1. Introduction
Cardiovascular diseases (CVDs) represent a major threat to global public health, with rapidly rising incidence rates fueled by modern lifestyles. As the leading cause of mortality worldwide for decades, CVDs claimed 20.5 million lives in 2021—accounting for nearly one-third of all global deaths, according to the World Heart Report [
1]. Cardiac arrhythmias—an important subset of cardiovascular diseases characterized by abnormal electrical activity manifesting as tachycardia, bradycardia, or irregular rhythms—pose significant diagnostic challenges due to their transient nature and morphologically diverse electrocardiographic signatures. The advent of electrocardiography (ECG) by Dutch physiologist Willem Einthoven in the early 20th century revolutionized cardiac diagnostics. Today, ECG remains the gold-standard tool for arrhythmia identification, offering temporal resolutions below 1 ms to capture fleeting electrical disturbances [
2].
Traditional ECG interpretation relies heavily on expert analysis, making it susceptible to subjectivity and inefficiency, with misdiagnosis rates reaching up to 15.2% in primary care [
3]. Recent advances in deep learning have significantly enhanced automated ECG analysis. Convolutional Neural Networks (CNNs) excel at morphological feature extraction [
4], while recurrent architectures like LSTMs and GRUs model temporal dynamics [
5,
6]. Hybrid CNN-RNN models have demonstrated superior classification performance (F1-scores up to 0.923) by capturing both spatial and temporal patterns.
Prior studies have proposed various deep learning solutions: Acharya et al. developed a nine-layer CNN for arrhythmia detection [
7]; Yildirim et al. added residual connections [
8]; Hannun et al. introduced a deep residual network trained on a large ECG dataset [
9]. Hybrid models, such as CNN-LSTM frameworks [
10,
11], and attention-enhanced networks [
12,
13], further improved performance. However, two core challenges persist: (1) performance degradation under class imbalance (F1-score drops of 12–18% for minority classes) and (2) sensitivity to noise and non-stationary waveforms, especially in ambulatory recordings [
14,
15,
16].
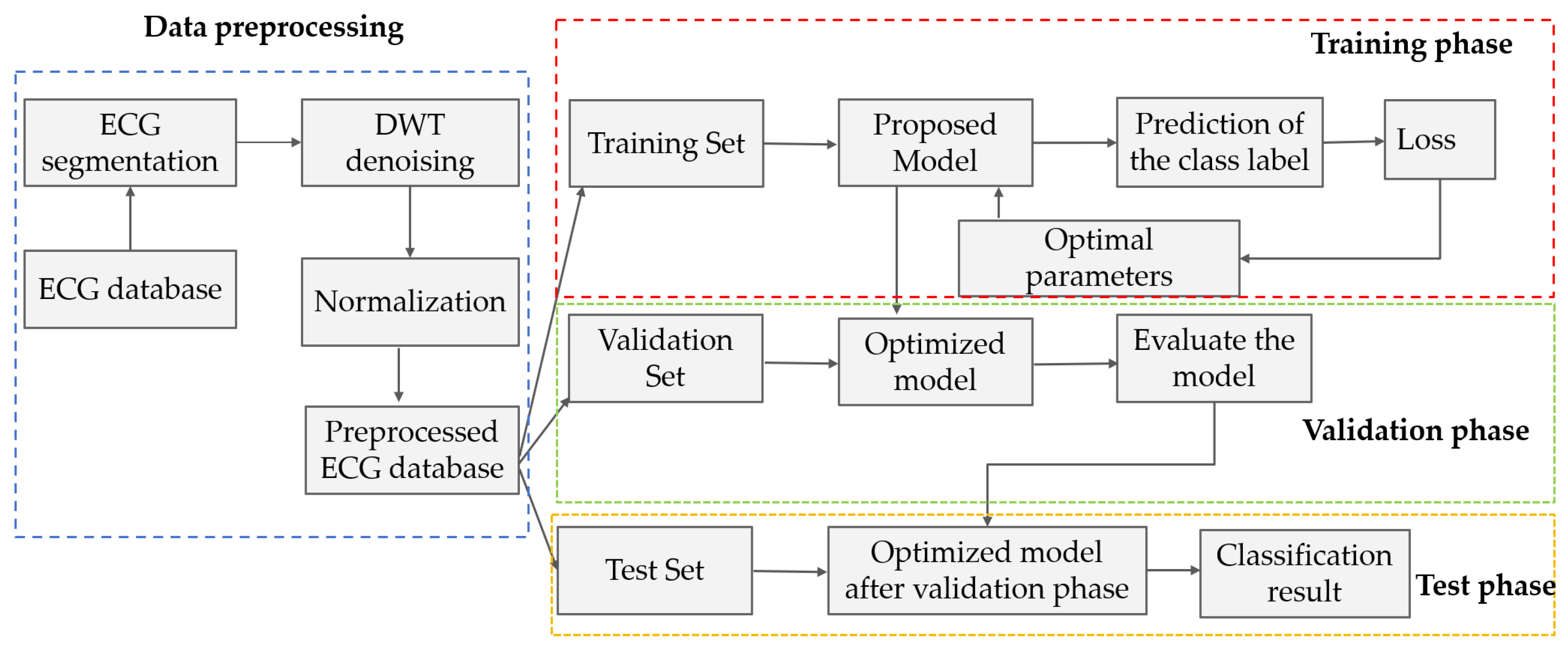
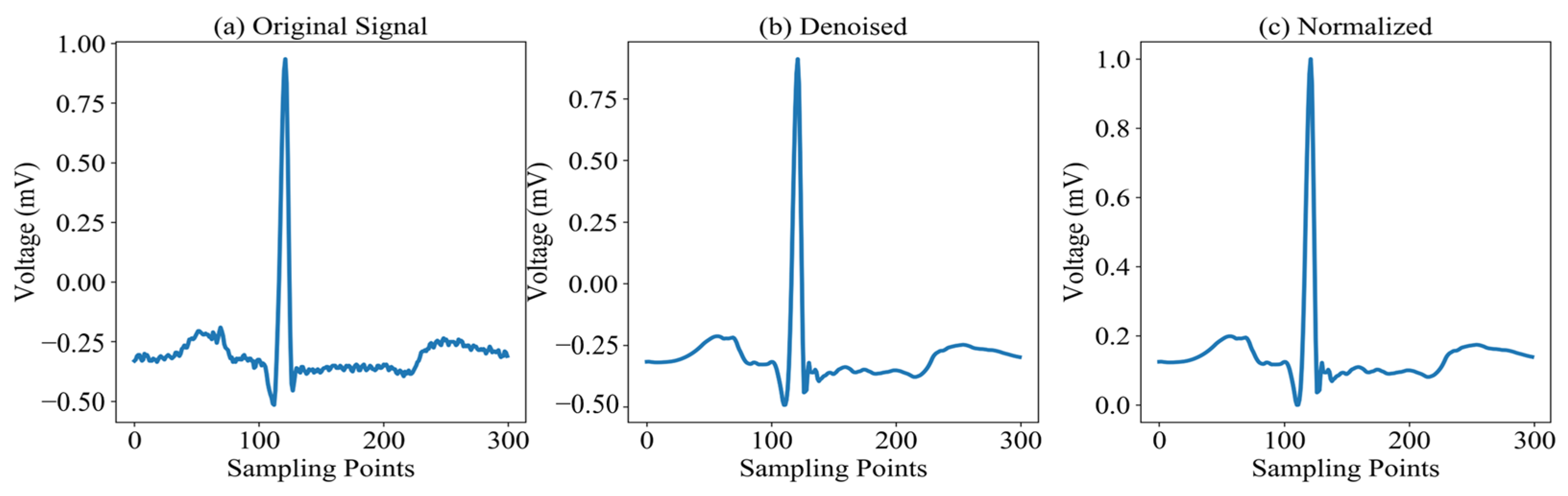
Building upon the aforementioned studies, we propose a multi-scale attentive Kolmogorov–Arnold network (MAK-Net)—a novel hybrid network model that leverages multi-scale convolution and attention mechanisms to extract rich feature information from ECG signals. The architecture consists of four main modules:
Multi-scale CNN Module: Utilizes residual convolution with various kernel sizes (e.g., 1 × 3, 1 × 11, 1 × 21, 1 × 31) to capture both local and global morphological features.
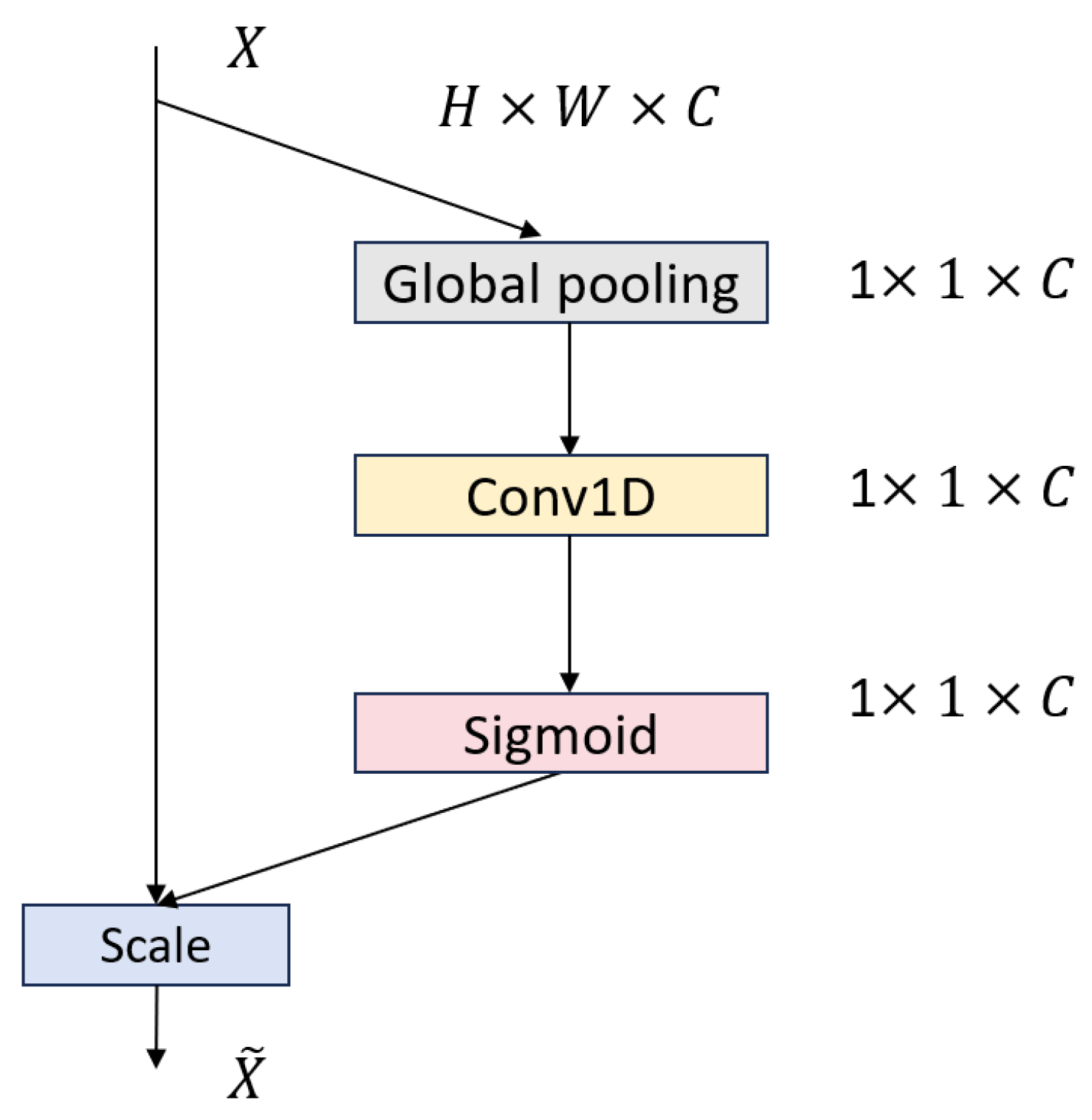
Attention Module: Integrates an enhanced MLPBlock with an Efficient Channel Attention (ECA) layer that uses adaptive 1D convolutions to generate channel-wise importance weights, balancing global context and local detail while adding minimal overhead.
BiGRU Module: Employs bidirectional Gated Recurrent Unit (GRU) layers to process ECG sequences in both forward and reverse directions, capturing long-range temporal dependencies and contextual features critical for accurate arrhythmia detection
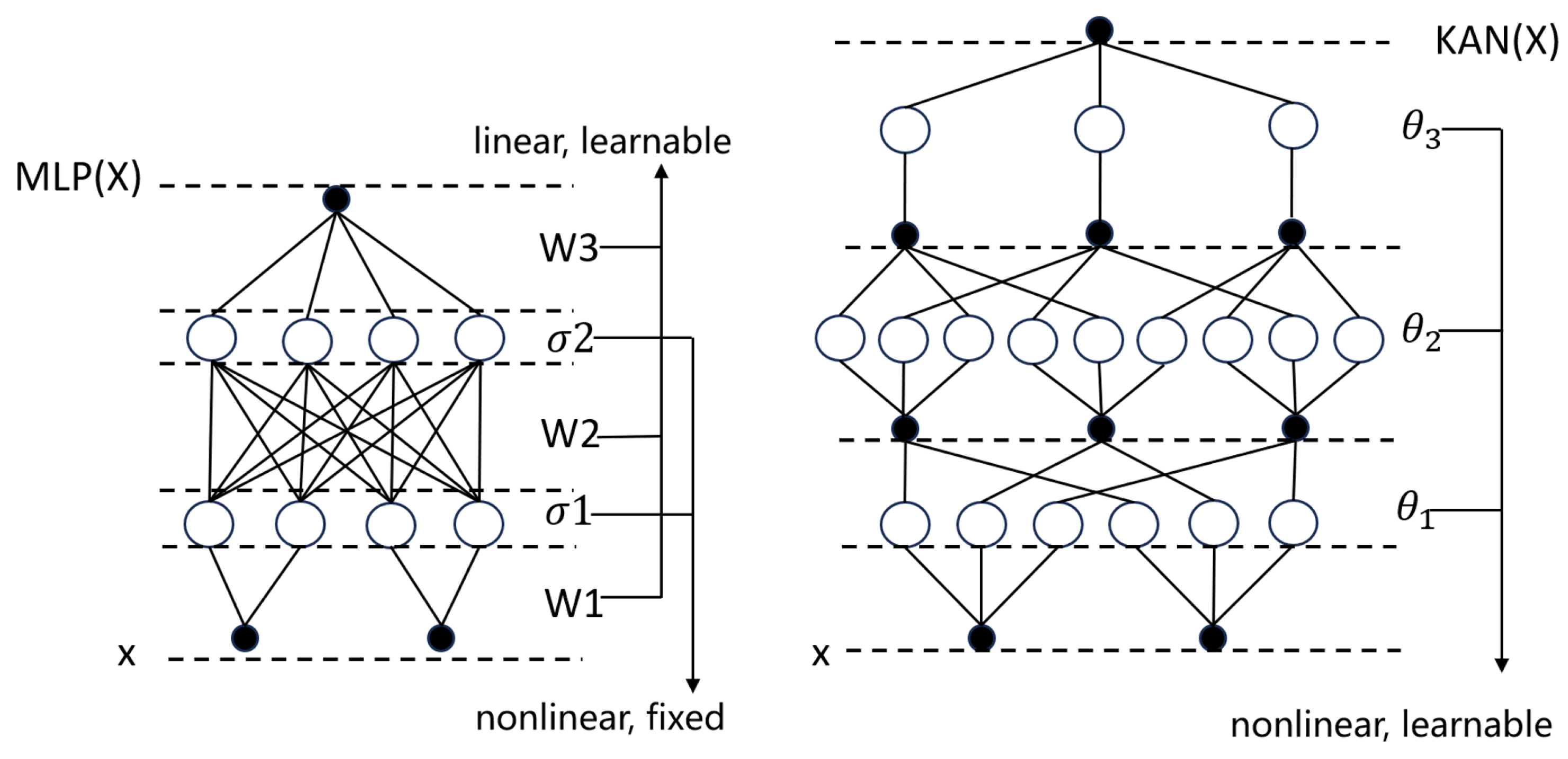
Kolmogorov–Arnold Network (KAN) Module: Replaces traditional MLPs to enhance nonlinear mapping.
Furthermore, the framework synergizes focal loss optimization with SMOTE-based oversampling, achieving dual mitigation of data imbalance.
The main contributions of this work are as follows:
Significant gains in minority-class detection: By integrating multiscale residual convolutions with dynamic attention, MAK-Net delivers state-of-the-art recall and F1-scores on imbalanced ECG datasets, overcoming the low sensitivity of existing approaches.
Parameter-efficient hybrid attention: Our attention module fuses the partial-convolution pruning of the 1D-conv MLPBlock with lightweight ECA channel weighting to capture both local waveform details and global context with minimal overhead, substantially boosting classification accuracy.
Interpretable nonlinear mapping via KAN: The KAN module replaces traditional MLP weight matrices with edge-based B-spline activation functions, yielding compact symbolic representations that enhance accuracy, interpretability, and cross-domain adaptability [
17].
Robust class-imbalance mitigation: We jointly apply SMOTE oversampling and focal loss (γ = 2.0) to balance training distributions and emphasize hard examples, resulting in markedly improved performance on under-represented arrhythmia classes.
The structure of this paper is organized as follows:
Section 2 provides a comprehensive overview of the MAK-Net architecture, details the preprocessing steps applied to the MIT-BIH dataset, and outlines the evaluation protocols employed.
Section 3 presents a comparative analysis of our results against state-of-the-art methods. Finally,
Section 4 discusses the clinical implications of our findings and offers concluding remarks.
4. Discussion
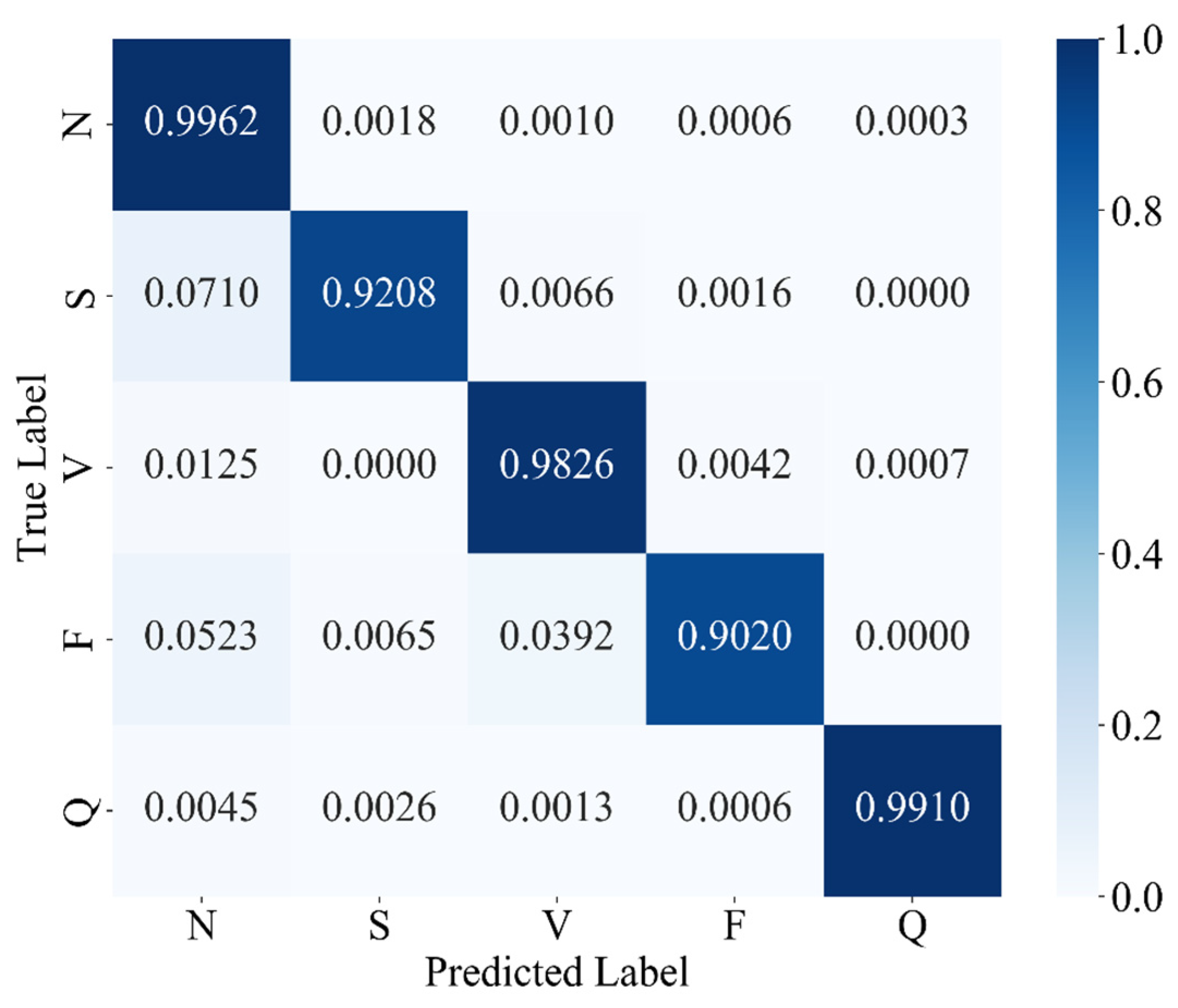
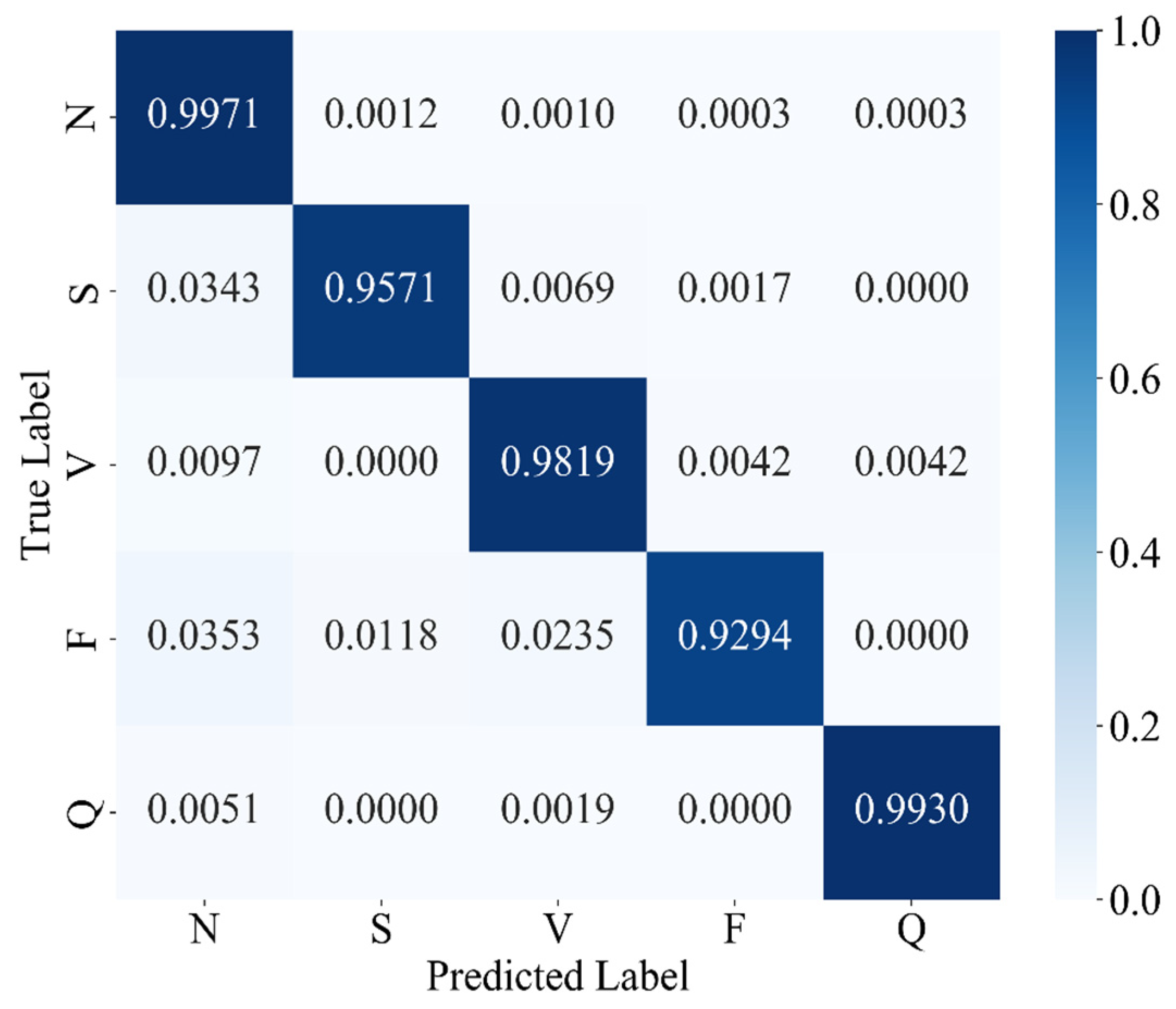
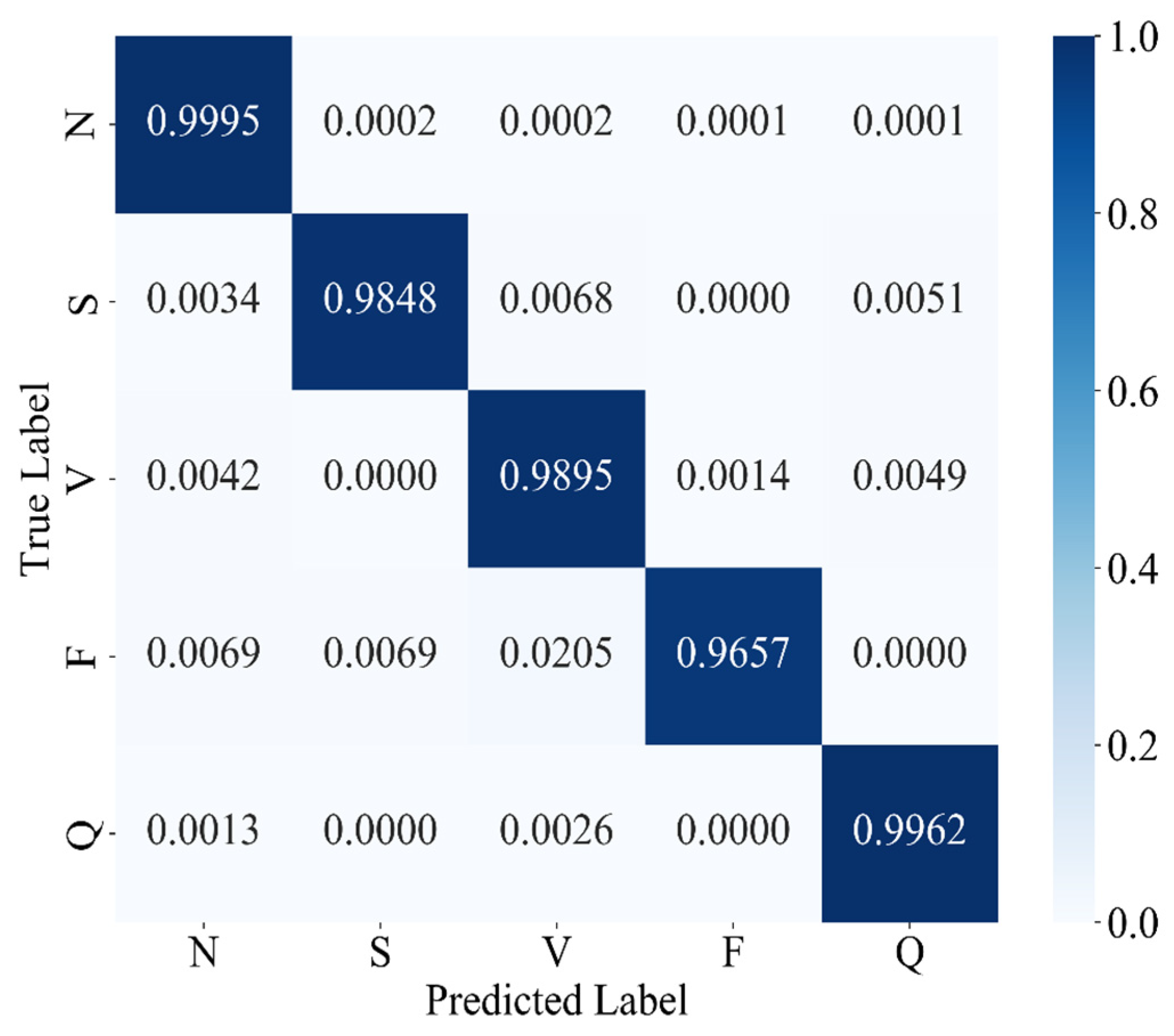
The experimental results confirm that MAK-Net delivers state-of-the-art ECG classification performance (accuracy 0.9980; F1-score 0.9888; specificity 0.9991), validating our design choices. The multiscale convolutional streams capture both fine-grained morphological features (e.g., QRS complexes) and broader rhythm patterns, while the ECA attention mechanism dynamically reweights diagnostically salient segments—analogous to a clinician’s focus on ST deviations and P-wave abnormalities [
39]. The BiGRU layers encode bidirectional temporal dependencies, essential for detecting arrhythmias that evolve over extended intervals [
50]. Furthermore, the KAN module’s learnable spline activations replace fixed nonlinearities, enabling post-hoc symbolic analysis of feature interactions and greatly enhancing interpretability—a critical requirement for clinical adoption [
51].
Our ablation and comparative studies also highlight the pivotal role of data-level balancing. SMOTE oversampling outperforms focal-loss reweighting alone, boosting minority-class recall and F1-scores by up to 3.8% over baseline models—consistent with prior findings on class-imbalance remedies in ECG tasks [
52]. This underscores that, even with advanced loss functions, equitable class representation in the training set remains indispensable for robust arrhythmia detection.
Clinically, MAK-Net’s near-perfect specificity (≥99.7%) minimizes false alarms for life-threatening conditions such as ventricular fibrillation, while its high recall (≥98.4% for rare beats) ensures reliable detection of critical but infrequent events like supraventricular ectopics. Moreover, the model’s interpretable attention maps and spline-based activations can help clinicians verify that the network’s focus aligns with established diagnostic markers, thereby fostering trust.
Nonetheless, there are still several challenges in further applying the MAK-Net in real clinical use. Generalizability to real-world ECG recordings—often corrupted by motion artifacts, electrode misplacement, and variable sampling rates—must be assessed on larger, more heterogeneous datasets [
52]. The hybrid architecture’s computational complexity may also hinder deployment on wearable or edge-computing platforms, motivating future work on model compression, pruning, and quantization to retain performance while reducing inference time [
53]. More specifically, we have measured MAK-Net’s computational footprint at 6.11 million parameters and an end-to-end inference time of 1.52 s for a batch of sixty-four ECG segments on a cloud server equipped with an NVIDIA RTX 3090 GPU and 14 vCPU Intel
® Xeon
® Platinum 8362 CPUs, corresponding to approximately 24 ms per segment. Although this complexity and latency preclude ultra-low-latency continuous monitoring, at this stage, MAK-Net is well suited for offline or semi-real-time applications requiring high diagnostic precision—such as detailed analysis of rare channelopathies (e.g., Brugada syndrome) or refined myocardial ischemia assessments. Finally, demographic biases in training data (e.g., underrepresentation of certain age or ethnic groups) necessitate rigorous fairness evaluations and bias-mitigation strategies before clinical rollout.
Looking forward, the modular nature of MAK-Net invites adaptation to other biosignal domains—such as EEG seizure detection or respiratory monitoring—where multiscale spatiotemporal modeling is equally vital. Incorporating causal inference frameworks could further refine arrhythmia precursor detection, transitioning from correlation-based classification to actionable diagnostic insights. To ensure comparability with the majority of existing studies using the MIT-BIH dataset, we adopted an intra-patient evaluation protocol, which is widely regarded as the standard approach given the dataset’s limited cohort size [
54,
55]. We acknowledge that inter-patient validation offers a more stringent test of generalizability, and we consider it a valuable direction for future work. In terms of the interpretability of MAK-Net, we have not included explicit visualizations or symbolic analyses of the learned spline activations; our interpretability claims are grounded in the architectural design of KANs, which employ learnable spline-based activation functions on edges, facilitating transparent and adaptable functional mappings [
28,
29,
30]. Thus, we intend to analyze the learned spline functions to extract symbolic representations, which could offer deeper insights into the model’s decision-making process and enhance its clinical utility.
We plan to extend MAK-Net’s validation to larger, publicly available ECG repositories—such as the PTB-XL 12-lead dataset and the CPSC 2018 challenge dataset—to robustly assess cross-center generalizability. In addition, we also plan to evaluate MAK-Net’s robustness using more diverse and noise-prone datasets, such as the MIT-BIH Noise Stress Test Database, which includes recordings with baseline wander, muscle artifacts, and electrode motion artifacts, and the Brno University of Technology ECG Quality Database (BUTQDB), which comprises long-term ECG recordings collected under free-living conditions with varying signal qualities. These evaluations will help assess and enhance the model’s performance in more realistic scenarios. Additionally, in collaboration with cardiologists at the First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China, we are curating a Brugada syndrome cohort, annotated for Type-1 and Type-2 ECG patterns, to evaluate MAK-Net’s sensitivity and specificity on this rare but clinically critical channelopathy. In sum, MAK-Net exemplifies how architectural innovation coupled with data-centric optimization can yield interpretable, reliable AI tools that augment, rather than replace, physician expertise in cardiovascular diagnostics.
5. Conclusions
This study introduces MAK-Net, a hybrid deep learning framework that combines multiscale convolutional streams, efficient channel attention, bidirectional GRUs, and a Kolmogorov–Arnold Network layer to deliver state-of-the-art ECG arrhythmia classification—achieving 0.9980 accuracy, a 0.9888 F1-score, 0.9871 recall, 0.9905 precision, and 0.9991 specificity on the MIT-BIH database. By integrating SMOTE-based oversampling with focal-loss optimization, MAK-Net effectively mitigates class imbalance, with SMOTE proving most impactful for minority-class detection. The model’s attention maps and spline-based activations enhance interpretability, aligning network focus with clinical markers, while its lightweight hybrid design suggests feasibility for real-time deployment. Nevertheless, further validation on heterogeneous, noisy ECG recordings and efforts in model compression and fairness assessment are essential before clinical translation. Future work will explore adapting MAK-Net’s modular architecture to other biosignals (e.g., EEG) and incorporating causal inference to advance from correlation to actionable diagnostic insights.