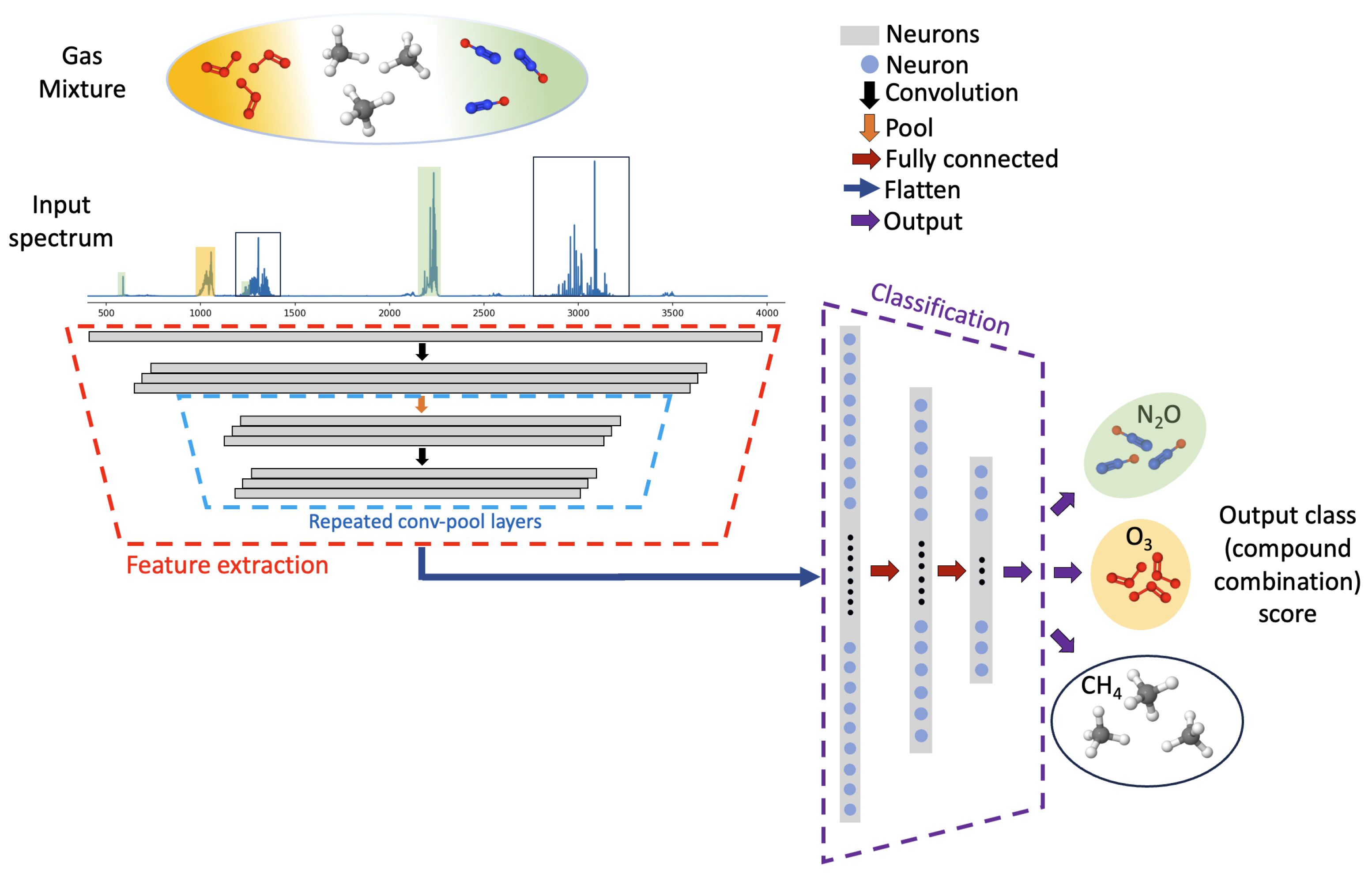
An overview of the deep learning classifier for infrared spectrum classification, developed herein, is shown in
Figure 1. Absorption spectra, containing up to three absorbers diluted in air, from ten possible species, are simulated and used for training the deep learning model. Spectral absorption is defined by the Beer–Lambert law as shown below:
where
I and
are the transmitted and incident intensities, respectively, at the frequency of interest;
is the absorbance for the mixture, which is the input to the deep neural network;
and
are the spectral absorption coefficient and concentration, respectively, of the
i-th species in the gas mixtures; and
l is the absorption pathlength.
During training, the model applies repeated convolution and pooling blocks to extract unique spectral information and downsample that information to reduce the dimensionality of the inputs to each layer of the network. The feature extraction and downsampling operations reduce the dimensionality of the problem from the original input absorbance values to a subset of extracted features. Following the feature extraction block, a flattening operation yields a single vector which enters a classification block containing three fully connected layers. The output layer of the classification block contains 175 neurons, each corresponding to a unique gas mixture or class. The model ultimately produces an integer output, providing prediction of the gas mixture, based on the input absorbance spectra.
2.1. Problem Formulation, Model Architecture, and Solution Approach
We consider the identification of components in a gas mixture by matching features in an unknown absorption spectrum to features in known reference spectra as a multi-label supervised classification problem. Hence, multiple labels are attributed to each spectra, one for each of the possible gaseous compounds present. We convert this multi-label classification problem to a multi-class classification problem using the label powerset method [
35].
For the full wavenumber (frequency) range considered, 400–4000 cm−1 (wavelength of 2.5–25 m) at 1 cm−1 resolution, the input absorbance is represented by a vector (, n is vector length). Smaller ranges were also considered: 500–2000 cm−1 (5–20 m, ); 1000–1500 cm−1 (6.67–10 m, ); 1250–1500 cm−1 (6.67–8 m, ); 1000–2000 cm−1 (5–10 m, ); and 2000–4000 cm−1 (2.5–5 m, ). Spectra for the smaller wavenumber ranges were considered at a resolution of 1 cm−1 and, hence, have smaller input absorbance vectors (length n) than the larger range. The model is flexible and allows for user-defined wavenumber ranges.
Each input absorbance vector is associated with a class-indicator integer
, where,
y is an integer from 0 to 174 representing a unique mixture composition. Mixtures containing one (pure), two, and three components are considered in this study. A deep neural network classification model is developed to obtain an approximate learned hypothesis function,
h, relating the spectrum to its class-indicator integer:
In this multi-class classification problem, the last layer of the deep neural network is activated using the softmax function
where
represents the raw score or logit for a specific class (unique gas mixture),
i. It is also the input to the softmax layer in the neural network (last layer). The numerator is the exponential of the raw score for class
i. The denominator is the sum of the exponentials of raw scores for all
k classes (unique combinations of mixture components). The softmax score is compatible with cross-entropy loss, offers stability in terms of model training, and highly penalizes the deep neural network for incorrect classifications.
The feature extraction block of the neural network (highlighted with dashed red lines in
Figure 1) applies repeated convolution and pooling operations to reduce the high-dimensional input to the network by capturing the component-distinctive spectral features. The convolution operation involves traversing a sliding window, a randomly initialized filter kernel, over its inputs, which captures important spectral feature information while marginally reducing the dimensionality. Each pooling layer substantially downsamples its inputs. The weights of the convolutional and pooling layers are trained by passing the input spectrum through each of these layers during a forward training pass. Then, a loss function is evaluated, and weights are updated during the backward propagation of error [
36]. The size of the input absorbance vector varies based on the frequency range; thus, the number of convolution and pooling blocks is varied to ensure that there are a sufficient number of neurons in the final layer to perform classification.
Each convolution converts the input vector to a new vector whose size is given by
, where
W,
,
P, and
S are the size of the input, kernel, padding, and the stride, respectively. We found three filters provided good classification performance and did not substantially reduce the input dimensionality in each layer. Consequently, for the full wavenumber range (400–4000 cm
−1), the output of the first convolutional layer with three filters is given by
, where the sizes of the input vector, kernel, and stride are 3601, 3, and 1, respectively, and valid (zero) padding was used. Each convolutional layers is followed by a pooling layer, where the output size is
. For a stride size of two and a pooling kernel size of two, the total output shape is
for the full range (400–4000 cm
−1). For details of convolution and pooling operations, see [
37].
The feature extraction block produces a matrix, due to the application of multiple filter kernels, containing the most relevant spectra-distinctive information from the input spectrum. This matrix is flattened to a vector and sent to a fully connected dense neural network (highlighted with dashed blue lines in
Figure 1) for classification. Each dense layer applies learned weights and biases followed by nonlinear ReLU activation. Here, the Adam optimizer was implemented to update weights and biases using a sparse categorical cross-entropy loss function [
38]. A batch size of 32 was used for training, and the network was trained for 40 epochs. With the application of a softmax layer, the final output from the neural network is a vector containing 175 softmax scores. The highest score indicates the class (mixture components) predicted by the neural network. The prediction is then processed to produce classification metrics for the assessment of model performance.
The final CNN architecture was selected through a grid-search hyperparameter tuning process, as outlined in our prior work [
22]. The architecture is flexible, with the user-defined and variable input frequency bands (variable input vector lengths) requiring variable degrees of downsampling in the feature extraction block to extract a low-dimensional representation of the input spectrum. A pool size and kernel size of 2 were chosen to halve the input feature space, controlling the depth of the feature extraction block in the network. Through hyperparameter tuning, the optimal number of filters and kernel size for convolutional layers was found to be 3. The tuning process revealed that adding a hidden layer between the output of the last convolutional layer and the final classification output layer improves classification accuracy. To enable Grad-CAM, a convolutional layer with trainable weights is essential. Therefore, a convolutional layer is consistently added before the reduced low-dimensional feature space undergoes classification in the fully connected dense neural network.
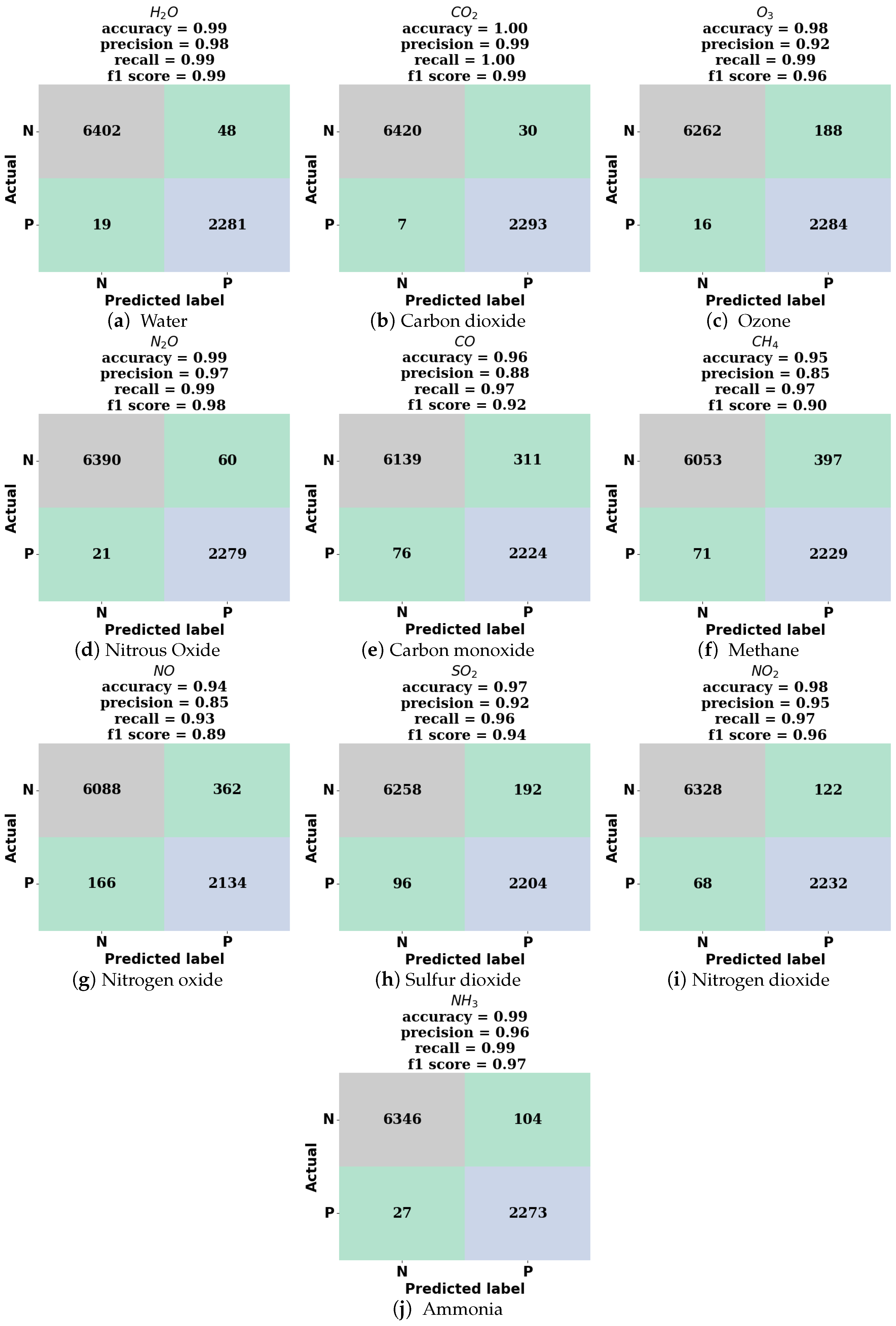
The classification model is implemented in Python version 3.10.12 using the following library packages: TensorFlow version 2.15, NumPy version 1.25, Pandas version 1.5, and scikit-learn version 1.2. The model code is executed on an Intel Xeon CPU with a clock speed of 2 GHz and 12 GB RAM. The training time for 40 epochs are approximately 20 min if the model is trained across the entire frequency region. Inference on the entire test spectra takes approximately 14 s.
2.2. Spectra Simulation for Training, Validation, and Testing
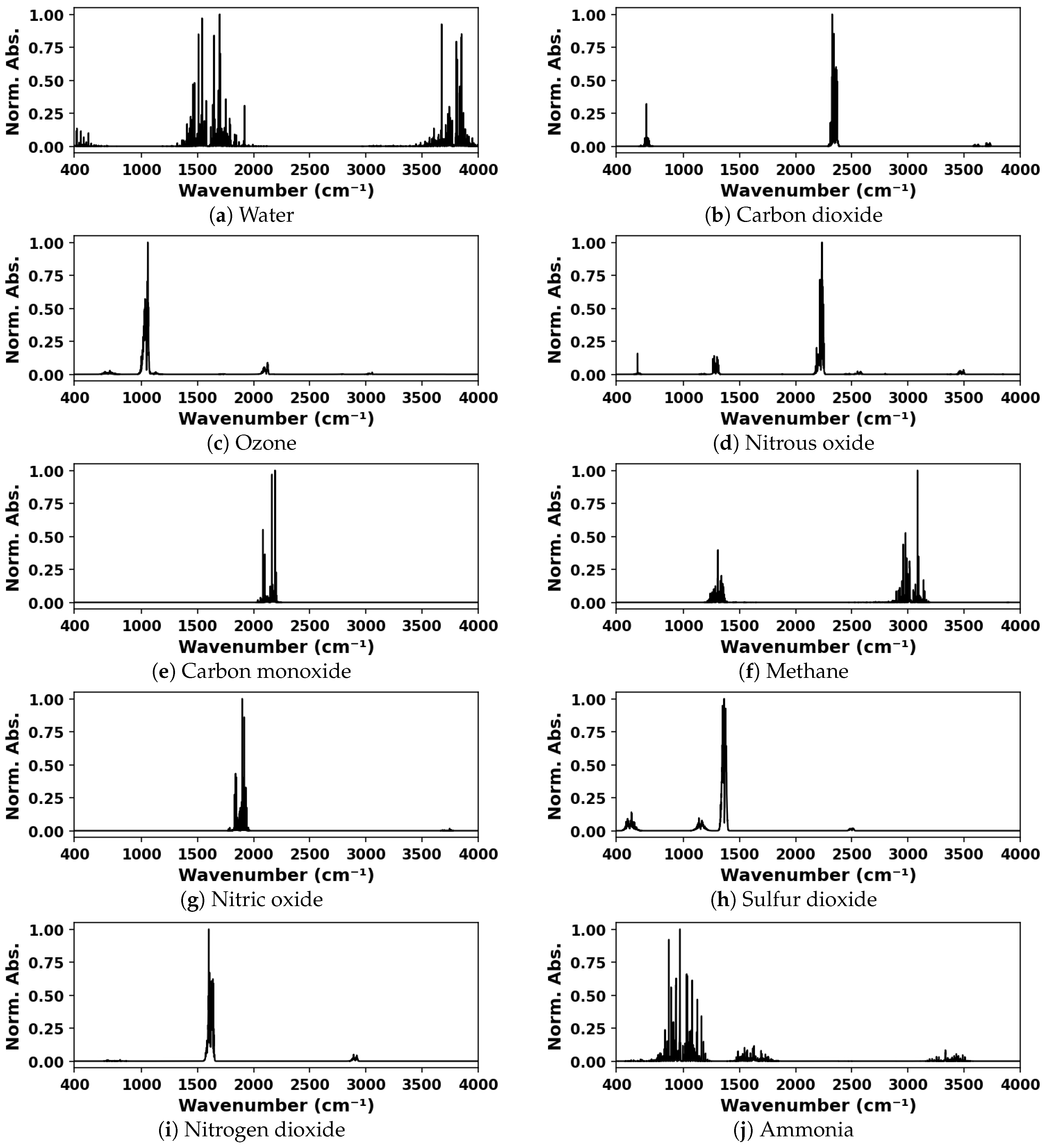
Simulated IR absorption spectra were used for training and validation of the deep neural network. Spectra for mixtures containing one, two, or three components, from a possible ten species, were generated from spectral lines found in the HITRAN database [
1] using the HAPI tool [
39]. The ten species considered were water vapor, carbon dioxide, ozone, nitrous oxide, carbon monoxide, methane, nitric oxide, sulfur dioxide, nitrogen dioxide, and ammonia. The spectra were generated for a standard thermodynamic condition, 297 K and 1 atm total pressure, and for a 10 cm absorption pathlength. The model was trained on various ranges of spectral data including and within the 400–4000 cm
−1 wavenumber range at a resolution of 1 cm
−1. The representative simulated spectra for each species considered are shown in
Figure 2.
For the ten species considered, there are 120 unique 3-component mixtures (
10), 45 2-component mixtures (
10), and 10 1-component (pure in air) mixtures. To produce training and validation datasets that are balanced, not favoring mixtures containing a larger number components, a larger number of discrete concentrations were considered for mixtures containing fewer components. Hence, for 3-component gas mixtures, we considered 5 discrete concentrations; for 2-component mixtures, we considered 12 discrete concentrations; and for 1-component mixtures, we considered 125 discrete concentrations (see
Table 1). Mixture spectra were obtained by a linear combination of pure gas spectra in air at 1 atm, which assumes that all collisional broadening is air dominated. To obtain a balanced set of training spectra, spectra were randomly sampled from each of the unique combinations of species to generate 15,000 3-component spectra, 5625 2-component spectra, and 1250 1-component spectra (21,875 total spectra). This set of simulated spectra was divided into training (13,125 spectra) and validation (8750 spectra) datasets using a stratified 60–40% split. The distribution of spectra in each dataset is shown in
Figure 3. The concentrations of absorbing gases considered in the dataset, given in
Table 1, were specified at mole fractions that are relevant to industrial and atmospheric gas sensing conditions. The maximum absorbance for each spectrum ranges from
to 4, which was chosen as a reasonable range for practical gas sensing applications. See
Figure 3 for a histogram of the maximum absorbance in the spectra contained in the training and validation datasets.
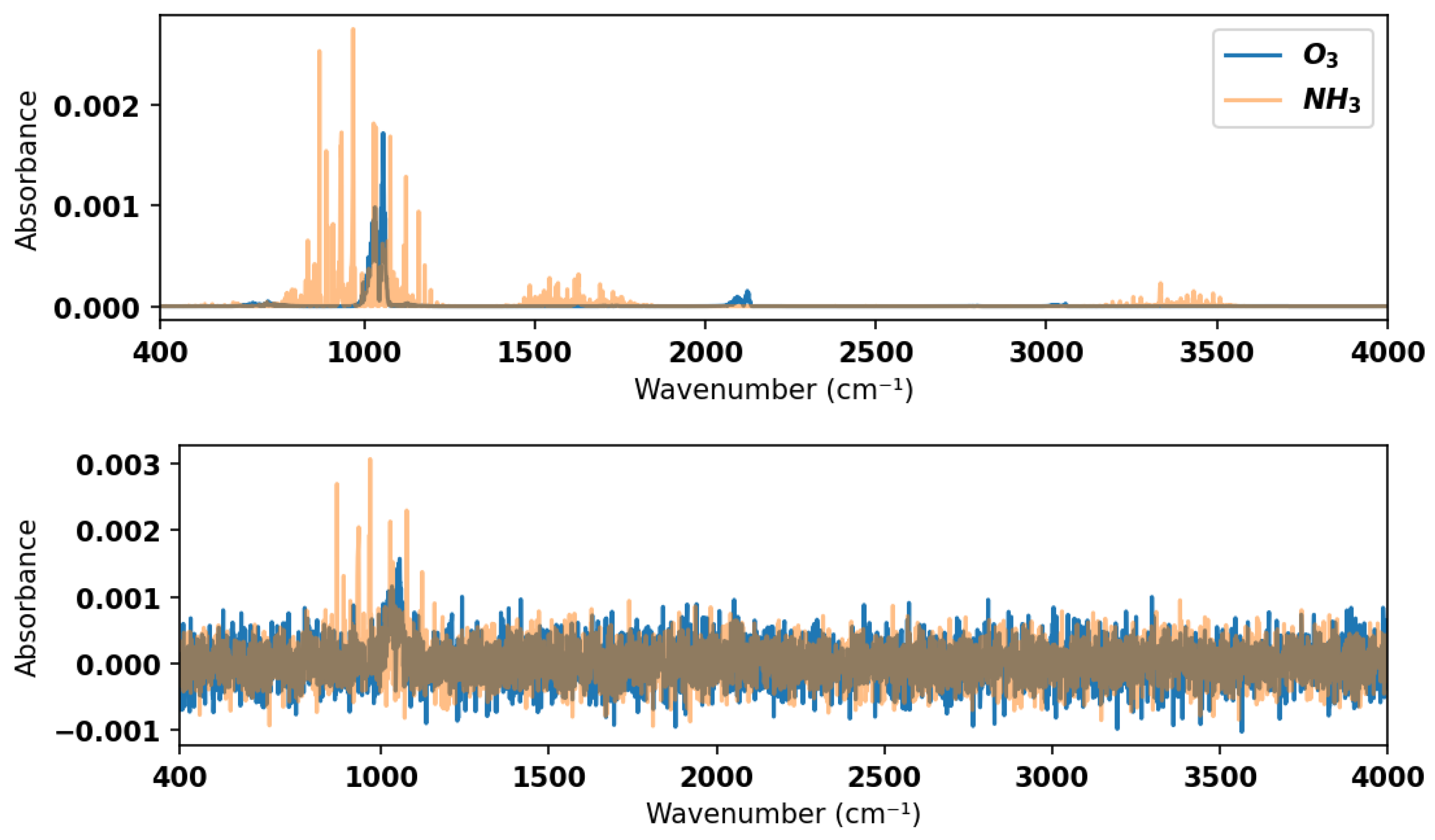
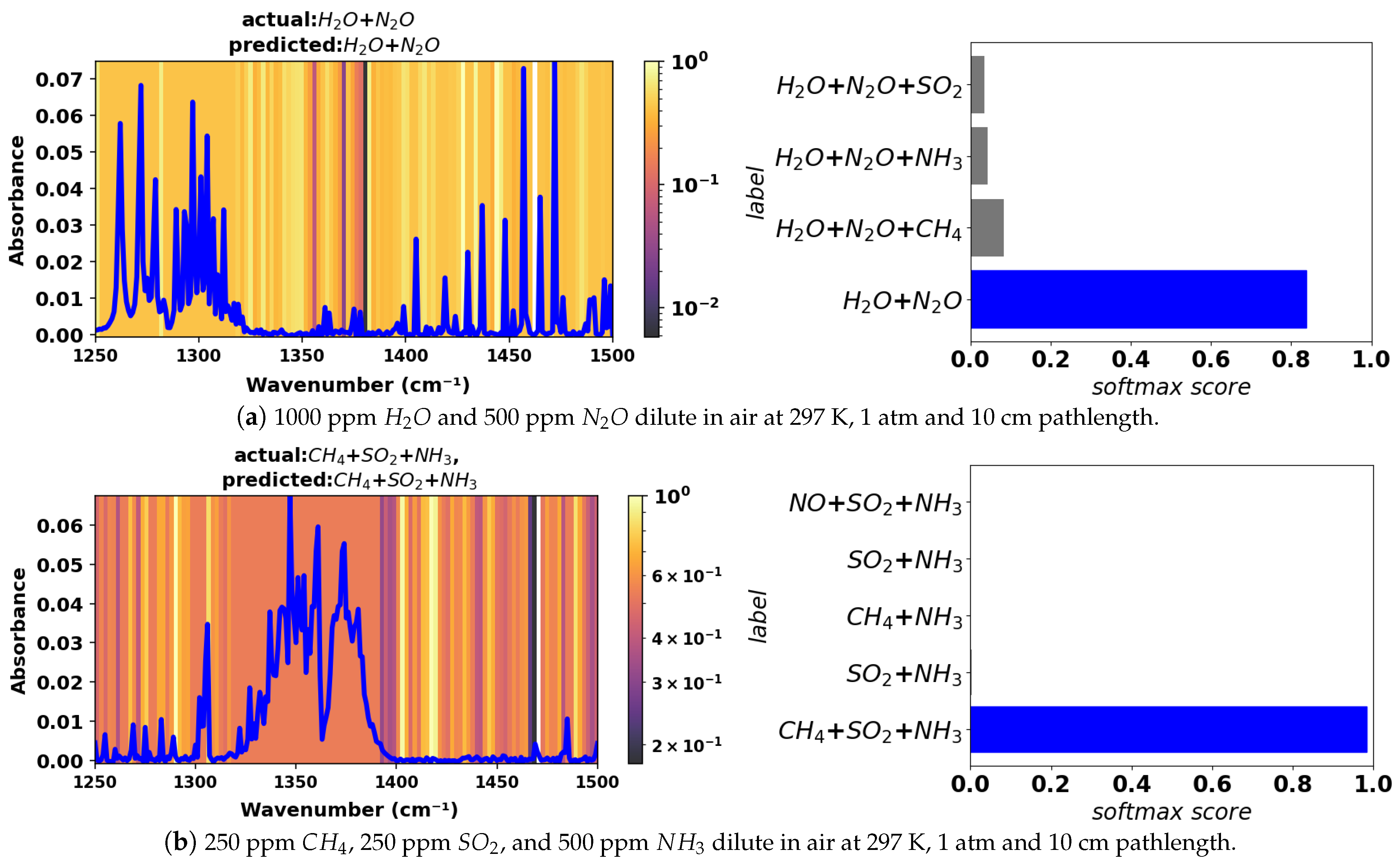
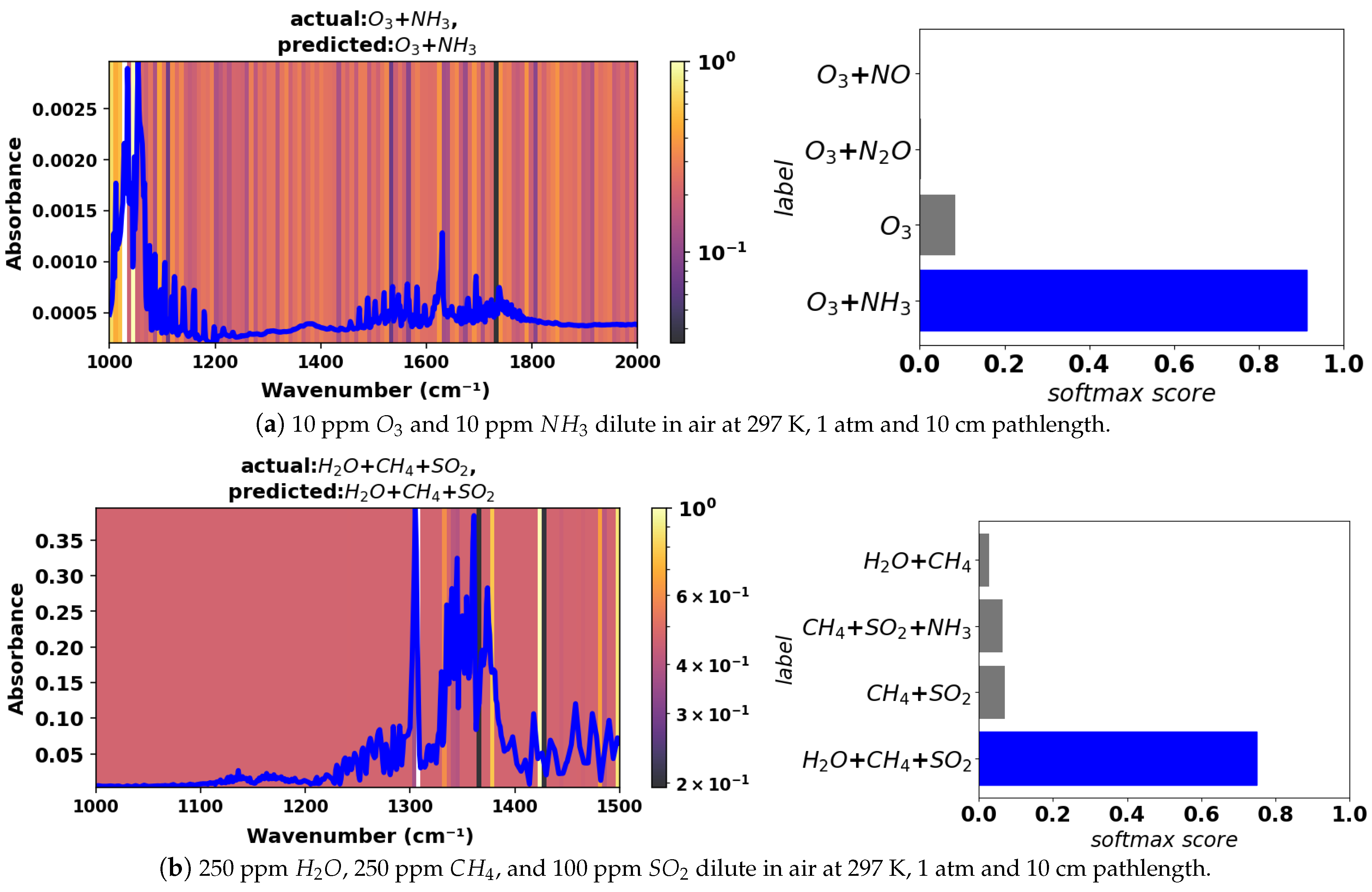
To ensure sufficient testing of the model, a test dataset comprising simulated spectra with added noise was generated. Noise in the experimental gas sensor signal is typically of the order of
or less in absorbance for direct absorption measurements without filtering or other efforts to improve the signal-to-noise ratio. Hence, we added random Gaussian noise with an amplitude of
in absorbance to the simulated mixture spectra contained in the validation dataset to form the test dataset. See
Figure 4 for an example of a noisy spectrum for a 2-component mixture with comparison to the noise-free equivalent. The model was tested against this noisy test dataset in the large wavenumber range in which it was trained (400–4000 cm
−1) as well the smaller ranges, which are more indicative of practical spectrometers. In the smaller wavenumber ranges, some species have a maximum absorbance below the
absorbance level and hence are always undetectable, irrespective of the classification method. Those species have not been considered in the performance evaluation for smaller wavenumber ranges.