Deep Learning Models for Multi-Part Morphological Segmentation and Evaluation of Live Unstained Human Sperm
Abstract
1. Introduction
2. Related Work
3. Methodology
3.1. Dataset
3.2. Pre-Processing
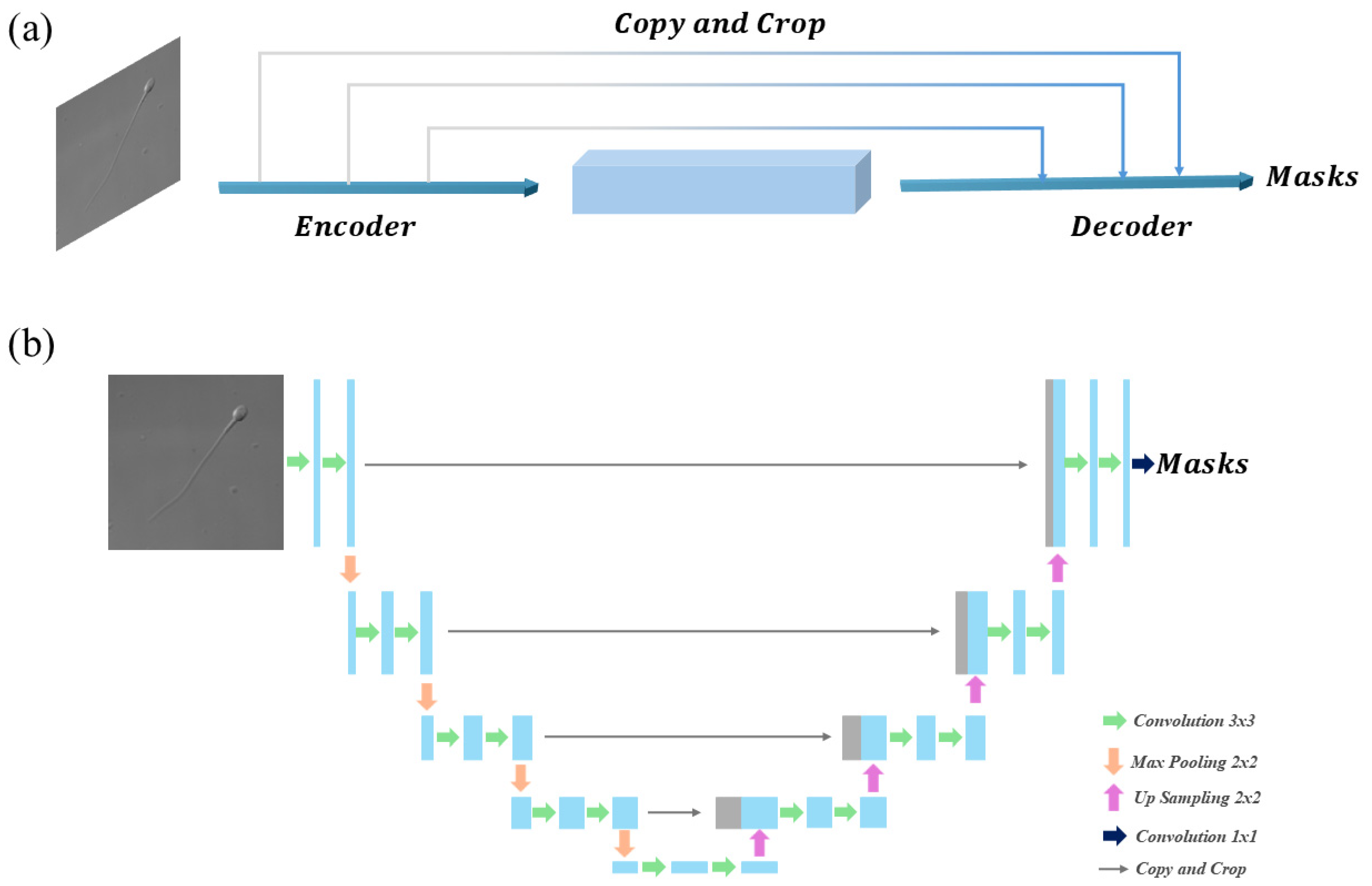
3.3. Overview of the Four Methods
3.4. Evaluation Metrics
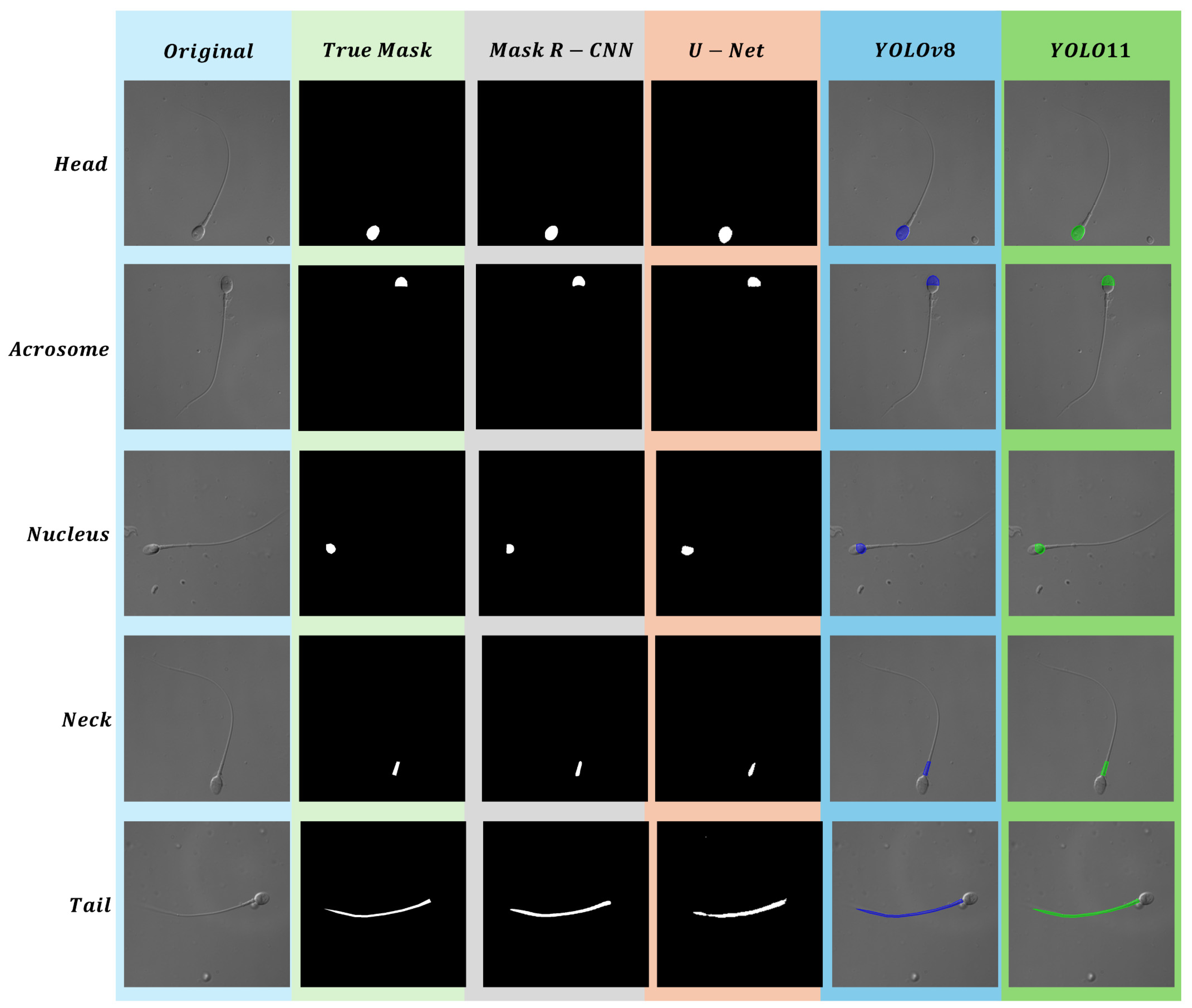
4. Results
4.1. Head
4.2. Acrosome
4.3. Nucleus
4.4. Neck
4.5. Tail
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Agarwal, A.; Mulgund, A.; Hamada, A.; Chyatte, M.R. A unique view on male infertility around the globe. Reprod. Biol. Endocrinol. 2015, 13, 37. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sauer, M.V. In vitro fertilization (IVF): A review of 3 decades of clinical innovation and technological advancement. Ther. Clin. Risk Manag. 2006, 2, 355–364. [Google Scholar] [CrossRef]
- Henkel, R. Sperm preparation: State-of-the-art—Physiological aspects and application of advanced sperm preparation methods. Asian J. Androl. 2012, 14, 260–269. [Google Scholar] [CrossRef]
- Barroso, G.; Mercan, R.; Ozgur, K.; Morshedi, M.; Kolm, P.; Coetzee, K.; Kruger, T.; Oehninger, S. Intra- and inter-laboratory variability in the assessment of sperm morphology by strict criteria: Impact of semen preparation, staining techniques and manual versus computerized analysis. Hum. Reprod. 1999, 14, 2036–2040. [Google Scholar] [CrossRef]
- Cherouveim, P.; Velmahos, C.; Bormann, C.L. Artificial intelligence for sperm selection—A systematic review. Fertil. Steril. 2023, 120, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Inge, G.B.; Brinsden, P.R.; Elder, K.T. Oocyte number per live birth in IVF: Were Steptoe and Edwards less wasteful? Hum. Reprod. 2005, 20, 588–592. [Google Scholar] [CrossRef] [PubMed]
- Rienzi, L.; Bariani, F.; Dalla Zorza, M.; Romano, S.; Scarica, C.; Maggiulli, R.; Costa, A.N.; Ubaldi, F.M. Failure mode and effects analysis of witnessing protocols for ensuring traceability during IVF. Reprod. Biomed. Online 2015, 31, 516–522. [Google Scholar] [CrossRef]
- Bodri, D.; Guillén, J.J.; Polo, A.; Trullenque, M.; Esteve, C.; Coll, O. Complications related to ovarian stimulation and oocyte retrieval in 4052 oocyte donor cycles. Reprod. Biomed. Online 2008, 17, 237–243. [Google Scholar] [CrossRef]
- Rose, B.I. Approaches to oocyte retrieval for advanced reproductive technology cycles planning to utilize in vitro maturation: A review of the many choices to be made. J. Assist. Reprod. Genet. 2014, 31, 1409–1419. [Google Scholar] [CrossRef]
- Castillo, J.; Amaral, A.; Oliva, R. Sperm nuclear proteome and its epigenetic potential. Andrology 2014, 2, 326–338. [Google Scholar] [CrossRef]
- Hirohashi, N.; Yanagimachi, R. Sperm acrosome reaction: Its site and role in fertilization†. Biol. Reprod. 2018, 99, 127–133. [Google Scholar] [CrossRef]
- Klingner, A.; Kovalenko, A.; Magdanz, V.; Khalil, I.S.M. Exploring sperm cell motion dynamics: Insights from genetic algorithm-based analysis. Comput. Struct. Biotechnol. J. 2024, 23, 2837–2850. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Laboratory Manual for the Examination and Processing of Human Semen; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Zhang, C.; Zhang, Y.; Chang, Z.; Li, C. Sperm YOLOv8E-TrackEVD: A Novel Approach for Sperm Detection and Tracking. Sensors 2024, 24, 3493. [Google Scholar] [CrossRef]
- Lu, J.C.; Huang, Y.F.; Lü, N.Q. Computer-aided sperm analysis: Past, present and future. Andrologia 2014, 46, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, S.T.; van der Horst, G.; Mortimer, D. The future of computer-aided sperm analysis. Asian J. Androl. 2015, 17, 545. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Zhang, Z.; Shan, G.; Chu, L.-T.; Huang, Z.; Moskovtsev, S.; Librach, C.; Jarvi, K.; Sun, Y. Advances in sperm analysis: Techniques, discoveries and applications. Nat. Rev. Urol. 2021, 18, 447–467. [Google Scholar] [CrossRef]
- Tomlinson, M.J.; Pooley, K.; Simpson, T.; Newton, T.; Hopkisson, J.; Jayaprakasan, K.; Jayaprakasan, R.; Naeem, A.; Pridmore, T. Validation of a novel computer-assisted sperm analysis (CASA) system using multitarget-tracking algorithms. Fertil. Steril. 2010, 93, 1911–1920. [Google Scholar] [CrossRef]
- Amann, R.P.; Waberski, D. Computer-assisted sperm analysis (CASA): Capabilities and potential developments. Theriogenology 2014, 81, 5–17.e3. [Google Scholar] [CrossRef]
- Gallagher, M.T.; Smith, D.J.; Kirkman-Brown, J.C. CASA: Tracking the past and plotting the future. Reprod. Fertil. Dev. 2018, 30, 867–874. [Google Scholar] [CrossRef]
- Pelzman, D.L.; Sandlow, J.I. Sperm morphology: Evaluating its clinical relevance in contemporary fertility practice. Reprod. Med. Biol. 2024, 23, e12594. [Google Scholar] [CrossRef]
- Menkveld, R.; El-Garem, Y.; Schill, W.-B.; Henkel, R. Relationship Between Human Sperm Morphology and Acrosomal Function. J. Assist. Reprod. Genet. 2003, 20, 432–438. [Google Scholar] [CrossRef]
- Menkveld, R.; Holleboom, C.A.; Rhemrev, J.P. Measurement and significance of sperm morphology. Asian J. Androl. 2011, 13, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Maalej, R.; Abdelkefi, O.; Daoud, S. Advancements in automated sperm morphology analysis: A deep learning approach with comprehensive classification and model evaluation. Multimed. Tools Appl. 2024, 1–34. [Google Scholar] [CrossRef]
- Hao, M.; Zhai, R.; Wang, Y.; Ru, C.; Yang, B. A Stained-Free Sperm Morphology Measurement Method Based on Multi-Target Instance Parsing and Measurement Accuracy Enhancement. Sensors 2025, 25, 592. [Google Scholar] [CrossRef]
- Lewandowska, E.; Węsierski, D.; Mazur-Milecka, M.; Liss, J.; Jezierska, A. Ensembling noisy segmentation masks of blurred sperm images. Comput. Biol. Med. 2023, 166, 107520. [Google Scholar] [CrossRef]
- Nissen, M.S.; Krause, O.; Almstrup, K.; Kjærulff, S.; Nielsen, T.T.; Nielsen, M. Convolutional Neural Networks for Segmentation and Object Detection of Human Semen. In Image Analysis; Sharma, P., Bianchi, F.M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 397–406. [Google Scholar] [CrossRef]
- Chaiya, J.; Vinayanuvattikhun, N.; Tanprasertkul, C.; Chaidarun, T.; Mebuathong, T.; Kaset, C. Effect of staining methods on human sperm morphometrics using HT CASA II. J. Gynecol. Obstet. Hum. Reprod. 2022, 51, 102322. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, N.; Zhang, Y.; Li, S.; Liang, P.; Zhao, Z.; Zhang, J.; Zha, X.; Zhou, Y.; Cao, Y.; Chen, D.Z. Shmc-Net: A Mask-Guided Feature Fusion Network for Sperm Head Morphology Classification. In Proceedings of the 2024 IEEE International Symposium on Biomedical Imaging (ISBI), Athens, Greece, 27–30 May 2024; pp. 1–5. [Google Scholar] [CrossRef]
- Lagares, M.d.A.; da Silva, G.C.; Cortes, S.F.; Moreira, F.H.M.; Neves, F.C.D.; Alves, N.d.C.; Viegas, R.N.; Diniz, T.F.; Lemos, V.S.; Rezende, A.S.C.; et al. L-carnitine added to post-thawed semen acts as an antioxidant and a stimulator of equine sperm metabolism. Andrologia 2022, 54, e14338. [Google Scholar] [CrossRef] [PubMed]
- Marín, R.; Chang, V. Impact of transfer learning for human sperm segmentation using deep learning. Comput. Biol. Med. 2021, 136, 104687. [Google Scholar] [CrossRef]
- Shaker, F.; Monadjemi, S.A.; Naghsh-Nilchi, A.R. Automatic detection and segmentation of sperm head, acrosome and nucleus in microscopic images of human semen smears. Comput. Methods Programs Biomed. 2016, 132, 11–20. [Google Scholar] [CrossRef]
- Movahed, R.A.; Mohammadi, E.; Orooji, M. Automatic segmentation of Sperm’s parts in microscopic images of human semen smears using concatenated learning approaches. Comput. Biol. Med. 2019, 109, 242–253. [Google Scholar] [CrossRef]
- Chen, W.; Song, H.; Dai, C.; Huang, Z.; Wu, A.; Shan, G.; Liu, H.; Jiang, A.; Liu, X.; Ru, C.; et al. CP-Net: Instance-aware part segmentation network for biological cell parsing. Med. Image Anal. 2024, 97, 103243. [Google Scholar] [CrossRef]
- Chang, V.; Saavedra, J.M.; Castañeda, V.; Sarabia, L.; Hitschfeld, N.; Härtel, S. Gold-standard and improved framework for sperm head segmentation. Comput. Methods Programs Biomed. 2014, 117, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Thambawita, V.; Hicks, S.A.; Storås, A.M.; Nguyen, T.; Andersen, J.M.; Witczak, O.; Haugen, T.B.; Hammer, H.L.; Halvorsen, P. VISEM-Tracking, a human spermatozoa tracking dataset. Sci. Data 2023, 10, 260. [Google Scholar] [CrossRef] [PubMed]
- Ghasemian, F.; Mirroshandel, S.A.; Monji-Azad, S.; Azarnia, M.; Zahiri, Z. An efficient method for automatic morphological abnormality detection from human sperm images. Comput. Methods Programs Biomed. 2015, 122, 409–420. [Google Scholar] [CrossRef]
- Zhang, Y. Animal sperm morphology analysis system based on computer vision. In Proceedings of the International Conference on Intelligent Control and Information Processing (ICICIP), Hangzhou, China, 3–5 November 2017; IEEE: New York, NY, USA, 2017; pp. 338–341. [Google Scholar] [CrossRef]
- Prasetyo, E.; Suciati, N.; Fatichah, C. A Comparison of YOLO and Mask R-CNN for Segmenting Head and Tail of Fish. In Proceedings of the 2020 4th International Conference on Informatics and Computational Sciences (ICICoS), Semarang, Indonesia, 10–11 November 2020; IEEE: New York, NY, USA, 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Lv, Q.; Yuan, X.; Qian, J.; Li, X.; Zhang, H.; Zhan, S. An Improved U-Net for Human Sperm Head Segmentation. Neural Process. Lett. 2022, 54, 537–557. [Google Scholar] [CrossRef]
- Chen, A.; Li, C.; Zou, S.; Rahaman, M.M.; Yao, Y.; Chen, H.; Yang, H.; Zhao, P.; Hu, W.; Liu, W.; et al. SVIA dataset: A new dataset of microscopic videos and images for computer-aided sperm analysis. Biocybern. Biomed. Eng. 2022, 42, 204–214. [Google Scholar] [CrossRef]
- Fraczek, A.; Karwowska, G.; Miler, M.; Lis, J.; Jezierska, A.; Mazur-Milecka, M. Sperm segmentation and abnormalities detection during the ICSI procedure using machine learning algorithms. In Proceedings of the 2022 15th International Conference on Human System Interaction (HSI), Melbourne, Australia, 29–31 July 2022; IEEE: New York, NY, USA, 2022; pp. 1–6. [Google Scholar] [CrossRef]
- Suleman, M.; Ilyas, M.; Lali, M.I.U.; Rauf, H.T.; Kadry, S. A review of different deep learning techniques for sperm fertility prediction. AIMS Math. 2023, 8, 16360–16416. [Google Scholar] [CrossRef]
- Yang, H.; Ma, M.; Chen, X.; Chen, G.; Shen, Y.; Zhao, L.; Wang, J.; Yan, F.; Huang, D.; Gao, H.; et al. Multidimensional morphological analysis of live sperm based on multiple-target tracking. Comput. Struct. Biotechnol. J. 2024, 24, 176–184. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where to Look for the Pancreas. arXiv 2018. [Google Scholar] [CrossRef]
- Chincholkar, V.; Srivastava, S.; Pawanarkar, A.; Chaudhari, S. Deep Learning Techniques in Liver Segmentation: Evaluating U-Net, Attention U-Net, ResNet50, and ResUNet Models. In Proceedings of the 2024 14th International Conference on Cloud Computing, Data Science & Engineering (Confluence), Noida, India, 18–19 January 2024; pp. 775–779. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021. [Google Scholar] [CrossRef]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-Like Pure Transformer for Medical Image Segmentation. In Computer Vision—Eccv 2022 Workshops; Karlinsky, L., Michaeli, T., Nishino, K., Eds.; Springer Nature: Cham, Switzerland, 2023; pp. 205–218. [Google Scholar] [CrossRef]
- Shahali, S.; Murshed, M.; Spencer, L.; Tunc, O.; Pisarevski, L.; Conceicao, J.; McLachlan, R.; O’bryan, M.K.; Ackermann, K.; Zander-Fox, D.; et al. Morphology Classification of Live Unstained Human Sperm Using Ensemble Deep Learning. Adv. Intell. Syst. 2024, 6, 2400141. [Google Scholar] [CrossRef]
- Clinically Labelled Live, Unstained Human Sperm Dataset 2024. Available online: https://bridges.monash.edu/articles/dataset/Clinically_labelled_live_unstained_human_sperm_dataset/25621500/1 (accessed on 23 May 2024).
- He, K.; Gkioxari, G.; Dollar, P.; Girshick, R. Mask R-CNN. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Lin, T.-Y.; Dollar, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature Pyramid Networks for Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Bouslama, A.; Laaziz, Y.; Tali, A. Diagnosis and precise localization of cardiomegaly disease using U-NET. Inform. Med. Unlocked 2020, 19, 100306. [Google Scholar] [CrossRef]
- Vijayakumar, A.; Vairavasundaram, S. YOLO-based Object Detection Models: A Review and its Applications. Multimed. Tools Appl. 2024, 83, 83535–83574. [Google Scholar] [CrossRef]
- Ali, M.L.; Zhang, Z. The YOLO Framework: A Comprehensive Review of Evolution, Applications, and Benchmarks in Object Detection. Computers 2024, 13, 336. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Liao, H.-Y.M.; Wu, Y.-H.; Chen, P.-Y.; Hsieh, J.-W.; Yeh, I.-H. CSPNet: A New Backbone That Can Enhance Learning Capability of CNN. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Seattle, WA, USA, 14–19 June 2020; pp. 390–391. [Google Scholar]
- Brar, K.K.; Goyal, B.; Dogra, A.; Mustafa, M.A.; Majumdar, R.; Alkhayyat, A.; Kukreja, V. Image segmentation review: Theoretical background and recent advances. Inf. Fusion 2025, 114, 102608. [Google Scholar] [CrossRef]
- Ali, S.; Ghatwary, N.; Jha, D.; Isik-Polat, E.; Polat, G.; Yang, C.; Li, W.; Galdran, A.; Ballester, M.G.; Thambawita, V.; et al. Assessing generalisability of deep learning-based polyp detection and segmentation methods through a computer vision challenge. Sci. Rep. 2024, 14, 2032. [Google Scholar] [CrossRef]
- Müller, D.; Soto-Rey, I.; Kramer, F. Towards a guideline for evaluation metrics in medical image segmentation. BMC Res. Notes 2022, 15, 210. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, E.; Zhu, Y. Image segmentation evaluation: A survey of methods. Artif. Intell. Rev. 2020, 53, 5637–5674. [Google Scholar] [CrossRef]
- Bharati, P.; Pramanik, A. Deep Learning Techniques—R-CNN to Mask R-CNN: A Survey. In Computational Intelligence in Pattern Recognition; Das, A.K., Nayak, J., Naik, B., Pati, S.K., Pelusi, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2020; pp. 657–668. [Google Scholar] [CrossRef]
- Azad, R.; Aghdam, E.K.; Rauland, A.; Jia, Y.; Avval, A.H.; Bozorgpour, A.; Karimijafarbigloo, S.; Cohen, J.P.; Adeli, E.; Merhof, D. Medical Image Segmentation Review: The Success of U-Net. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 10076–10095. [Google Scholar] [CrossRef]
- Williams, C.; Falck, F.; Deligiannidis, G.; Holmes, C.C.; Doucet, A.; Syed, S. A Unified Framework for U-Net Design and Analysis. Adv. Neural Inf. Process. Syst. 2023, 36, 27745–27782. [Google Scholar]
- Ghafari, M.; Mailman, D.; Hatami, P.; Peyton, T.; Yang, L.; Dang, W.; Qin, H. A Comparison of YOLO and Mask-RCNN for Detecting Cells from Microfluidic Images. In Proceedings of the 2022 International Conference on Artificial Intelligence in Information and Communication (ICAIIC), Jeju Island, Repbulic of Korea, 21–24 February 2022; pp. 204–209. [Google Scholar] [CrossRef]
- Sapkota, R.; Ahmed, D.; Karkee, M. Comparing YOLOv8 and Mask R-CNN for instance segmentation in complex orchard environments. Artif. Intell. Agric. 2024, 13, 84–99. [Google Scholar] [CrossRef]
- Wang, W.; Meng, Y.; Li, S.; Zhang, C. HV-YOLOv8 by HDPconv: Better lightweight detectors for small object detection. Image Vis. Comput. 2024, 147, 105052. [Google Scholar] [CrossRef]
- Perazzi, F.; Pont-Tuset, J.; McWilliams, B.; Van Gool, L.; Gross, M.; Sorkine-Hornung, A. A Benchmark Dataset and Evaluation Methodology for Video Object Segmentation. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; IEEE: New York, NY, USA, 2016; pp. 724–732. [Google Scholar] [CrossRef]
- Gustafson, L.; Rolland, C.; Ravi, N.; Duval, Q.; Adcock, A.; Fu, C.-Y. FACET: Fairness in Computer Vision Evaluation Benchmark. In Proceedings of the 2023 IEEE/CVF International Conference on Computer Vision (ICCV), Paris, France, 1–6 October 2023; IEEE: New York, NY, USA, 2023; pp. 20313–20325. [Google Scholar] [CrossRef]
- Wan, Z.; Wang, Z.; Chung, C.; Wang, Z. A Survey of Dataset Refinement for Problems in Computer Vision Datasets. ACM Comput. Surv. 2024, 56, 172:1–172:34. [Google Scholar] [CrossRef]
- Fabbrizzi, S.; Papadopoulos, S.; Ntoutsi, E.; Kompatsiaris, I. A survey on bias in visual datasets. Comput. Vis. Image Underst. 2022, 223, 103552. [Google Scholar] [CrossRef]
- Shaker, F.; Monadjemi, S.A.; Alirezaie, J.; Naghsh-Nilchi, A.R. A dictionary learning approach for human sperm heads classification. Comput. Biol. Med. 2017, 91, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Javadi, S.; Mirroshandel, S.A. A novel deep learning method for automatic assessment of human sperm images. Comput. Biol. Med. 2019, 109, 182–194. [Google Scholar] [CrossRef]
- Hicks, S.A.; Andersen, J.M.; Witczak, O.; Thambawita, V.; Halvorsen, P.; Hammer, H.L.; Haugen, T.B.; Riegler, M.A. Machine Learning-Based Analysis of Sperm Videos and Participant Data for Male Fertility Prediction. Sci. Rep. 2019, 9, 16770. [Google Scholar] [CrossRef]
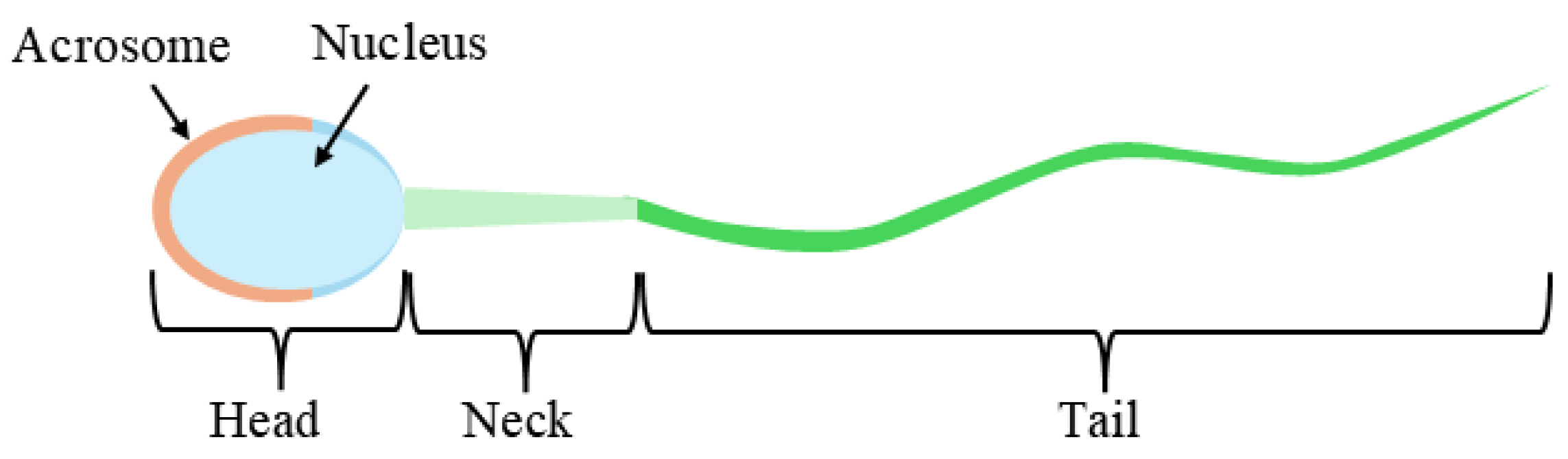


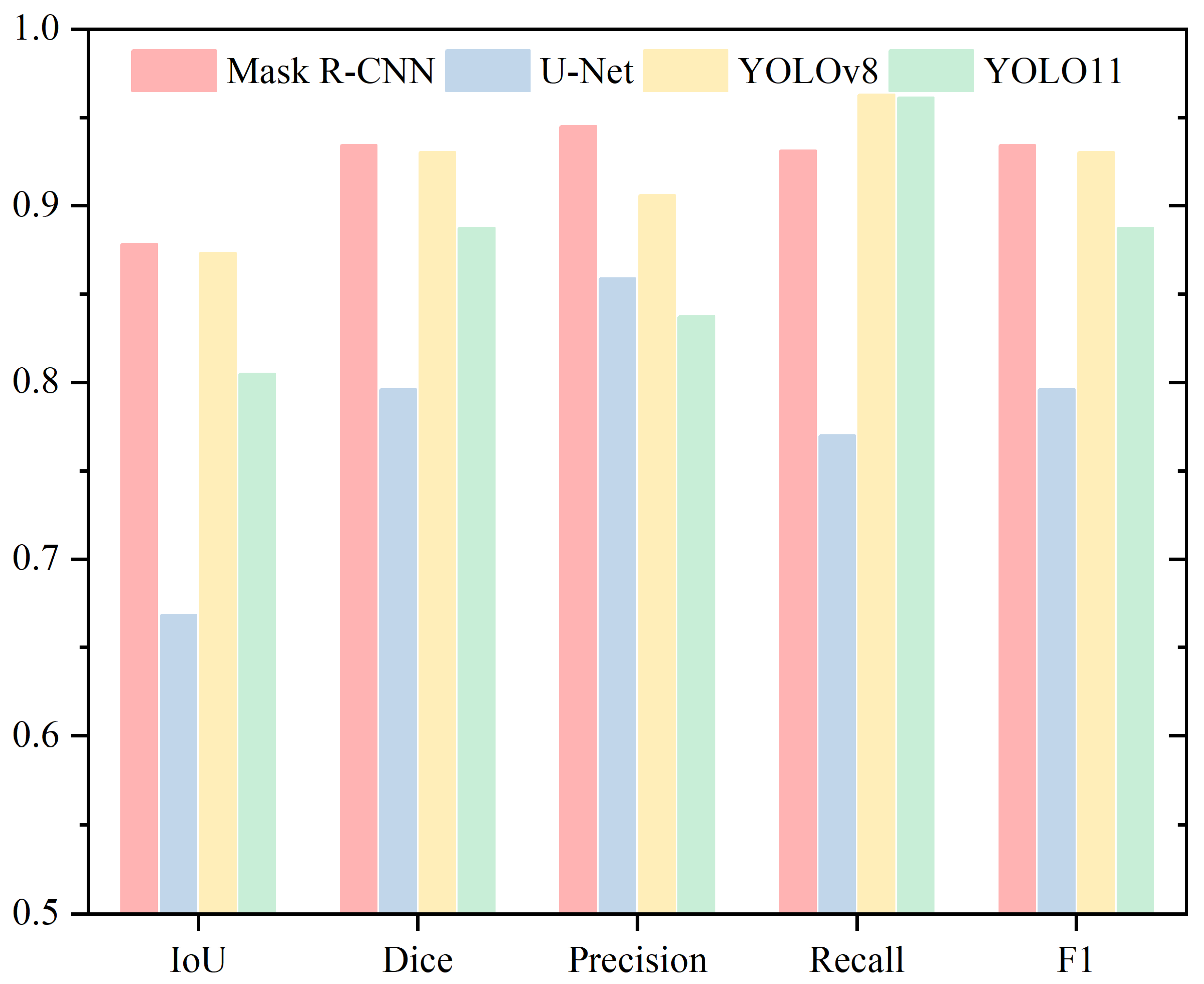


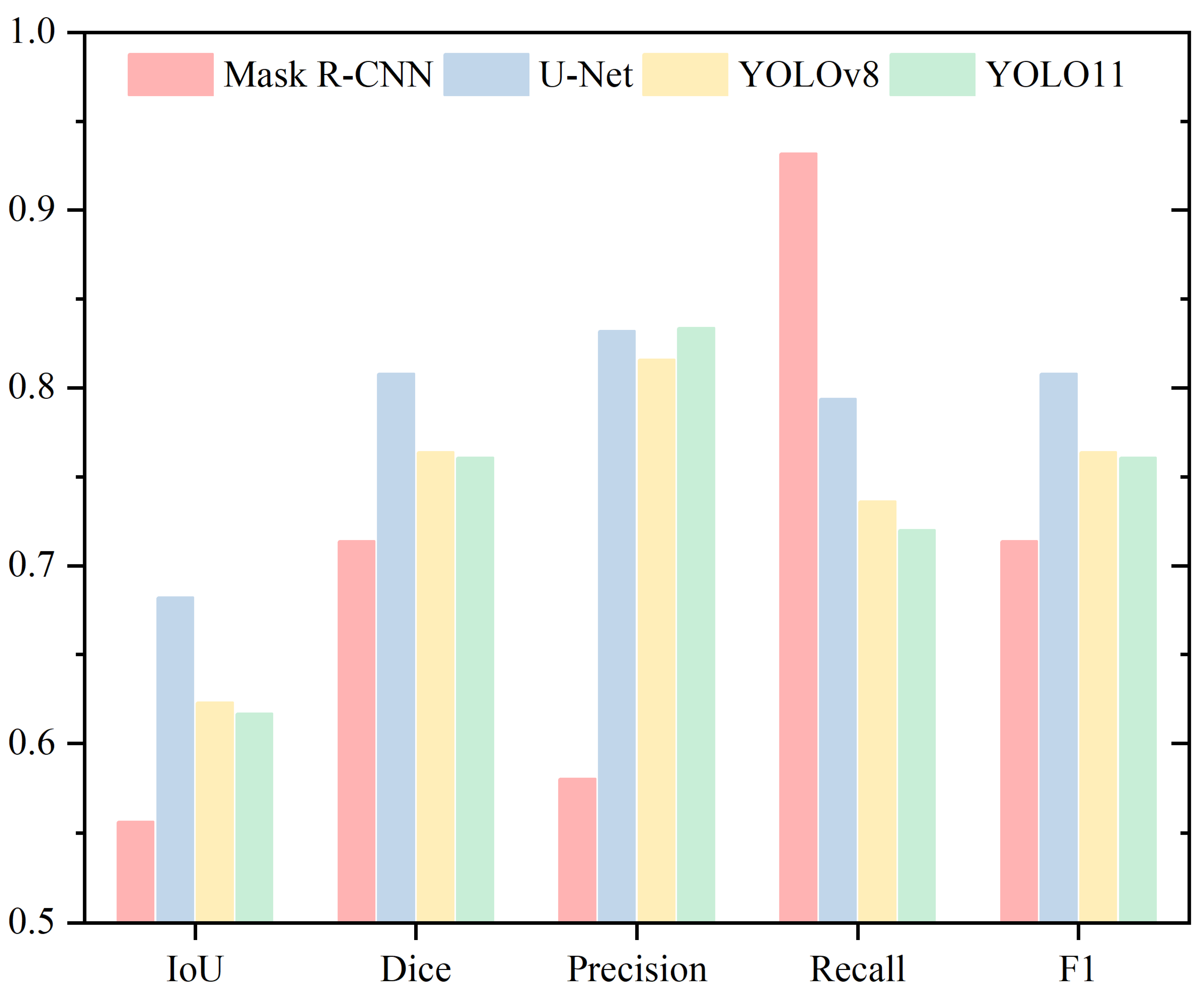
| Head | ||||
|---|---|---|---|---|
| IoU | Dice | Precision | Recall | |
| Mask R-CNN | 0.8783 | 0.9342 | 0.9451 | 0.9312 |
| U-Net | 0.6682 | 0.7959 | 0.8589 | 0.7701 |
| YOLOv8 | 0.8731 | 0.9305 | 0.9060 | 0.9628 |
| YOLO11 | 0.8049 | 0.8875 | 0.8374 | 0.9613 |
| Acrosome | ||||
|---|---|---|---|---|
| IoU | Dice | Precision | Recall | |
| Mask R-CNN | 0.7641 | 0.8648 | 0.8194 | 0.9243 |
| U-Net | 0.6920 | 0.8142 | 0.8681 | 0.7793 |
| YOLOv8 | 0.7284 | 0.8390 | 0.8056 | 0.9088 |
| YOLO11 | 0.7487 | 0.8531 | 0.8097 | 0.9213 |
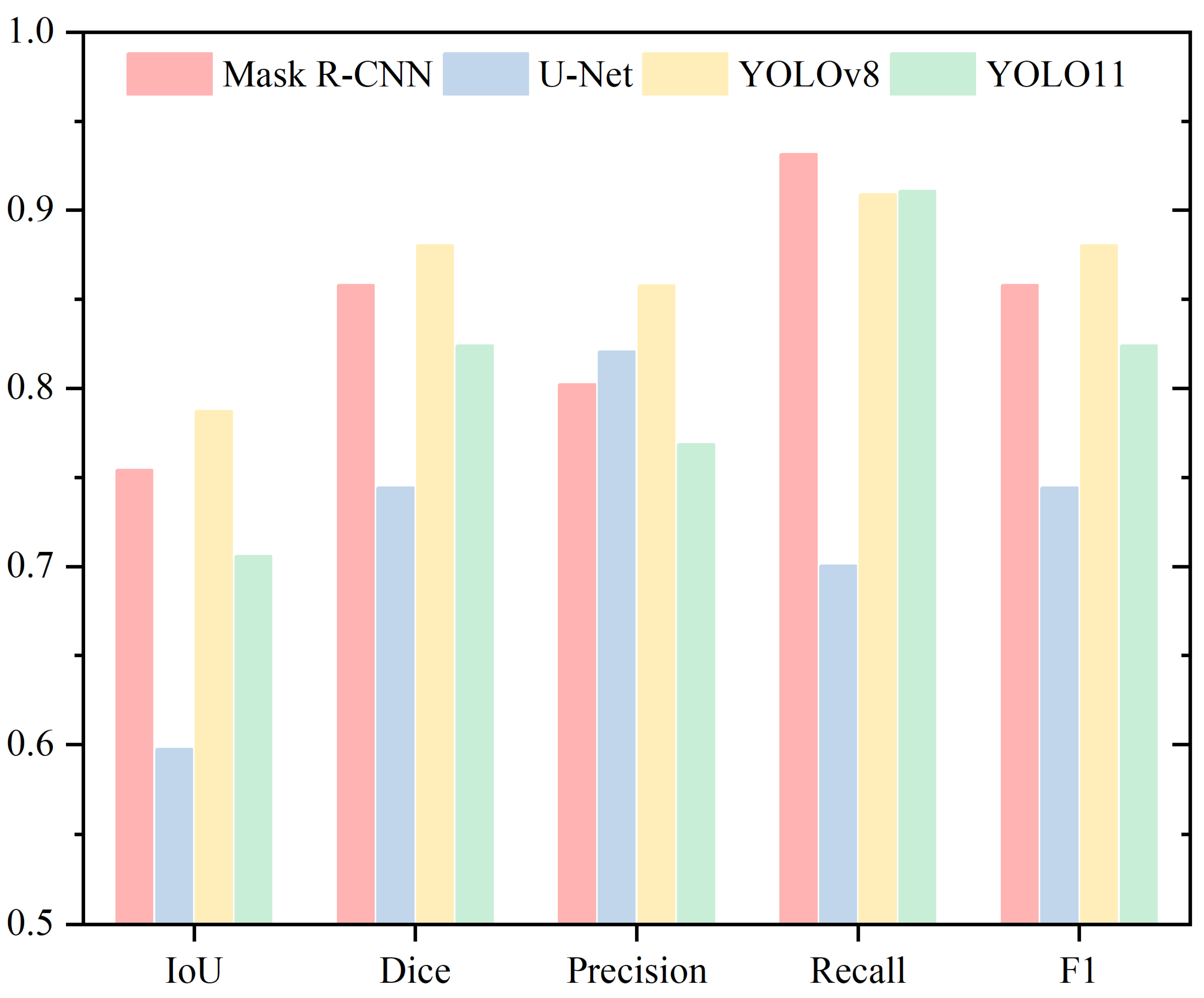
| Nucleus | ||||
|---|---|---|---|---|
| IoU | Dice | Precision | Recall | |
| Mask R-CNN | 0.7275 | 0.8408 | 0.8811 | 0.8163 |
| U-Net | 0.6026 | 0.7502 | 0.7466 | 0.7742 |
| YOLOv8 | 0.7224 | 0.8373 | 0.7875 | 0.9182 |
| YOLO11 | 0.6921 | 0.8142 | 0.7453 | 0.9251 |
| Neck | ||||
|---|---|---|---|---|
| IoU | Dice | Precision | Recall | |
| Mask R-CNN | 0.7542 | 0.8579 | 0.8023 | 0.9315 |
| U-Net | 0.5976 | 0.7443 | 0.8207 | 0.7006 |
| YOLOv8 | 0.7872 | 0.8803 | 0.8577 | 0.9088 |
| YOLO11 | 0.7058 | 0.8241 | 0.7685 | 0.9107 |
| Tail | ||||
|---|---|---|---|---|
| IoU | Dice | Precision | Recall | |
| Mask R-CNN | 0.5563 | 0.7138 | 0.5802 | 0.9318 |
| U-Net | 0.6821 | 0.8079 | 0.8319 | 0.7937 |
| YOLOv8 | 0.6229 | 0.7638 | 0.8159 | 0.7360 |
| YOLO11 | 0.6168 | 0.7606 | 0.8337 | 0.7200 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, P.; Saadat, M.; Hassani, M.G.; Shu, C. Deep Learning Models for Multi-Part Morphological Segmentation and Evaluation of Live Unstained Human Sperm. Sensors 2025, 25, 3093. https://doi.org/10.3390/s25103093
Lei P, Saadat M, Hassani MG, Shu C. Deep Learning Models for Multi-Part Morphological Segmentation and Evaluation of Live Unstained Human Sperm. Sensors. 2025; 25(10):3093. https://doi.org/10.3390/s25103093
Chicago/Turabian StyleLei, Peiran, Mozafar Saadat, Mahdieh Gol Hassani, and Chang Shu. 2025. "Deep Learning Models for Multi-Part Morphological Segmentation and Evaluation of Live Unstained Human Sperm" Sensors 25, no. 10: 3093. https://doi.org/10.3390/s25103093
APA StyleLei, P., Saadat, M., Hassani, M. G., & Shu, C. (2025). Deep Learning Models for Multi-Part Morphological Segmentation and Evaluation of Live Unstained Human Sperm. Sensors, 25(10), 3093. https://doi.org/10.3390/s25103093






