Missing Data Statistics Provide Causal Insights into Data Loss in Diabetes Health Monitoring by Wearable Sensors
Abstract
1. Introduction
2. Materials and Methods
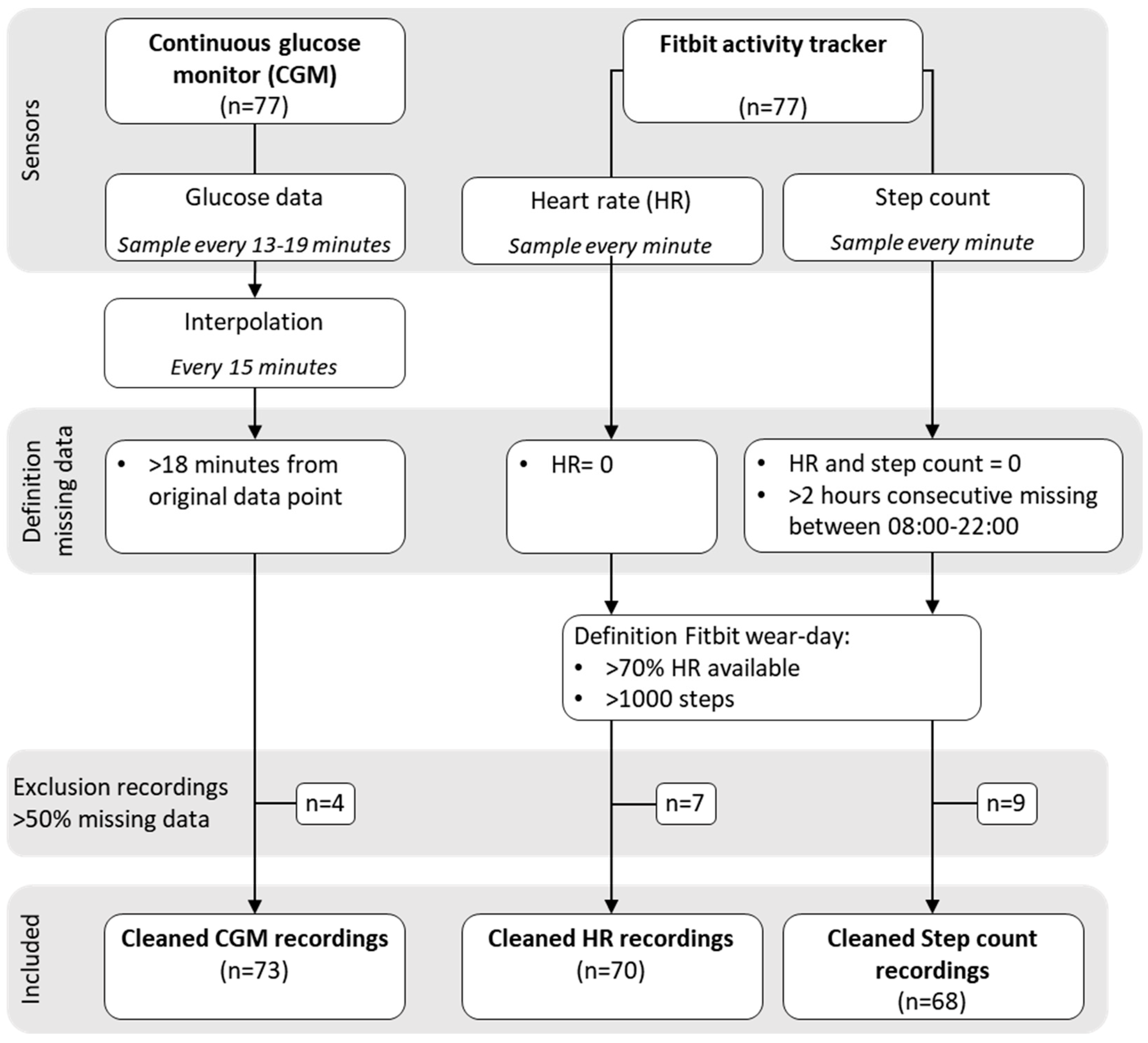
2.1. Data Set
2.2. Data Pre-Processing
2.3. Data Analysis
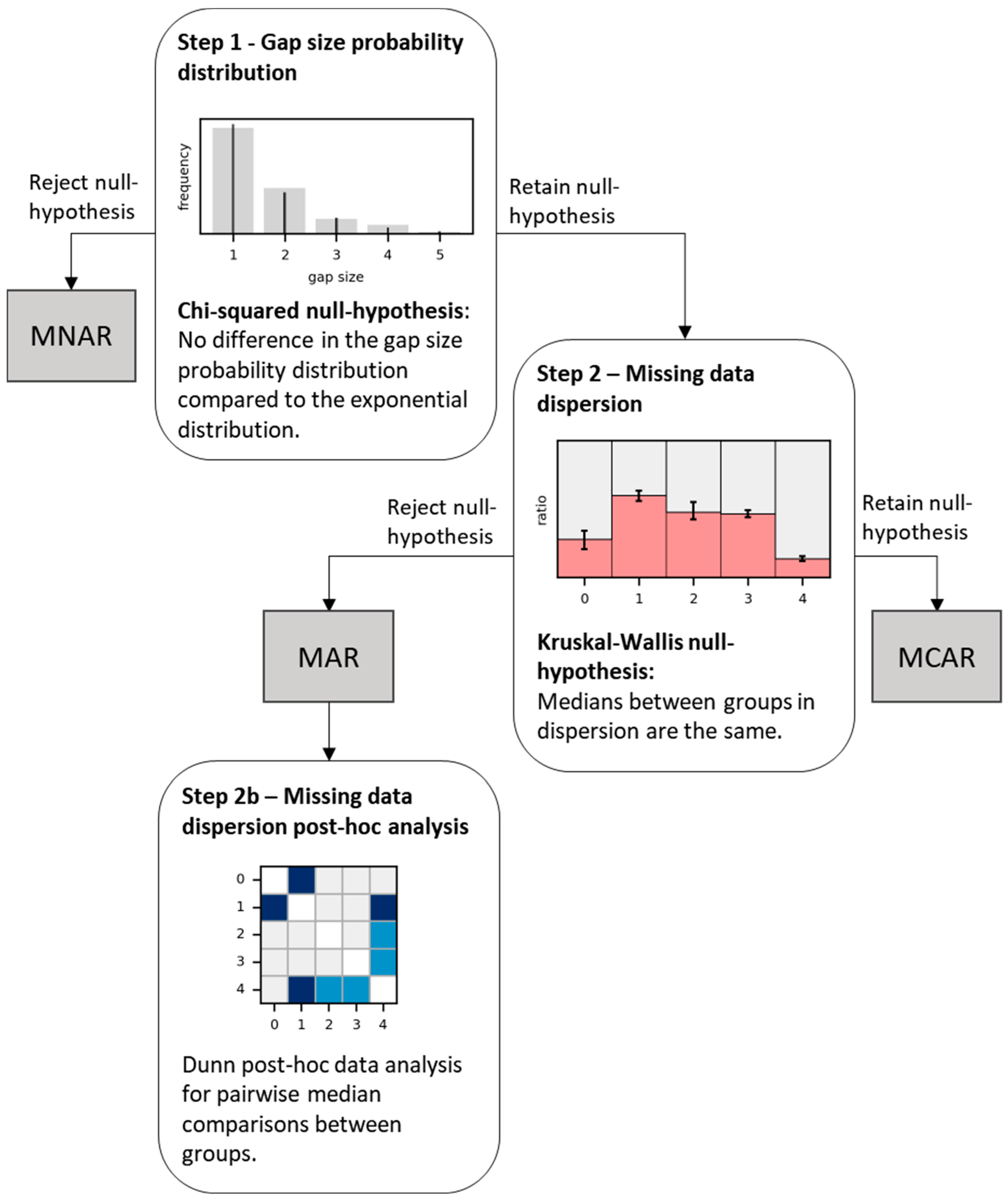
2.4. Statistics of Missing Data and Missing Mechanisms
2.4.1. Step 1—Gap Size Probability Distribution
2.4.2. Step 2—Missing Data Dispersion
2.5. Subgroup Analysis
2.5.1. Subgroups Based on Missing Data
2.5.2. Subgroups Based on Descriptive Data
2.6. Statistics
3. Results
3.1. Exclusion of Recordings
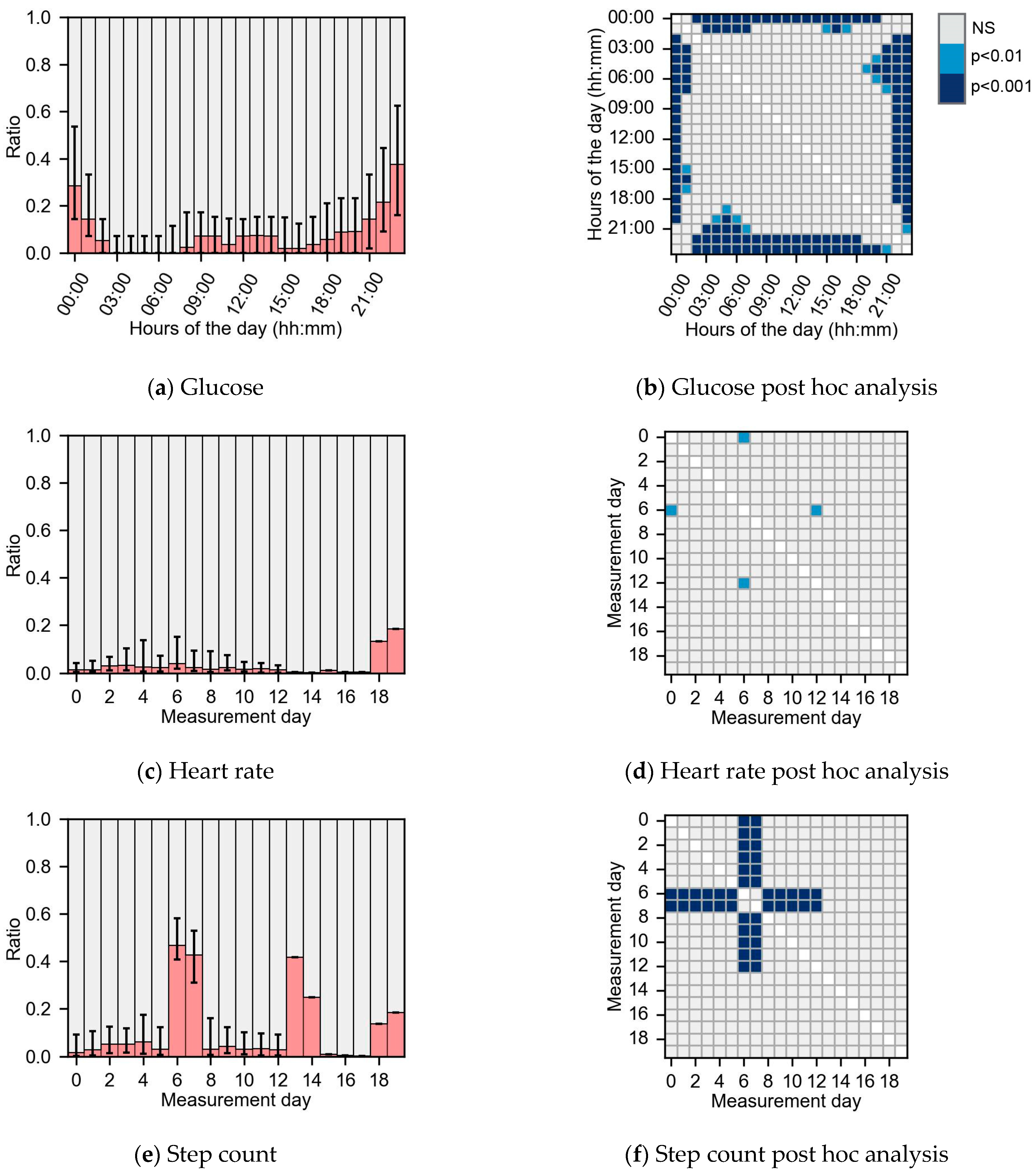
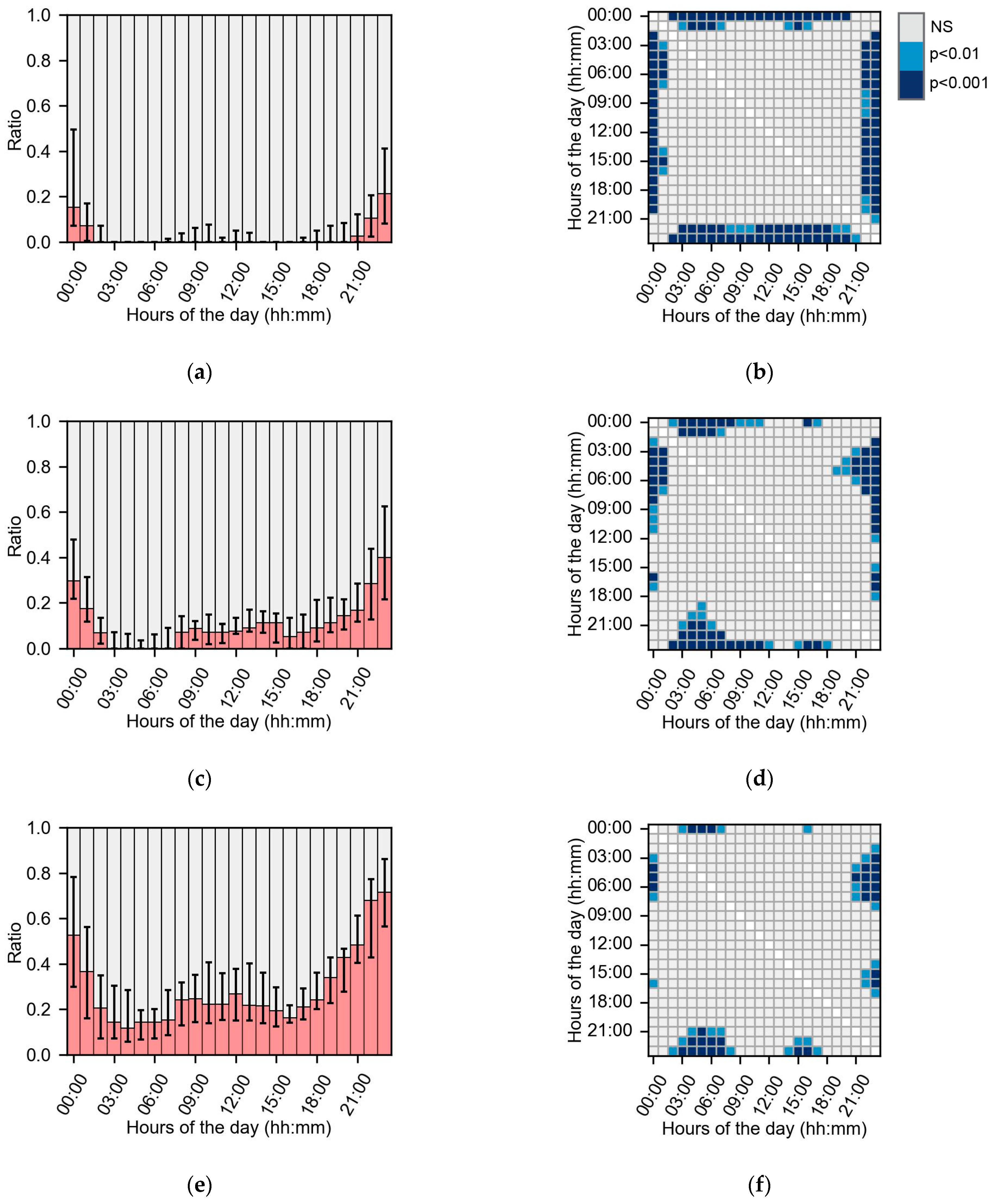
3.2. Gap Frequency
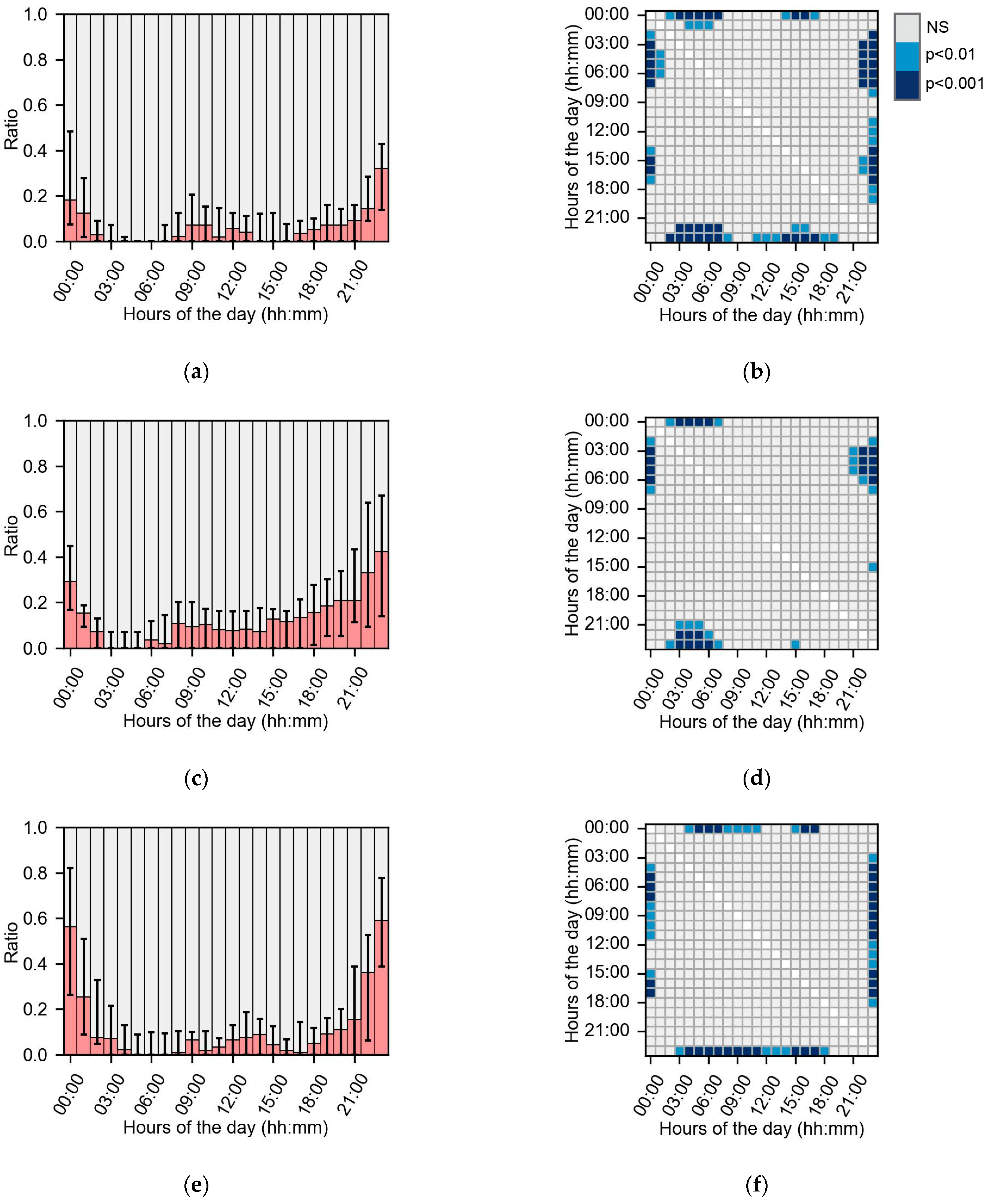
3.3. Missing Dispersion
3.4. Subgroups Analysis
3.4.1. Subgroups Based on Missing Data
3.4.2. Subgroups Based on Descriptive Data
4. Discussion
4.1. Limitations
4.2. Future Research
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Michie, S.; Abraham, C.; Whittington, C.; McAteer, J.; Gupta, S. Effective Techniques in Healthy Eating and Physical Activity Interventions: A Meta-Regression. Health Psychol. 2009, 28, 690–701. [Google Scholar] [CrossRef]
- Phillips, S.M.; Cadmus-Bertram, L.; Rosenberg, D.; Buman, M.P.; Lynch, B.M. Wearable Technology and Physical Activity in Chronic Disease: Opportunities and Challenges. Am. J. Prev. Med. 2018, 54, 144–150. [Google Scholar] [CrossRef]
- Peters, A.; Cohen, N.; Calhoun, P.; Ruedy, K.J.; Beck, R.W.; Martens, T.W.; Bao, S.; Njeru, N.M.; Beck, S.E.; Price, D.A. Glycaemic Profiles of Diverse Patients with Type 2 Diabetes Using Basal Insulin: MOBILE Study Baseline Data. Diabetes Obes. Metab. 2021, 23, 631–636. [Google Scholar] [CrossRef] [PubMed]
- Maher, C.; Ryan, J.; Ambrosi, C.; Edney, S. Users’ Experiences of Wearable Activity Trackers: A Cross-Sectional Study. BMC Public Health 2017, 17, 880. [Google Scholar] [CrossRef]
- Matthews, C.E.; Hagströmer, M.; Pober, D.M.; Bowles, H.R. Best Practices for Using Physical Activity Monitors in Population-Based Research. MSSE 2012, 44, S68–S76. [Google Scholar] [CrossRef]
- Yao, J.; Tan, C.S.; Lim, N.; Tan, J.; Chen, C.; Müller-Riemenschneider, F. Number of Daily Measurements Needed to Estimate Habitual Step Count Levels Using Wrist-Worn Trackers and Smartphones in 212,048 Adults. Sci. Rep. 2021, 11, 9633. [Google Scholar] [CrossRef]
- Battelino, T.; Danne, T.; Bergenstal, R.M.; Amiel, S.A.; Beck, R.; Biester, T.; Bosi, E.; Buckingham, B.A.; Cefalu, W.T.; Close, K.L.; et al. Clinical Targets for Continuous Glucose Monitoring Data Interpretation: Recommendations from the International Consensus on Time in Range. Diabetes Care 2019, 42, 1593–1603. [Google Scholar] [CrossRef]
- Riddlesworth, T.D.; Beck, R.W.; Gal, R.L.; Connor, C.G.; Bergenstal, R.M.; Lee, S.; Willi, S.M. Optimal Sampling Duration for Continuous Glucose Monitoring to Determine Long-Term Glycemic Control. Diabetes Technol. Ther. 2018, 20, 314–316. [Google Scholar] [CrossRef] [PubMed]
- Compernolle, S.; DeSmet, A.; Poppe, L.; Crombez, G.; De Bourdeaudhuij, I.; Cardon, G.; Van Der Ploeg, H.P.; Van Dyck, D. Effectiveness of Interventions Using Self-Monitoring to Reduce Sedentary Behavior in Adults: A Systematic Review and Meta-Analysis. Int. J. Behav. Nutr. Phys. Act. 2019, 16, 63. [Google Scholar] [CrossRef]
- Breteler, M.J.M.; Huizinga, E.; Van Loon, K.; Leenen, L.P.H.; Dohmen, D.A.J.; Kalkman, C.J.; Blokhuis, T.J. Reliability of Wireless Monitoring Using a Wearable Patch Sensor in High-Risk Surgical Patients at a Step-down Unit in the Netherlands: A Clinical Validation Study. BMJ Open 2018, 8, e020162. [Google Scholar] [CrossRef]
- Johansson, D.; Malmgren, K.; Alt Murphy, M. Wearable Sensors for Clinical Applications in Epilepsy, Parkinson’s Disease, and Stroke: A Mixed-Methods Systematic Review. J. Neurol. 2018, 265, 1740–1752. [Google Scholar] [CrossRef]
- Allet, L.; Knols, R.H.; Shirato, K.; Bruin, E.D.D. Wearable Systems for Monitoring Mobility-Related Activities in Chronic Disease: A Systematic Review. Sensors 2010, 10, 9026–9052. [Google Scholar] [CrossRef]
- Haescher, M.; Matthies, D.J.C.; Krause, S.; Bieber, G. Presenting a Data Imputation Concept to Support the Continuous Assessment of Human Vital Data and Activities. In Proceedings of the 12th ACM International Conference on PErvasive Technologies Related to Assistive Environments, Rhodes, Greece, 5–7 June 2019; ACM: New York, NY, USA, 2019; pp. 587–592. [Google Scholar]
- Yang, S.; Kalpakis, K.; Mackenzie, C.F.; Stansbury, L.G.; Stein, D.M.; Scalea, T.M.; Hu, P.F. Online Recovery of Missing Values in Vital Signs Data Streams Using Low-Rank Matrix Completion. In Proceedings of the 2012 11th International Conference on Machine Learning and Applications, Boca Raton, FL, USA, 12–15 December 2012; IEEE: Boca Raton, FL, USA, 2012; pp. 281–287. [Google Scholar]
- Chakrabarti, S.; Biswas, N.; Karnani, K.; Padul, V.; Jones, L.D.; Kesari, S.; Ashili, S. Binned Data Provide Better Imputation of Missing Time Series Data from Wearables. Sensors 2023, 23, 1454. [Google Scholar] [CrossRef]
- Lin, S.; Wu, X.; Martinez, G.; Chawla, N.V. Filling Missing Values on Wearable-Sensory Time Series Data. In Proceedings of the 2020 SIAM International Conference on Data Mining (SDM), Cincinnati, OH, USA, 7–9 May 2020; Society for Industrial and Applied Mathematics: Philadelphia, PA, USA, 2020; pp. 46–54. [Google Scholar]
- Wu, X.; Mattingly, S.; Mirjafari, S.; Huang, C.; Chawla, N.V. Personalized Imputation on Wearable-Sensory Time Series via Knowledge Transfer. In Proceedings of the 29th ACM International Conference on Information & Knowledge Management, Virtual, 19–23 October 2020; Association for Computing Machinery: New York, NY, USA, 2020; pp. 1625–1634. [Google Scholar]
- Haveman, M.E.; Van Rossum, M.C.; Vaseur, R.M.E.; Van Der Riet, C.; Schuurmann, R.C.L.; Hermens, H.J.; De Vries, J.-P.P.M.; Tabak, M. Continuous Monitoring of Vital Signs with Wearable Sensors During Daily Life Activities: Validation Study. JMIR Form. Res. 2022, 6, e30863. [Google Scholar] [CrossRef]
- Syversen, A.; Dosis, A.; Jayne, D.; Zhang, Z. Wearable Sensors as a Preoperative Assessment Tool: A Review. Sensors 2024, 24, 482. [Google Scholar] [CrossRef]
- Staffini, A.; Svensson, T.; Chung, U.; Svensson, A.K. A Disentangled VAE-BiLSTM Model for Heart Rate Anomaly Detection. Bioengineering 2023, 10, 683. [Google Scholar] [CrossRef]
- Staffini, A.; Svensson, T.; Chung, U.; Svensson, A.K. Heart Rate Modeling and Prediction Using Autoregressive Models and Deep Learning. Sensors 2022, 22, 34. [Google Scholar] [CrossRef]
- Jeon, J.; Leimbigler, P.J.; Baruah, G.; Li, M.H.; Fossat, Y.; Whitehead, A.J. Predicting Glycaemia in Type 1 Diabetes Patients: Experiments in Feature Engineering and Data Imputation. J. Health Inf. Res. 2020, 4, 71–90. [Google Scholar] [CrossRef]
- Gashi, S.; Alecci, L.; Gjoreski, M.; Di Lascio, E.; Mehrotra, A.; Musolesi, M.; Debus, M.E.; Gasparini, F.; Santini, S. Handling Missing Data for Sleep Monitoring Systems. In Proceedings of the 2022 10th International Conference on Affective Computing and Intelligent Interaction (ACII), Nara, Japan, 18–21 October 2022; pp. 1–8. [Google Scholar]
- Lee, P.H. Data Imputation for Accelerometer-Measured Physical Activity: The Combined Approach. Am. J. Clin. Nutr. 2013, 97, 965–971. [Google Scholar] [CrossRef]
- Chinapaw, M.J.M.; de Niet, M.; Verloigne, M.; De Bourdeaudhuij, I.; Brug, J.; Altenburg, T.M. From Sedentary Time to Sedentary Patterns: Accelerometer Data Reduction Decisions in Youth. PLoS ONE 2014, 9, e111205. [Google Scholar] [CrossRef]
- Altenburg, T.M.; de Niet, M.; Verloigne, M.; De Bourdeaudhuij, I.; Androutsos, O.; Manios, Y.; Kovacs, E.; Bringolf-Isler, B.; Brug, J.; Chinapaw, M.J.M. Occurrence and Duration of Various Operational Definitions of Sedentary Bouts and Cross-Sectional Associations with Cardiometabolic Health Indicators: The ENERGY-Project. Prev. Med. 2015, 71, 101–106. [Google Scholar] [CrossRef]
- Orstad, S.L.; Gerchow, L.; Patel, N.R.; Reddy, M.; Hernandez, C.; Wilson, D.K.; Jay, M. Defining Valid Activity Monitor Data: A Multimethod Analysis of Weight-Loss Intervention Participants’ Barriers to Wear and First 100 Days of Physical Activity. Informatics 2021, 8, 39. [Google Scholar] [CrossRef]
- Akturk, H.K.; Herrero, P.; Oliver, N.; Wise, H.; Eikermann, E.; Snell-Bergeon, J.; Shah, V.N. Impact of Different Types of Data Loss on Optimal Continuous Glucose Monitoring Sampling Duration. Diabetes Technol. Ther. 2022, 24, 749–753. [Google Scholar] [CrossRef]
- Chen, R.; Jankovic, F.; Marinsek, N.; Foschini, L.; Kourtis, L.; Signorini, A.; Pugh, M.; Shen, J.; Yaari, R.; Maljkovic, V.; et al. Developing Measures of Cognitive Impairment in the Real World from Consumer-Grade Multimodal Sensor Streams. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Anchorage, AK, USA, 4–8 August 2019; ACM: New York, NU, USA, 2019; pp. 2145–2155. [Google Scholar]
- Rubin, D.B. Inference and Missing Data. Biometrika 1976, 63, 581–592. [Google Scholar] [CrossRef]
- Moritz, S. Comparison of Different Methods for Univariate Time Series Imputation in R. arXiv 2015, arXiv:1510.03924. [Google Scholar] [CrossRef]
- Zhou, N.; Patel, D.; Iyengar, A.; Shrivastava, S.; Bhamidipaty, A. A Verifiable Imputation Analysis for Univariate Time Series and Enabling Package. In Proceedings of the 2020 IEEE International Conference on Big Data (Big Data), Atlanta, GA, USA, 10–13 December 2020; IEEE: Piscataway, NL, USA, 2020; pp. 1764–1773. [Google Scholar]
- Lee, M.; An, J.; Lee, Y. Missing-Value Imputation of Continuous Missing Based on Deep Imputation Network Using Correlations among Multiple IoT Data Streams in a Smart Space. IEICE Trans. Inf. Syst. 2019, E102.D, 289–298. [Google Scholar] [CrossRef]
- Che, Z.; Purushotham, S.; Cho, K.; Sontag, D.; Liu, Y. Recurrent Neural Networks for Multivariate Time Series with Missing Values. Sci. Rep. 2018, 8, 6085. [Google Scholar] [CrossRef]
- Gant, C.; Binnenmars, S.; Berg, E.; Bakker, S.; Navis, G.; Laverman, G. Integrated Assessment of Pharmacological and Nutritional Cardiovascular Risk Management: Blood Pressure Control in the DIAbetes and LifEstyle Cohort Twente (DIALECT). Nutrients 2017, 9, 709. [Google Scholar] [CrossRef]
- Den Braber, N.; Vollenbroek-Hutten, M.; Oosterwijk, M.; Gant, C.; Hagedoorn, I.; Van Beijnum, B.-J.; Hermens, H.; Laverman, G. Requirements of an Application to Monitor Diet, Physical Activity and Glucose Values in Patients with Type 2 Diabetes: The Diameter. Nutrients 2019, 11, 409. [Google Scholar] [CrossRef]
- Claudel, S.E.; Tamura, K.; Troendle, J.; Andrews, M.R.; Ceasar, J.N.; Mitchell, V.M.; Vijayakumar, N.; Powell-Wiley, T.M. Comparing Methods to Identify Wear-Time Intervals for Physical Activity with the Fitbit Charge 2. J. Aging Phys. Act. 2020, 29, 529–535. [Google Scholar] [CrossRef]
- Wing, D.; Godino, J.G.; Baker, F.C.; Yang, R.; Chevance, G.; Thompson, W.K.; Reuter, C.; Bartsch, H.; Wilbur, A.; Straub, L.K.; et al. Recommendations for Identifying Valid Wear for Consumer-Level Wrist-Worn Activity Trackers and Acceptability of Extended Device Deployment in Children. Sensors 2022, 22, 9189. [Google Scholar] [CrossRef]
- Hilden, P.; Schwartz, J.E.; Pascual, C.; Diaz, K.M.; Goldsmith, J. How Many Days Are Needed? Measurement Reliability of Wearable Device Data to Assess Physical Activity. PLoS ONE 2023, 18, e0282162. [Google Scholar] [CrossRef]
- Matlary, R.E.D.; Holme, P.A.; Glosli, H.; Rueegg, C.S.; Grydeland, M. Comparison of Free-living Physical Activity Measurements between ActiGraph GT3X-BT and Fitbit Charge 3 in Young People with Haemophilia. Haemophilia 2022, 28, e172–e180. [Google Scholar] [CrossRef]
- Tamura, K.; Vijayakumar, N.P.; Troendle, J.F.; Curlin, K.; Neally, S.J.; Mitchell, V.M.; Collins, B.S.; Baumer, Y.; Gutierrez-Huerta, C.A.; Islam, R.; et al. Multilevel Mobile Health Approach to Improve Cardiovascular Health in Resource-Limited Communities with Step It Up: A Randomised Controlled Trial Protocol Targeting Physical Activity. BMJ Open 2020, 10, e040702. [Google Scholar] [CrossRef]
- Hardcastle, S.J.; Jiménez-Castuera, R.; Maxwell-Smith, C.; Bulsara, M.K.; Hince, D. Fitbit Wear-Time and Patterns of Activity in Cancer Survivors throughout a Physical Activity Intervention and Follow-up: Exploratory Analysis from a Randomised Controlled Trial. PLoS ONE 2020, 15, e0240967. [Google Scholar] [CrossRef]
- Schober, P.; Boer, C.; Schwarte, L.A. Correlation Coefficients: Appropriate Use and Interpretation. Anesth. Analg. 2018, 126, 1763. [Google Scholar] [CrossRef]
- Gallager, R.G. Stochastic Processes: Theory for Applications; Cambridge University Press: Cambridge, UK, 2013; ISBN 978-1-107-03975-9. [Google Scholar]
- Statistical Functions (Scipy.Stats)—SciPy v1.11.1 Manual. Available online: https://docs.scipy.org/doc/scipy/reference/stats.html#discrete-distributions (accessed on 30 June 2023).
- McHugh, M.L. The Chi-Square Test of Independence. Biochem. Med. 2013, 23, 143–149. [Google Scholar] [CrossRef]
- Weir, C.B.; Jan, A. BMI Classification Percentile and Cut Off Points. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- ElSayed, N.A.; Aleppo, G.; Aroda, V.R.; Bannuru, R.R.; Brown, F.M.; Bruemmer, D.; Collins, B.S.; Hilliard, M.E.; Isaacs, D.; Johnson, E.L.; et al. 6. Glycemic Targets: Standards of Care in Diabetes—2023. Diabetes Care 2023, 46, S97–S110. [Google Scholar] [CrossRef]
- Martinez, G.J.; Mattingly, S.M.; Robles-Granda, P.; Saha, K.; Sirigiri, A.; Young, J.; Chawla, N.; Choudhury, M.D.; D’Mello, S.; Mark, G.; et al. Predicting Participant Compliance with Fitness Tracker Wearing and Ecological Momentary Assessment Protocols in Information Workers: Observational Study. JMIR mHealth uHealth 2021, 9, e22218. [Google Scholar] [CrossRef]
- Khandelwal, D.; Dutta, D.; Chittawar, S.; Kalra, S. Sleep Disorders in Type 2 Diabetes. Indian. J. Endocrinol. Metab. 2017, 21, 758–761. [Google Scholar] [CrossRef]

| Median or n | [IQR] or % | |
|---|---|---|
| Male | 46 | 60.0% |
| Age (years) | 64 | [56–71] |
| BMI (kg/m2) | 33 | [29.0–35.9] |
| 17.5–25 | 4 | 5.2% |
| 25–30 | 22 | 28.6% |
| 30–40 | 45 | 58.4% |
| >40 | 7 | 9.1% |
| Hb1Ac (mmol/mol) | 58 | [52–64] |
| ≤53 | 27 | 36.5% |
| >53 | 50 | 64.9% |
| Years since diagnosis (years) | 14 | [7–20] |
| Use of type 2 diabetes medication | 74 | 100% |
| Oral | 69 | 89.6% |
| Insulin | 43 | 55.8% |
| Other | 30 | 39.0% |
| n | Recording Length (Days, Hours: Minutes) | Missing Data (%) | |||||
|---|---|---|---|---|---|---|---|
| (%) | Median | [IQR] | [Range] | Median | [IQR] | [Range] | |
| Glucose | 73 (95%) | 13, 19:15 | [11, 21:15–13, 21:15] | [6, 22:15–20, 17:00] | 11.1 | [6.2–17.6] | [0.2–47.2] |
| HR | 70 (91%) | 13, 00:00 | [12, 00:00–13, 00:00] | [2, 00:00–20, 00:00] | 9.4 | [3.3–18.0] | [1.2–44.6] |
| Steps count | 68 (88%) | 12, 12:53 | [11, 21:51–12, 15:16] | [1, 11:44–19, 15:04] | 15.4 | [12.5–22.4] | [6.8–42.1] |
| Recordings in Subgroups Based on Missing Data | |||
|---|---|---|---|
| <10% | 10–20% | >20% | |
| Glucose | 34 (47%) | 23 (32%) | 16 (22%) |
| HR | 37 (53%) | 20 (29%) | 13 (19%) |
| Step count | 15 (22%) | 32 (47%) | 21 (31%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Braem, C.I.R.; Yavuz, U.S.; Hermens, H.J.; Veltink, P.H. Missing Data Statistics Provide Causal Insights into Data Loss in Diabetes Health Monitoring by Wearable Sensors. Sensors 2024, 24, 1526. https://doi.org/10.3390/s24051526
Braem CIR, Yavuz US, Hermens HJ, Veltink PH. Missing Data Statistics Provide Causal Insights into Data Loss in Diabetes Health Monitoring by Wearable Sensors. Sensors. 2024; 24(5):1526. https://doi.org/10.3390/s24051526
Chicago/Turabian StyleBraem, Carlijn I. R., Utku S. Yavuz, Hermie J. Hermens, and Peter H. Veltink. 2024. "Missing Data Statistics Provide Causal Insights into Data Loss in Diabetes Health Monitoring by Wearable Sensors" Sensors 24, no. 5: 1526. https://doi.org/10.3390/s24051526
APA StyleBraem, C. I. R., Yavuz, U. S., Hermens, H. J., & Veltink, P. H. (2024). Missing Data Statistics Provide Causal Insights into Data Loss in Diabetes Health Monitoring by Wearable Sensors. Sensors, 24(5), 1526. https://doi.org/10.3390/s24051526






