight CNN model for crop disease identification. Multimed. Tools Appl. 2023, 82, 497–520. [Google Scholar] [CrossRef]
- Xiang, S.; Liang, Q.; Sun, W.; Zhang, D.; Wang, Y. L-CSMS: Novel lightweight network for plant disease severity recognition. J. Plant Dis. Prot. 2021, 128, 557–569. [Google Scholar] [CrossRef]
- Ametefe, D.S.; Sarnin, S.S.; Ali, D.M.; Caliskan, A.; Caliskan, I.T.; Aliu, A.A.; John, D. Enhancing leaf disease detection accuracy through synergistic integration of deep transfer learning and multimodal techniques. Inf. Process. Agric. 2024, in press. [Google Scholar]
- Zhou, J.; Li, J.; Wang, C.; Wu, H.; Zhao, C.; Teng, G. Crop disease identification and interpretation method based on multimodal deep learning. Comput. Electron. Agric. 2021, 189, 106408. [Google Scholar] [CrossRef]
- Li, H.; Tan, B.; Sun, L.; Liu, H.; Zhang, H.; Liu, B. Multi-Source Image Fusion Based Regional Classification Method for Apple Diseases and Pests. Appl. Sci. 2024, 14, 7695. [Google Scholar] [CrossRef]
- Lee, H.; Park, Y.S.; Yang, S.; Lee, H.; Park, T.J.; Yeo, D. A Deep Learning-Based Crop Disease Diagnosis Method Using Multimodal Mixup Augmentation. Appl. Sci. 2024, 14, 4322. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Z.; Hu, H.; Chen, Z.; Wang, K.; Wang, K.; Lian, S. A Multimodal Benchmark Dataset and Model for Crop Disease Diagnosis. In Proceedings of the European Conference on Computer Vision, Milan, Italy, 29 September–4 October 2024; Springer: Cham, Switzerland, 2024; pp. 157–170. [Google Scholar]
- Radford, A.; Kim, J.W.; Hallacy, C.; Ramesh, A.; Goh, G.; Agarwal, S.; Sastry, G.; Askell, A.; Mishkin, P.; Clark, J.; et al. Learning transferable visual models from natural language supervision. In Proceedings of the International Conference on Machine Learning, PMLR, Virtual, 18–24 July 2021; pp. 8748–8763. [Google Scholar]
- Brooks, T.; Holynski, A.; Efros, A.A. Instructpix2pix: Learning to follow image editing instructions. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 18–22 June 2023; pp. 18392–18402. [Google Scholar]
- Touvron, H.; Lavril, T.; Izacard, G.; Martinet, X.; Lachaux, M.A.; Lacroix, T.; Rozière, B.; Goyal, N.; Hambro, E.; Azhar, F.; et al. Llama: Open and efficient foundation language models. arXiv 2023, arXiv:2302.13971. [Google Scholar]
- Li, J.; Li, D.; Xiong, C.; Hoi, S. Blip: Bootstrapping language-image pre-training for unified vision-language understanding and generation. In Proceedings of the International Conference on Machine Learning, PMLR, Baltimore, MD, USA, 17–23 July 2022; pp. 12888–12900. [Google Scholar]
- Li, J.; Li, D.; Savarese, S.; Hoi, S. Blip-2: Bootstrapping language-image pre-training with frozen image encoders and large language models. In Proceedings of the International Conference on Machine Learning, PMLR, Honolulu, HI, USA, 23–29 July 2023; pp. 19730–19742. [Google Scholar]
- Ramesh, A.; Pavlov, M.; Goh, G.; Gray, S.; Voss, C.; Radford, A.; Chen, M.; Sutskever, I. Zero-shot text-to-image generation. In Proceedings of the International Conference on Machine Learning, PMLR, Virtual, 18–24 July 2021; pp. 8821–8831. [Google Scholar]
- Ramesh, A.; Dhariwal, P.; Nichol, A.; Chu, C.; Chen, M. Hierarchical text-conditional image generation with clip latents. arXiv 2022, arXiv:2204.06125. [Google Scholar]
- Achiam, J.; Adler, S.; Agarwal, S.; Ahmad, L.; Akkaya, I.; Aleman, F.L.; Almeida, D.; Altenschmidt, J.; Altman, S.; Anadkat, S.; et al. Gpt-4 technical report. arXiv 2023, arXiv:2303.08774. [Google Scholar]
- Bai, Y.; Zhang, J.; Lv, X.; Zheng, L.; Zhu, S.; Hou, L.; Dong, Y.; Tang, J.; Li, J. Longwriter: Unleashing 10,000+ word generation from long context llms. arXiv 2024, arXiv:2408.07055. [Google Scholar]
- Zamir, S.W.; Arora, A.; Khan, S.; Hayat, M.; Khan, F.S.; Yang, M.H. Restormer: Efficient transformer for high-resolution image restoration. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 5728–5739. [Google Scholar]
- Bevers, N.; Sikora, E.; Hardy, N. Soybean disease identification using original field images and transfer learning with convolutional neural networks. Comput. Electron. Agric. 2022, 203, 107449. [Google Scholar] [CrossRef]
- Hughes, D.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
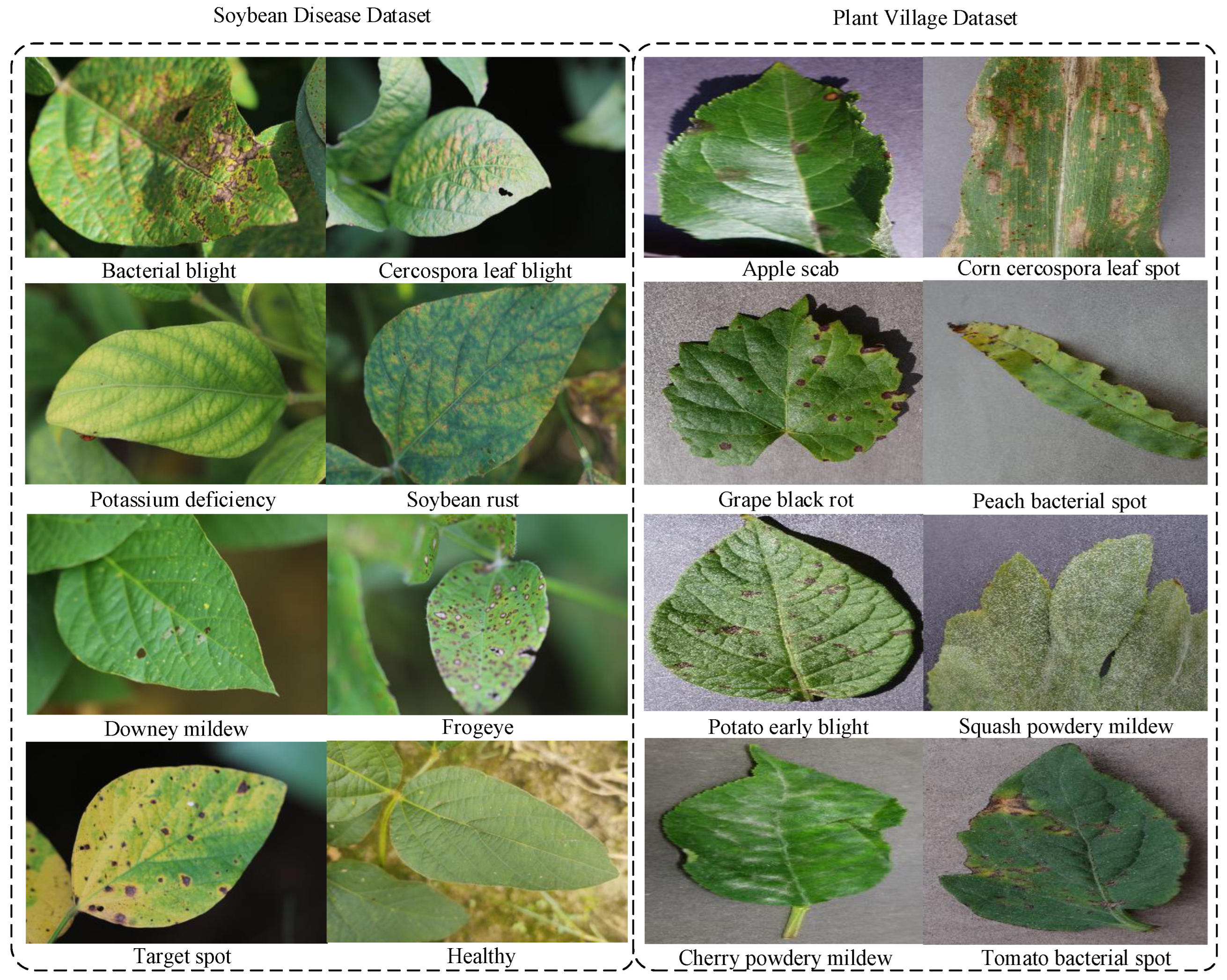
| Dataset | Number of Images | Number of Categories | Train | Test |
|---|---|---|---|---|
| Soybean Disease Dataset [38] | 10,722 | 8 | 7505 | 3217 |
| AI Challenge 2018 Dataset | 35,000 | 59 | 24,500 | 10,500 |
| Plant Village Dataset [39] | 54,305 | 36 | 38,013 | 16,292 |
| Dataset | Configurations | Accuracy (%) | ||
|---|---|---|---|---|
| Global Description | Local Lesion Description | Color-Texture description | ||
| Soybean Disease Dataset | 97.99 | |||
| ✓ | 98.43 | |||
| ✓ | ✓ | 98.69 | ||
| ✓ | ✓ | ✓ | 98.74 | |
| AI Challenge 2018 Dataset | 85.99 | |||
| ✓ | 87.09 | |||
| ✓ | ✓ | 87.32 | ||
| ✓ | ✓ | ✓ | 87.64 | |
| Plant Village Dataset | 97.81 | |||
| ✓ | 98.60 | |||
| ✓ | ✓ | 98.84 | ||
| ✓ | ✓ | ✓ | 99.08 | |
| Description | Soybean Disease Dataset | AI Challenge 2018 Dataset | Plant Village | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Params () | Acc (%) | Pre (%) | F1 (%) | R (%) | Params () | Acc (%) | Pre (%) | F1 (%) | R (%) | Params () | Acc (%) | Pre (%) | F1 (%) | R (%) | |
| ResNet-18 [40] | 11.18 | 98.05 | 97.83 | 97.96 | 98.11 | 11.20 | 86.77 | 83.54 | 82.30 | 82.06 | 11.19 | 98.79 | 98.69 | 98.70 | 98.72 |
| ResNet-34 [40] | 21.28 | 97.81 | 97.60 | 97.69 | 97.79 | 21.31 | 86.68 | 83.83 | 82.58 | 82.23 | 21.30 | 98.65 | 98.56 | 98.61 | 98.66 |
| ResNet-50 [40] | 23.52 | 97.84 | 97.61 | 97.72 | 97.83 | 23.62 | 86.92 | 84.88 | 83.78 | 83.82 | 23.58 | 98.61 | 98.55 | 98.61 | 98.67 |
| MobileNet-v1 [18] | 3.21 | 97.60 | 97.41 | 97.53 | 97.65 | 3.26 | 86.68 | 85.46 | 83.97 | 83.96 | 3.24 | 96.32 | 96.12 | 96.18 | 96.25 |
| MobileNet-v2 [19] | 2.23 | 97.71 | 97.60 | 97.64 | 97.69 | 2.29 | 86.53 | 83.95 | 83.04 | 83.07 | 2.27 | 96.92 | 96.68 | 96.70 | 96.72 |
| GhostNet [20] | 3.97 | 97.25 | 96.58 | 96.85 | 97.16 | 3.98 | 86.54 | 85.91 | 83.75 | 83.44 | 3.98 | 98.30 | 98.05 | 98.07 | 98.10 |
| ShuffleNet-v2 [17] | 4.72 | 97.95 | 97.78 | 97.83 | 97.89 | 4.87 | 85.68 | 85.02 | 82.23 | 81.70 | 4.81 | 96.59 | 96.33 | 96.42 | 96.52 |
| CMDF-VLM | 1.14 | 98.74 | 98.56 | 98.64 | 98.72 | 1.20 | 87.64 | 86.18 | 85.63 | 85.09 | 1.17 | 99.08 | 98.94 | 98.98 | 99.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Wu, G.; Wang, H.; Ren, F. Cross-Modal Data Fusion via Vision-Language Model for Crop Disease Recognition. Sensors 2025, 25, 4096. https://doi.org/10.3390/s25134096
Liu W, Wu G, Wang H, Ren F. Cross-Modal Data Fusion via Vision-Language Model for Crop Disease Recognition. Sensors. 2025; 25(13):4096. https://doi.org/10.3390/s25134096
Chicago/Turabian StyleLiu, Wenjie, Guoqing Wu, Han Wang, and Fuji Ren. 2025. "Cross-Modal Data Fusion via Vision-Language Model for Crop Disease Recognition" Sensors 25, no. 13: 4096. https://doi.org/10.3390/s25134096
APA StyleLiu, W., Wu, G., Wang, H., & Ren, F. (2025). Cross-Modal Data Fusion via Vision-Language Model for Crop Disease Recognition. Sensors, 25(13), 4096. https://doi.org/10.3390/s25134096







