1. Introduction
With the continuous development of science and technology, the volume of data is increasing exponentially, which creates “the curse of dimensionality” [
1]. In the domain of feature selection, researchers have introduced numerous approaches to address the diversity and complexity of data characteristics. However, many features of the vast databases are nonessential or even redundant. To cope with this issue, researchers have turned to feature selection (FS) as a common tool. By removing irrelevant and redundant features, FS obtains an optimal subset with very few features and high classification accuracies to achieve dimensionality reduction [
2]. There are applications of FS techniques in many fields, including text classification [
3], bioinformatics [
4], spectral-spatial classification [
5], and others.
FS has four main operations, generating feature subsets, evaluating feature subsets, stopping criterion, and evaluation [
6]. The search methods and evaluation criterion are the most essential parts of the FS algorithms. There are three main search strategies, a complete search strategy, sequence search strategy, and heuristic search strategy. A complete search necessitates traversing all the potential feature subset combinations, such as an exhaustive search [
7]. While this strategy assures that the optimal subsets can be obtained, it comes with a considerable computational cost for large datasets. Based on model assessment feedback, the sequence search continuously adds or removes features from the candidate feature subsets to form the best feature subset. Common approaches are sequential backward selection (SBS) [
8], sequential forward selection (SFS) [
9], sequential floating backward selection (SFBS) [
10], and sequential floating forward selection (SFFS) [
11]. Lakshmi and Mohanaiah proposed a Multi-Support Vector Neural Network (MultiSVNN) method for FER based on a hybrid Whale Optimization Algorithm-Teaching Learning Based Optimization (WOA-TLBO) [
12]. The sequence search algorithms reduce computing costs effectively, but it is easy to fall into the local optimum. A heuristic search gradually employs metaheuristics to approximate the global best subset. The metaheuristics can thoroughly consider the feature interactions during exploration and find the position closer to the target. Hence, a heuristic search is better suited than the first two methods for dealing with complicated and high-dimensional real-world problems. These heuristic algorithms include adaptively balanced grey wolf optimization [
13], bare-bones particle swarm optimization [
14], and a cultural algorithm and differential evolution [
15], etc.
The Whale Optimization Algorithm (WOA) is one of the typical heuristic algorithms that is designed based on the feeding process of humpback whales [
16]. The WOA is known for its efficient local exploitation ability and only needs to tune two main internal parameters. With these benefits, WOA has been implemented in a range of areas, including facial emotion recognition [
17], medical diagnosis problems [
18], and feature selection [
19]. To achieve superior feature combinations, researchers frequently improve WOA from many perspectives. To tackle the challenge of feature selection in imbalanced classification, Lin Sun and colleagues proposed a two-stage feature subset selection scheme that integrates the Fuzzy Multi-Granularity Rough Set (FMRS) and a Binary Whale Optimization Algorithm (BWOA) [
20], for instance, maintaining the balance of exploration and exploitation [
21], getting rid of the local optimum [
22,
23], improving the global search ability [
24,
25,
26], speeding up convergence [
27], and promoting population diversity [
28].
This study focuses on three aspects of the challenges in WOA-based feature selection. First, the global best individual of a canonical WOA is the primary directing factor in the search process. Individuals may likely cluster together early in this instance, resulting in limited global exploration capabilities and a premature convergence. Second, as the distance between individuals gradually decreases, the original search approach cannot guarantee that individuals continue to explore alternative locations, which could cause the search to stall. Finally, the population explores everywhere, and individual quality is poor in the initial stage, leading to low search efficiency.
In the context of fall detection, our proposed algorithm demonstrates the effectiveness of the feature selection (FS) method while primarily contributing to advancing FS research. Koo et al. introduced an artificial neural network (ANN) approach integrating ranking algorithms for post-fall detection, which enhances classification performance and balances accuracy with computational efficiency [
29]. Their innovative use of ranking algorithms significantly improves dimensionality reduction, offering valuable insights for FS optimization. Similarly, Le et al. compared machine learning classifiers in fall detection, showing that FS techniques improve model performance, reduce computational complexity, and refine feature sets [
30]. This study highlighted the synergistic effects of classifiers and FS methods, providing critical guidance for addressing fall-detection challenges. Moreover, widely adopted datasets, such as those in [
31] and the influential dataset by Jang et al. [
32], have significantly supported research efforts. These works underscore the importance of effective FS and robust datasets in advancing fall-detection applications and FS research.
Therefore, a multispiral whale optimization algorithm (MSWOA) with three improvements is presented to enhance the global search capability and jump out of the local optima trap. The following are the key theoretical contributions of this paper:
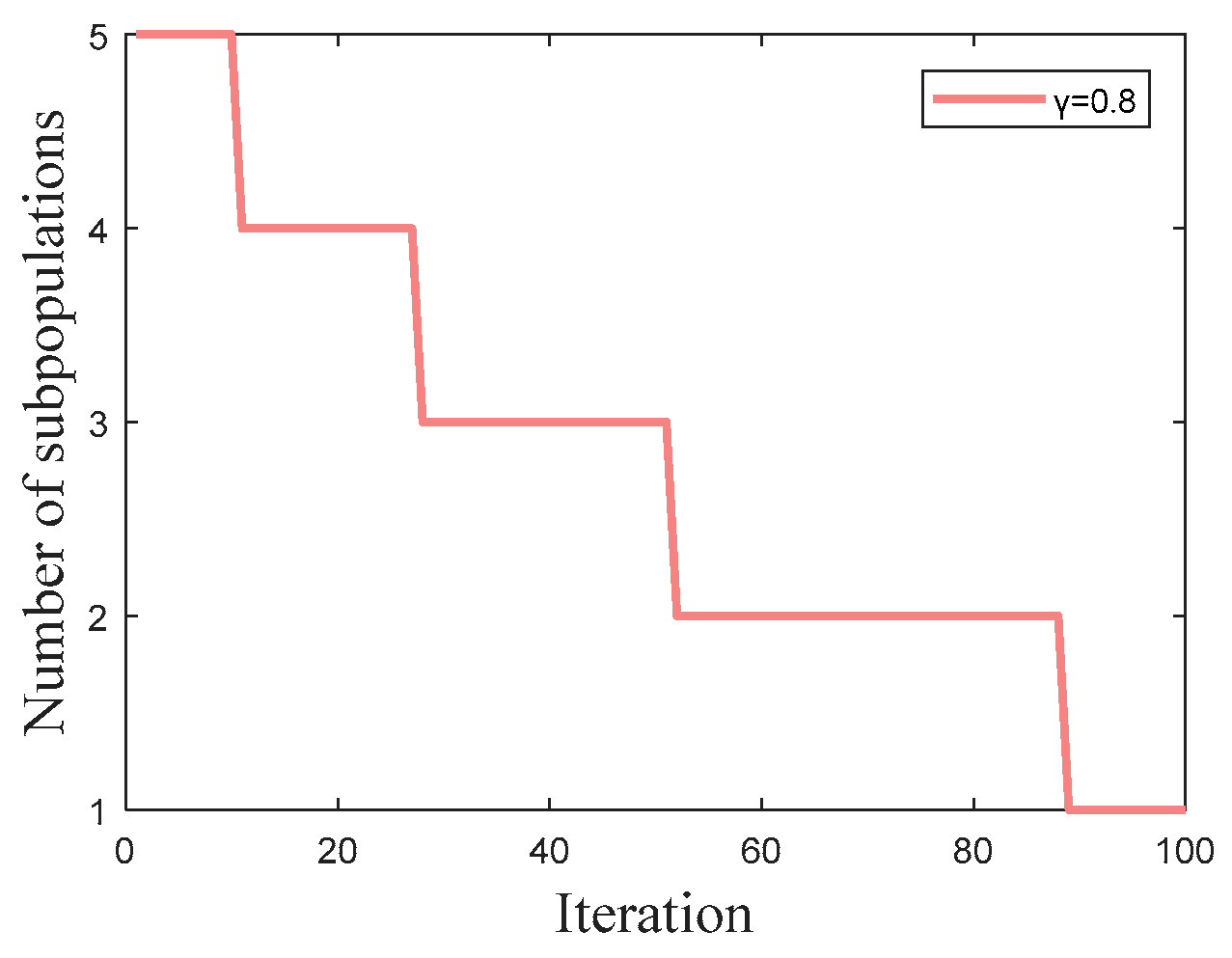
A new Adaptive Multipopulation merging strategy (AMS) separates a population into several subpopulations based on a particular exponential function, which can generate more candidate solutions and expand the exploration space. Each subpopulation selects individuals according to a fitness-distance criterion, which enhances global exploration capability and prevents premature convergence.
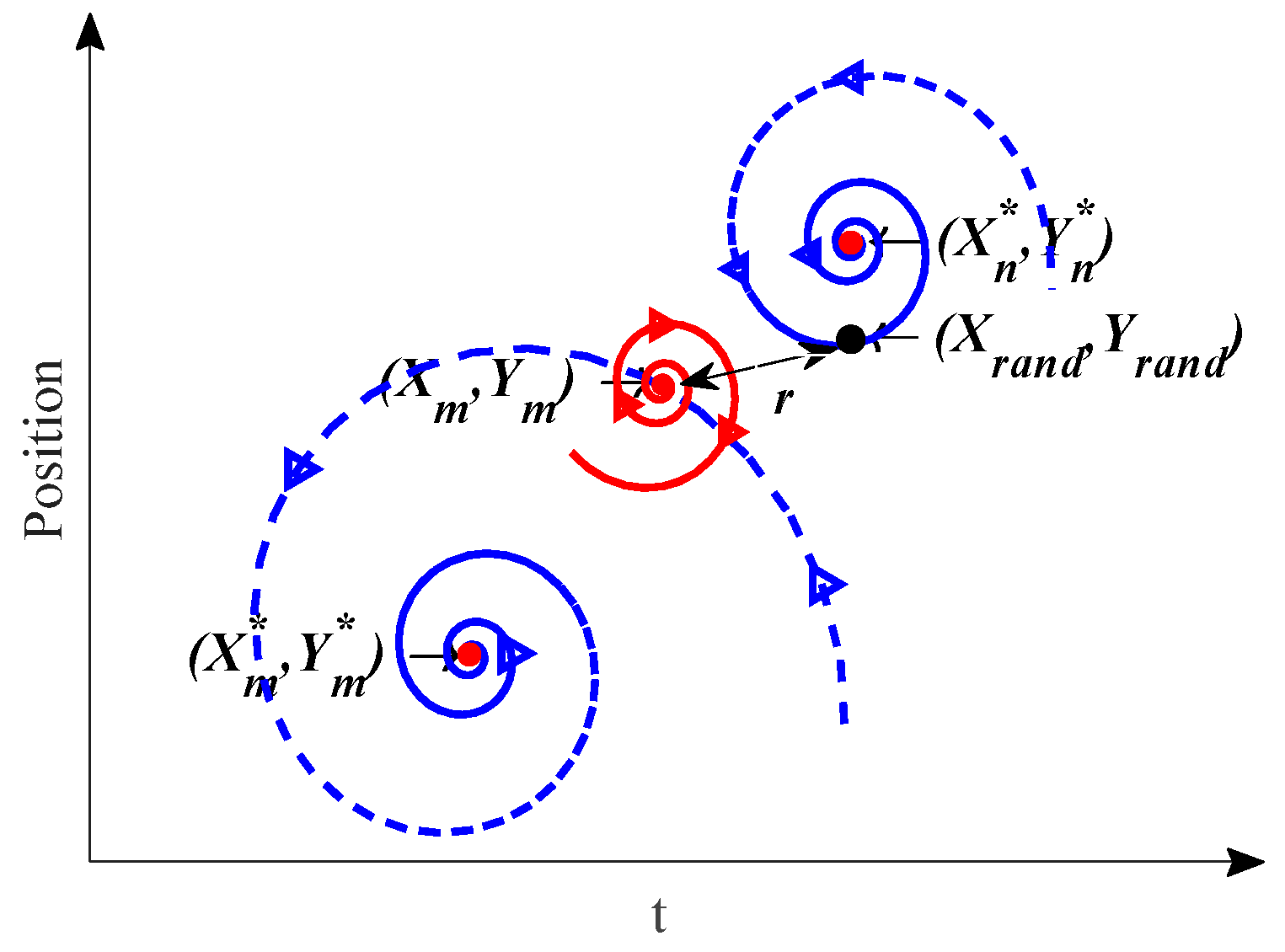
A Double Spiral updating Strategy (DSS) based on the golden spiral is designed to update the position of individuals along a specific spiral path. The DSS and the bubble-net attacking method use different spiral properties to achieve inward exploitation and outward exploration, which can address the issue of stalling to the local optimum solutions.
A Baleen neighborhood Exploitation Strategy (BES) combines quadratic interpolation (QI) with baleen-assisted predation behavior to enhance the candidate solutions and accelerate convergence. This strategy explores more prospective areas by focusing on the area near the best location, which provides a good direction for the population in the early optimization process.
The performance of the MSWOA-based feature selection algorithm is tested on 20 UCI classification datasets and a realistic classification optimization problem.
The MSWOA is tested against six variants of the WOA and six other advanced swarm intelligence optimization approaches. Extensive feature selection experiments indicate that MSWOA obtains a more promising overall performance compared with the comparison algorithms on most datasets.
The remaining organization of this paper is as follows:
Section 2 provides the various works related to feature selection.
Section 3 describes an introduction to the original WOA.
Section 4 details the improvement points of the MSWOA approach.
Section 5 compares MSWOA with other algorithms and analyzes the reported results from the experiments on various benchmark datasets and real-world problems.
Section 6 summarizes this paper and suggests the subsequent research options.
2. Related Work
The feature subset chosen using feature selection approaches contains valuable information, which reduces the training time and facilitates the classification accuracy. Thus, FS can cope with the challenges of high-dimensional data with large quantities of information and complex feature interaction. FS approaches are broadly categorized into filter-based, wrapper-based, and embedding-based methods [
33].
Filter-based methods sort features according to different criteria and then select features that meet the threshold and generate a feature subset. This method is independent of the classification algorithm, which gives it the advantage of being computationally efficient. There are several commonly used filtering methods. Eiras-Franco et al. [
34] described a new version of ReliefF that simplifies the most expensive phase by approximating the nearest neighbor graph with locality-sensitive hashing (LSH). Song et al. [
35] presented a new feature ranking approach on the basis of Fisher discriminant analysis (FDA) and an F-score, which obtained a suitable subset by maximizing the proportion of the average between-class distance to the relative within-class scatter. Other improved approaches in this category include minimal redundancy maximal new classification information (MR-MNCI) [
36] and information gain (IG) [
37]. However, the selected feature subset using filters has low prediction accuracies due to the lack of a specific learning algorithm guiding the feature selection phase [
38].
Wrapper-based methods introduce the learning algorithm to evaluate the quality of feature subsets and consider the complex interactions between the features. A learning algorithm is used in each iteration to assess whether the candidate feature subset is superior to the current best subset [
39]. As learning algorithms for evaluating features, the decision tree (DT), K-nearest neighbor (KNN), naïve Bayes (NB) and support vector machine (SVM) have been frequently utilized. In this case, wrappers have better classification accuracy than filters. Recursive feature elimination (RFE) is a traditional wrapper FS method that employs a machine learning model for training and constantly removes features based on the weight value. Han et al. [
40] presented a dynamic recursive feature elimination (dRFE) framework. Another typical wrapper feature selection method is the Las Vegas wrapper (LVW) [
41]. However, the adoption of learning algorithms incurs a considerable computational cost if the search space is vast [
42]. The conventional optimization techniques that search for the optimal feature subset becomes extremely difficult. Therefore, with their powerful global search ability, metaheuristic approaches have been successfully employed in feature selection, these include the ant colony optimization (ACO) and the grasshopper optimization algorithm (GOA).
Embedded-based approaches combine the model construction of the learning algorithm with the feature selection process. The regularization method is one of the most common embedded methods. Based on multimodal neuroimaging, Zhang et al. [
43] introduced a new multiclass categorization framework for Alzheimer’s disease (AD) with the introduction of feature selection and fusion, which used a l2,1-norm regularization term and a lp-norm (1 <
p < ∞) regularization term. Other algorithms also include the particle swarm optimization-based multiobjective memetic algorithm (PSOMMFS) [
44], classification and regression trees (CARTs) [
45], decision tree ID3 algorithm [
46], etc. Although embedded methods have good classification performance and efficiency, their approach structure is more complicated and ignores feature interactions. Therefore, this paper utilizes a wrapper method based on a metaheuristic algorithm to address the feature-selection problem.
Among the existing approaches, metaheuristics are employed in feature selection in various forms, with hybrid algorithms being one of the most used. Amini and Hu [
47] presented a novel hybrid two-level FS method, which combines a GA and EN to construct an appropriate subset of predictors to improve computational efficiencies and prediction performances. Ibrahim et al. [
48] integrated the SSA with PSO to boost exploitation abilities and diversity, helping to swiftly obtain the optimal feature subset. This paper presented a novel algorithm for finding high-quality solutions for feature-selection tasks by incorporating the SSA into the Harris hawks optimization (HHO) algorithm [
49]. Additionally, academics frequently introduce different strategies in metaheuristics to improve classification performance. Li et al. [
50] suggested an enhanced PSO algorithm for feature selection. The personal best position was updated with genetic operators in this technique to avoid the local optimum. Chen et al. [
51] presented a novel PSO-based feature selection approach that uses information about the swarm to produce more potential solutions. An improved discretization-based PSO for FS (IDPSO-FS) in [
52] was developed to identify each selected feature index accurately. The authors of [
53] suggested an improved version of the salp swarm algorithm (ISSA) to solve feature selection problems and to select the optimal subset of the features in wrapper mode. In [
54], an improved binary global harmony search algorithm (IBGHS) with a modified improvisation step was presented. Furthermore, several recent articles have combined filter approaches and metaheuristic-based wrapper approaches. Ghosh et al. [
55] developed a wrapper-filter combination on the basis of ACO, in which a filter approach is used to evaluate a subset to reduce the computing complexity. In the same year, Moslehi and Haeri [
56] presented a hybrid filter-wrapper method that integrates evolutionary-based GA and PSO; an artificial neural network was included into the fitness function.
The WOA is an evolutionary algorithm with simple principles and an easy implementation. Moreover, the WOA exhibits superior performance in solving FS problems. Tubishat et al. [
57] incorporated IG filter-feature-reduction techniques with the WOA to capitalize on its advantages and compensate for its shortcomings. The IWOA used the EOBL technique to improve the diversity of the initial solutions and DE evolutionary operators to improve the solutions after each iteration. The IWOA outperformed the other approaches (i.e., WOA, DE, PSO, GA, ALO) on four datasets. To achieve automatic lung tumor detection, a hybrid bioinspired algorithm (WOA_APSO) was developed that combined the benefits of the WOA and APSO to select the optimized feature subset [
58]. The experimental results revealed that it outperformed the other advanced methods and achieved 97.18%, 97%, and 98.66% accuracy, sensitivity, and specificity, respectively. In [
59], a wrapper-based FS method that hybridized GWO and WOA was developed, aggregating the advantages of their exploration and exploitation. The results showed that HSGW outperforms several well-known feature selection methods (i.e., BGOA, BGSA, GA, and PSO). Mafarja et al. [
60] presented a new WOA-based FS by introducing both V-shaped and S-shaped transfer functions, which were adopted in IoT attack detections. This paper integrated five natural selection operators into WOA to enhance the search efficacy in dealing with FS tasks [
61]. The proposed wrapper approach was used to determine the most valuable features from the software fault prediction (SFP) datasets. To find the best subset, two binary variants of the WOA were presented [
62], which used the sigmoid transfer function and the sigmoid transfer function, respectively. There is also a substantial body of research describing the different WOA variants used in feature selection [
63,
64,
65].
Based on the no-free lunch (NFL) theorem [
66], no single strategy can solve all the WOA issues. Thus, there is still some room for further improvement. Focusing on the problem of insufficient global search capability and frequent search stagnation, this paper proposes an innovative wrapper approach called the multispiral whale optimization algorithm (MSWOA). We divide the population into many subpopulations using the novel multipopulation technique, which may enlarge the search space and yield more candidate subsets. In addition, a new stochastic exploration strategy is provided to assist individuals in breaking free from local optima. Furthermore, to maximize the quality of the optimal individual, we apply an exploitation method based on humpback whale behaviors. The MSWOA is tested on 20 UCI datasets and in two group contrast trials. The results show that MSWOA can obtain feature subsets with higher classification accuracies and fewer features on most datasets.
Section 4 will go through the MSWOA in further detail.
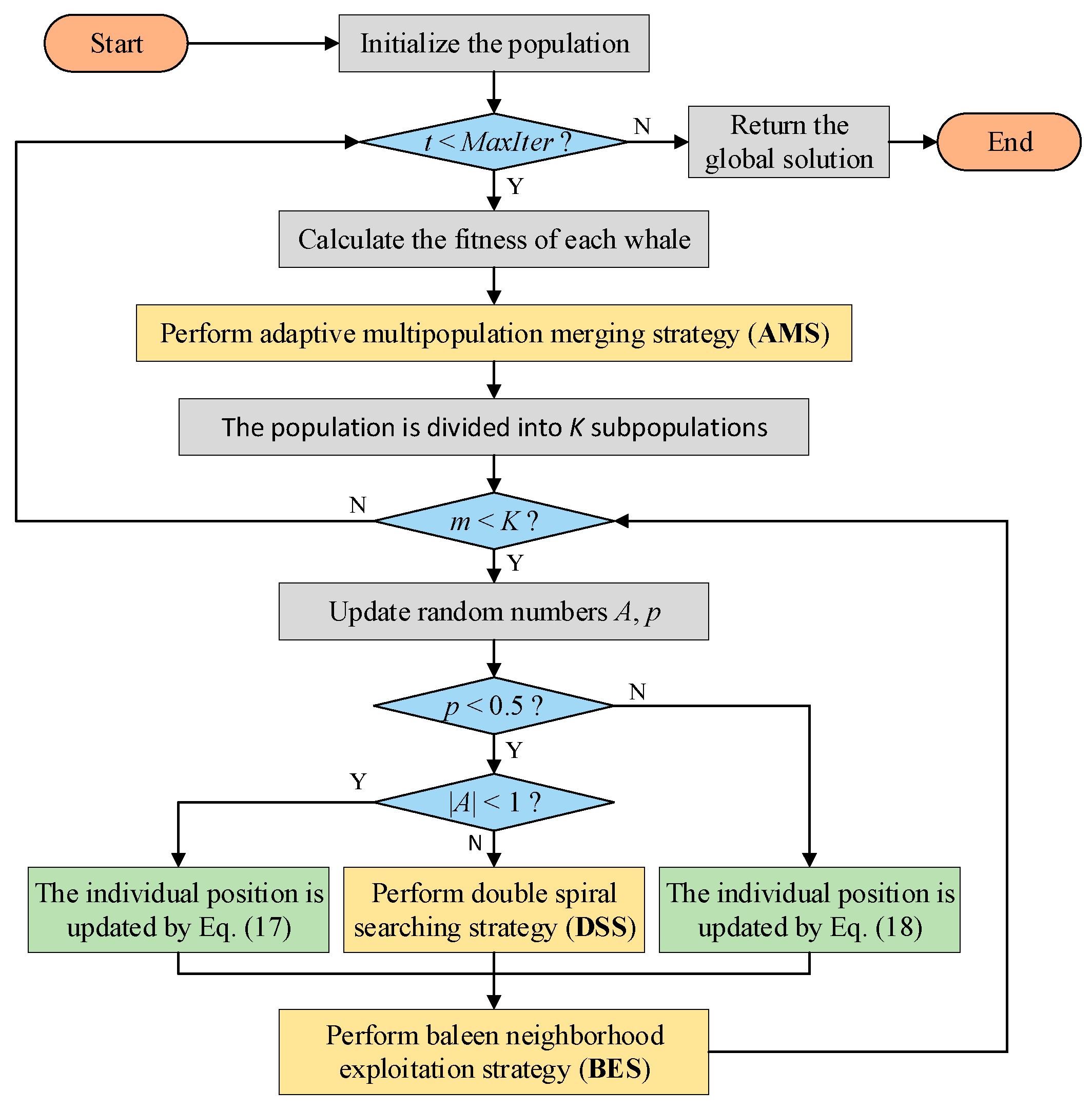
5. Framework of MSWOA
The pseudocode and flow chart of MSWOA are shown in Algorithm 4 and
Figure 3, respectively. The optimization process mainly includes three stages: dividing the population, updating the positions and enhancing the optimal value of the subpopulation. The MSWOA starts with an initial population and then divides it into
K subpopulations using the AMS approach. Each subswarm has an optimal individual
Xm*. Accordingly, Equations (2) and (6) are revised to Equations (17) and (18).
| Algorithm 4. MSWOA |
| 1 Initializing the population Xi (i = 1, 2…, n) |
| 2 while (t < MaxIter) |
| 3 Divide population using AMS |
| 4 for each subpopulation |
| 5 for each solution of subpopulation |
| 6 Update a, l, p, A, and C |
| 7 If (p < 0.5) |
| 8 If (|A| < 1) |
| 9 Calculate the new position of the current individual according to Equation (17) |
| 10 else if (|A| > 1) |
| 11 Randomly select a solution (Xrand) |
| 12 Calculate the new position of the current individual according to Equation (13) |
| 13 end |
| 14 else if (p > 0.5) |
| 15 Calculate the new position of the current individual according to Equation (18) |
| 16 end |
| 17 end |
| 18 Update Xm* by BES |
| 19 end |
| 20 Obtain the global optimal solution X* by comparing Xm* (m = 1, 2…, K) |
| 21 t = t + 1 |
| 22 end |
| 23 return X* |
In the second step, according to the values of the random parameters p and A, individuals are updated using one formula from Equations (13), (17), or (18). After updating each subpopulation, the BES strategy is conducted to purposefully explore the optimal value. If the fitness value of the obtained solution is better, Xm* will be updated. The optimal values of K subpopulations are compared at the end of each iteration, and the global optimal solution is the search agent with the best fitness value.
The computational complexity of MSWOA depends on the maximum number of iterations (MaxIter), subpopulation size (M), subpopulation number (K), dimension size (d), and population size (N). The time complexity of the improved algorithm mainly relies on four parts: initialization, Adaptive Multipopulation merging Strategy (AMS), Double Spiral searching Strategy (DSS), and Baleen neighborhood Exploitation Strategy (BES). The initial stage is O(N × d). The complexity under AMS is O(N). The adaptability of the DSS is assessed to be O(K × M × d). The solution update mechanism of BES is O(K × M × d + K × M). The total time complexity is O(MSWOA) = O(N × d) +O(N + K × M × d + K × M).
5.1. Fitness Function
The evaluation of feature subsets is a crucial aspect of wrapper-based approaches. Generally, the evaluation metrics are classification accuracy and the number of features. In this work, the classification accuracy is calculated by the k-nearest Neighbor (
k-NN) classifier (where
k = 5) [
70]. Since the classification accuracy needs to be maximized while the number of features needs to be minimized, the classification error rate is chosen as an evaluation criterion. In this paper, the fitness function combines the classification error rate and the number of selected features, as shown in Equation (19). Therefore, the criterion is to minimize the fitness value [
71].
where
ErrorRate is the classification error rate,
R is the selected features, and
N is the full set of features.
α and
β are the parameters that regulate the ratio of that classification rate and length of a subset, and
and
. In this work, we set
α = 0.99 [
72].
5.2. Experimental Results and Discussion
Each experiment is completed using MATLAB2017. The machine used to conduct the experiments is configured with an Intel Core i5-10400 2.90 GHz CPU and 16.0 GB RAM. The evaluator is the
k-NN classifier, and each dataset is randomly partitioned into a training set and a test set according to the ratio of 80% and 20% [
73].
Cross-validation is a more comprehensive model evaluation method that makes effective use of the dataset to enhance the stability and reliability of the model [
74]. In comparison to a single train–test split, 5-fold cross-validation divides the entire dataset into five equal parts. In each iteration, one part is used as the validation set, while the remaining four parts are used for training. This process is repeated five times, with a different part serving as the validation set in each iteration. The final model performance is determined by aggregating the results from these five validation rounds. This method helps mitigate the potential evaluation bias introduced by a single validation set and provides a more stable and thorough assessment of model performance [
75].
Moreover, cross-validation improves the generalization ability of the model, ensuring that it performs well across different data subsets, thereby reducing the risk of overfitting [
76,
77,
78]. Additionally, it maximizes the utility of each data sample, making it particularly beneficial in cases where the dataset is limited in size. Finally, cross-validation reduces the variability in evaluation results due to the randomness of data splits by repeatedly validating the model on different data partitions, leading to more reliable performance metrics.
For all trials, 5-fold cross-validation is utilized to verify the performance of the selected feature subset to reduce the overfitting issue.
5.3. Datasets and Evaluation Metrics
Twenty different benchmark datasets are gathered from the University of California at Irvine (UCI) [
79] to assess the performance of the proposed MSWOA. The details of all the datasets are presented in
Table 1. The dimensions of these datasets range from 60 to 22,283, and the datasets were collected from various fields, such as biological/gene expression (Lung_Cancer, Leukemia, Prostate_Tumor), physic (Sonar), speech recognition (Isolet), and face images (Yale, orlraws10P, warpPIE10P). Hence, the performance of the MSWOA can be thoroughly investigated.
The experiments use the following evaluation metrics: average fitness values, average classification accuracy, the number of selected features, convergence curves, and run time. The primary performance indicator for assessing the algorithms is the classification accuracy. In addition, we conduct a significance test and use the symbols ‘+’ ‘−’ ‘≈’ to show various differences in the table, where ‘+’ indicates the MSWOA outperforms the competitor algorithm significantly, ‘−’ indicates the competing algorithm greatly outperforms the MSWOA, and ‘≈’ indicates there is no significant difference. The population size is set according to the corresponding papers [
72,
80,
81,
82]. The maximum number of iterations is 100 and the experiments are executed 30 times on each dataset. The feature selection threshold is set to 0.5, which means that if the value is more than 0.5, the feature is chosen; otherwise, the feature is not chosen.
5.4. Comparison Between the WOA-Based Approaches
To verify the superiority of the proposed MSWOA, it is compared with the original WOA and five recent WOA-based methods, including the RDWOA [
83], SBWOA [
84], QWOA [
85], WOASA [
86], and WOACM [
87].
Table 2 displays several main parameters of the algorithms that have been well tuned.
The average classification accuracy and standard deviations of seven algorithms for the test sets are reported in
Table 3, with the numbers in bold indicating the best results. As seen from
Table 3, MSWOA outperforms the other variants on 17 out of the 20 datasets. On the PDC, Nci9, and GLIOMA datasets, WOASA exceeds the other approaches and MSWOA achieves the second-highest classification accuracy. On the SRBCT and Leukemia datasets, QWOA, WOACM, and MSWOA all attain 100% classification accuracy. For the Lung_discrete dataset, MSWOA achieves an accuracy of 94.29%, outperforming the second-best result of 92.38%. Furthermore, significance testing indicates that MSWOA holds significant advantages over competing algorithms on most datasets, including Yale, Lung, and Lung_Cancer. In addition, the significance test results indicate that the MSWOA has apparent advantages over the competitive algorithms on most datasets, such as Yale, Lung, and Lung_Cancer. The excellent performance of MSWOA is mainly due to the proposed AMS technique, which boosts the global search capability to explore the high-performance space. Simultaneously, the BES strategy enhances individual quality in the first 20 iterations, providing a better exploration direction for the population.
The average number of selected features of seven methods on 20 datasets is provided in
Table 4. The experiment indicates that MSWOA selects fewer features than other algorithms in 15 of the 20 datasets. RDWOA performs second only to MSWOA, with the best results on 3 datasets (i.e., PDC, warpPIE10P, and Arcene). SBWOA performs best on 2 datasets (i.e., Semion and Isolet). Moreover, the average number of selected features of MSWOA is 364.3767 and it ranks first, followed by RDWOA, SBWOA, WOASA, WOA, WOACM, and QWOA. The AMS strategy is the primary rationale for selecting the smallest number of features. Using the proposed AMS, we can evaluate numerous candidate feature subsets rather than just one in each loop, increasing the probability of finding the best answer. To some extent, the AMS also helps to enhance the classification accuracy. For example, on the GLI_85 dataset, MSWOA selects the fewest features compared to the other algorithms and ranks first with a classification accuracy of 100%. In conclusion, the suggested MSWOA determines the best feature combination with high classification accuracy and very few features.
The fitness value can be used to deeply investigate the robustness of the MSWOA.
Table 5 shows the standard deviations and average fitness of the seven methods, from which we can see that the MSWOA achieves a better performance than other six methods on 17 out of the 20 datasets. WOASA obtains the smallest fitness values in the remaining three datasets, and MSWOA is slightly worse. WOACM comes in second out of the 6 datasets and is quite close to MSWOA. In addition, the significance test results demonstrate that MSWOA performs better than other methods on most datasets, particularly when compared with WOA, QWOA, and WOACM. MSWOA performs best on the Semion, Lung, 9_Tumors, and Lung_Cancer datasets, with the other algorithms trailing far behind. From the overall results, MSWOA has outstanding performance on different dimensional datasets, which verifies the excellent robustness of MSWOA.
To demonstrate the convergence capability of the proposed MSWOA, the variation curves of the fitness of the seven algorithms on 17 datasets are depicted in
Figure 4, from which we can see that the fitness of MSWOA converges fast and achieves the best results in most cases. To explain the change in the mean fitness values, several datasets are presented as examples. For instance, on the Lung_discrete, the fitness value of MSWOA drops to approximately 0.09 after around 25 iterations, whereas the best fitness value among the other six algorithms remains at approximately 0.12. On the 11_Tumors, Isolet, and Lung_discrete datasets, MSWOA converges much faster than the other methods in the entire procedure. On the 9_Tumors, Orlraws10P, and Semion datasets, some algorithms (e.g., WOA, RDWOA, QWOA, SBWOA) perform slightly better than MSWOA in the early phase, but MSWOA can seek better positions and quickly exceeds the others by using the BES strategy. On the Colon, Lung_Cancer, and Yale datasets, MSWOA continuously eliminates search stalls, where the other algorithms are no longer updated. The Leukemia dataset shows that although the other algorithms can also handle the stagnant dilemma, their final fitness values are worse than MSWOA. This is due to the AMS strategy’s enhanced exploration ability, which discoveries additional potential locations.
Training time is a crucial metric for assessing the efficiency of FS algorithms. The average training time for all competing approaches is shown in
Table 6. According to the experimental results, the execution time of MSWOA increases slightly when compared with WOA except for the Isolet dataset, because the BES strategy requires one additional estimation of the fitness value. The WOA and RDWOA attain the lowest average training time on nine datasets, respectively. The SBWOA consumes less time on two datasets. According to the average training time, WOA has the fastest calculation speed, followed by SBWOA, RDWOA, MSWOA, WOACM, QWOA, and WOASA. Although RDWOA has the shortest training time on nine datasets, it performs poorly on high-dimensional datasets. In contrast, MSWOA is competitive with the other algorithms on high-dimensional datasets. For example, on the orlraws10P dataset, MSWOA is less than RDWOA, QWOA, WOASA, and WOACM and comparable to SBWOA. Furthermore, it is far superior to the comparison methods in the accuracy, fitness, and screening feature. As a result, MSWOA obtains a more comprehensive feature subset at a reduced cost, especially for high-dimensional datasets.
5.5. Comparisons with Other Well-Known Optimization Algorithms
In this subsection, MSWOA is compared with six different metaheuristic methods to further validate its effectiveness, including MPSO [
88], HGWOP [
89], HGSO [
81], DSSA [
90], BGOA [
80], and HBBOG [
91].
Table 7 displays several main parameters of the methods that have been well tuned.
The average classification accuracy rates, standard deviations, and rankings of MPSO, HGWOP, HGSO, DSSA, BGOA, HBBOG, and MSWOA are presented in
Table 8, where the numbers in bold indicate the optimal results. As shown in
Table 8, MSWOA has the highest classification accuracy on 15 out of the 20 datasets. On the Sonar, Semion, and Isolet datasets, HGWOP takes first place and the significance test results show little difference between HGWOP and MSWOA. HGSO slightly outperforms MSWOA on the Lung_Cancer datasets. BGOA rounds out the top on the Lung dataset, while the proposed MSWOA ranks second. On the SRBCT and GLI_85 datasets, HGSO and BGOA reach the same level of accuracy as MSWOA, respectively. On the Arcene dataset, MSWOA, HGSO, BGOA, and HBBOG achieve close classification accuracies. Furthermore, the results of the significance tests reveal that MSWOA performs better than other comparison methods on most datasets. For example, when compared to DSSA, MSWOA remained significantly different on 17 datasets and performed similarly on 3 datasets.
According to
Table 9, MSWOA produces a smaller number of features than other approaches in 15 out of the 20 datasets. HGSO performs slightly better than MSWOA on the warpPIE10P, 9_Tumors, and Prostate_Tumor datasets. On the PDC and GLIOMA datasets, BGOA is superior to MSWOA. The average number of features selected by MSWOA is 362.2713, which is less than that of any other algorithm. Observing the final ranking, the MSWOA achieves the best results, followed by HGSO, BGOA, MPSO, HBBOG, HGWOP, and DSSA. For high-dimensional datasets, the effect of MSWOA is more pronounced. For example, on the Arcene dataset, the average number of features selected by MSWOA is 600.7333, barely 6% of the total features. As a whole, MSWOA can achieve a higher classification accuracy while using fewer features on high-dimensional and complex datasets. The result also demonstrates that the MSWOA has a significant advantage over the other algorithms in terms of the convergence speed. Considering all the results above, MSWOA is a very competitive feature-selection algorithm. Although MSWOA demonstrates advantages in various aspects, such as achieving higher classification accuracy on 15 out of 20 datasets and significantly reducing the number of selected features, its limitations warrant deeper exploration.
Firstly, while the algorithm’s performance generally surpasses other methods, it did not show substantial superiority over HGSO and BGOA on specific datasets like Lung, SRBCT, and GLI_85. This suggests that MSWOA may have room for further optimization when handling high-dimensional data and complex feature distributions.
5.6. MSWOA for Human Fall Recognition
5.6.1. Problem Description
In this section, the proposed MSWOA is employed as an FS method in a human fall-detection system to enhance the classification recognition. The fall-detection system allows the elderly to obtain the necessary assistance in time after an accidental fall, reducing injuries caused by falls. Human activities are complex composites composed of various simple motions, influenced by diverse contexts, environments, and individual differences, which contribute to variations in behavior [
92,
93,
94]. Consequently, machine learning models trained on incomplete samples and limited features typically exhibit poor generalization performance, making it difficult to accurately differentiate between fall incidents and routine daily activities. To ensure an adequate dataset and diverse feature types, this chapter outlines the design of a data-collection scheme for fall detection.
The current literature divides fall-detection systems into three broad classes, i.e., wearable device, computer vision, and environmental sensor systems [
95]. Nevertheless, the latter two systems have the disadvantage of a limited monitoring range, which results in fall detection failing when the user is not within the sensor coverage region. In addition, a vision-based fall-detection system will bring users a huge risk of privacy disclosure. Therefore, the wearable system is selected for a fall-detection experiment because of the benefits of a low cost, simplicity, and low privacy disclosure risk.
Figure 5 illustrates the overall flow of the human fall-detection scheme, including the hardware circuitry, data collection, feature extraction and selection, classification, and results transmission. The hardware consists of a microprocessor, a battery, an inertial sensor module (i.e., MPU6050), a wireless serial module, and an alarm module (i.e., buzzer). MPU6050 has an accelerometer and a gyroscope, which can capture acceleration and angular velocity values and automatically calculate three angles (i.e., pitch, yaw and roll). The sensor’s accelerometer has a measurement range of ±16 g, while the gyroscope’s range is ±2000°/s. The angular range for the X and Z axes is ±180°, and for the Y axis, it is ±90°. The measurement accuracy of the posture angles is 0.05° when static and can reach 0.1° in dynamic conditions, ensuring high measurement precision. To form the original dataset, 13 time-domain features, 6 frequency-domain features, and 8 time-frequency features are extracted from the raw data. Furthermore, the feature subset containing the most information is selected as the input data for the classifier (SVM) using a feature selection method. If the recognition result is a fall, an alert signal is transmitted to both the guardian’s phone and the wearer’s device.
5.6.2. Results and Discussion
The dataset used in the experiment is called SFall, which consists of activities of daily living (ADLs) and falls performed by volunteers. The experiment included seven participants—two females and five males. Due to the inherent risk of fall activities, elderly volunteers were not involved, and all participants were students with an average age of 22 years, an average height of 174.7 cm, and an average weight of 65.2 kg. The fall detection system was configured with a sampling frequency of 20 Hz, and each sampling session was set to a duration of 10 s. The SFall dataset comprises 157 collected samples, each with 200 data points and 243 features derived from acceleration, angular velocity, and angular displacement data.
Table 10 shows six falls and eight ADLs. These features include 13 time-domain, 6 frequency-domain, and 8 time-frequency domain metrics, extracted across three axes (x, y, z) from MPU6050 sensor data. The volunteers fix the equipment at the waist and perform a set of activities to collect data.
Figure 6 depicts the experimental scenario of some of the actions. The sampling frequency for the data-collection experiment is set to 20 Hz. The acquisition time is 10 s for all falls, and recalibration was performed after every session. As shown in
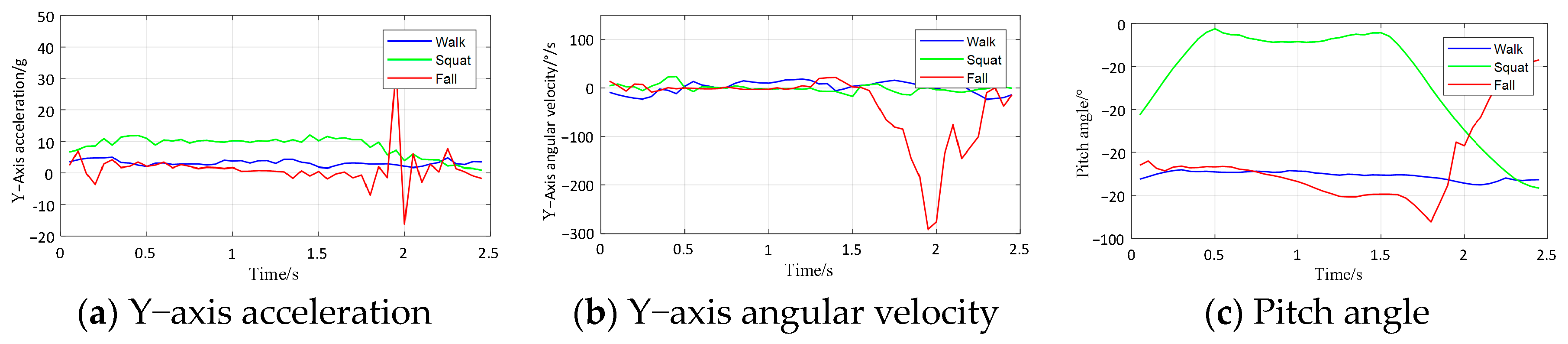
Figure 7, the data signals of the Y-axis acceleration, Y-axis angular velocity, and pitch angle for the three behaviors (walk, squat, and fall) have different magnitudes of variation, which indicates that the collected data can extract effective features to separate falls from ADLs.
On the SFall dataset, MSWOA is experimented against three advanced methods to evaluate their performance, including SASS [
96], COLSHADE [
97], and sCMAgES [
98]. To obtain stable results, the test is run 30 times, and the classifier is Support Vector Machine (SVM). The evaluation metrics are the F1-score (FS) and confusion matrix. As shown in
Table 11, TP is the number of falls correctly predicted as falls, FP is the number of ADLs incorrectly predicted as falls, FN is the number of falls incorrectly predicted as ADLs, and TN is the number of ADLs correctly predicted as ADLs. According to the experimental results in
Table 12, MSWOA has obtained the best performance. Based on the F1-score analysis, MSWOA achieved a score of 89.6567%, which is 1.5 percentage points higher than the second-ranked SASS algorithm. According to the table, the accuracy of MSWOA is the highest, which is 20% higher than sCMAgE. Based on the confusion matrix, MSWOA predicts the highest number of correct falls with 12.2 and the least number of overall false predictions (FN+FP) with 2.8, indicating that the algorithm possesses robust recognition capabilities for both fall and non-fall behaviors. Thus, the MSWOA-enabled human fall detection system has more robust recognition performance, which proves that the MSWOA method has promising applications in realistic optimization problems.
6. Conclusions
Because of the insufficient randomness of the exploration formula and the single leader, WOA-based feature-selection techniques often suffer from premature convergence and entrapment in local optimal solutions. Therefore, a multispiral WOA (MSWOA) for FS is proposed in this paper to address these dilemmas. The MSWOA includes three major improved modifications. Initially, we utilized an Adaptive Multipopulation merging Strategy (AMS) with a novel allocation method for population delimitation. Each subpopulation chooses its best individual to be the leader, which expands the exploration space and diversity. Additionally, using the Double Spiral updating Strategy (DSS), individuals take their current position as a starting point and randomly explore outward along the spiral trajectory. In this case, the search process is not limited by the distance between the individuals, which prevents search stagnation in the later iterations. Finally, a Baleen neighborhood Exploitation Strategy (BES) with quadratic interpolation was incorporated into the early search phase to obtain better positions and facilitate convergence by searching for regions near the optimal search agent. The proposed MSWOA approach was evaluated against the original WOA and 11 other recent algorithms, including 5 WOA-based and 6 metaheuristic variants on 20 UCI datasets. The results demonstrated that the MSWOA has advantages in classification accuracy, the number of selected features, and the fitness values on most datasets. In addition, MSWOA exhibited a faster convergence speed when compared to the competitive methods. Moreover, the MSWOA is employed for feature selection in a human fall-detection task, and experimental results indicated that it is capable of solving real-world problems.
There is further work that can be performed in the future, for example, combining the multipopulation technique with other updating mechanisms or utilizing different classifiers (e.g., neural networks) to further evaluate the algorithm’s effectiveness. Moreover, the temporal complexity of the algorithm needs to be further reduced to achieve greater computational efficiency. The new MSWOA algorithm can also be applied to other engineering fields, such as fault diagnosis, image segmentation, and path planning.