Hybrid Reconstruction Approach for Polychromatic Computed Tomography in Highly Limited-Data Scenarios
Abstract
1. Introduction
2. Methods
2.1. L2-PICCS Method
2.2. Split Bregman Algorithm
2.3. DeepBH
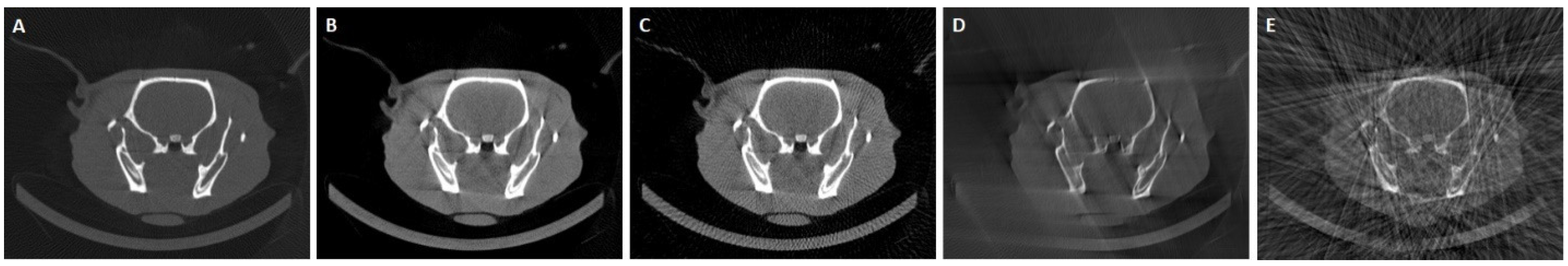
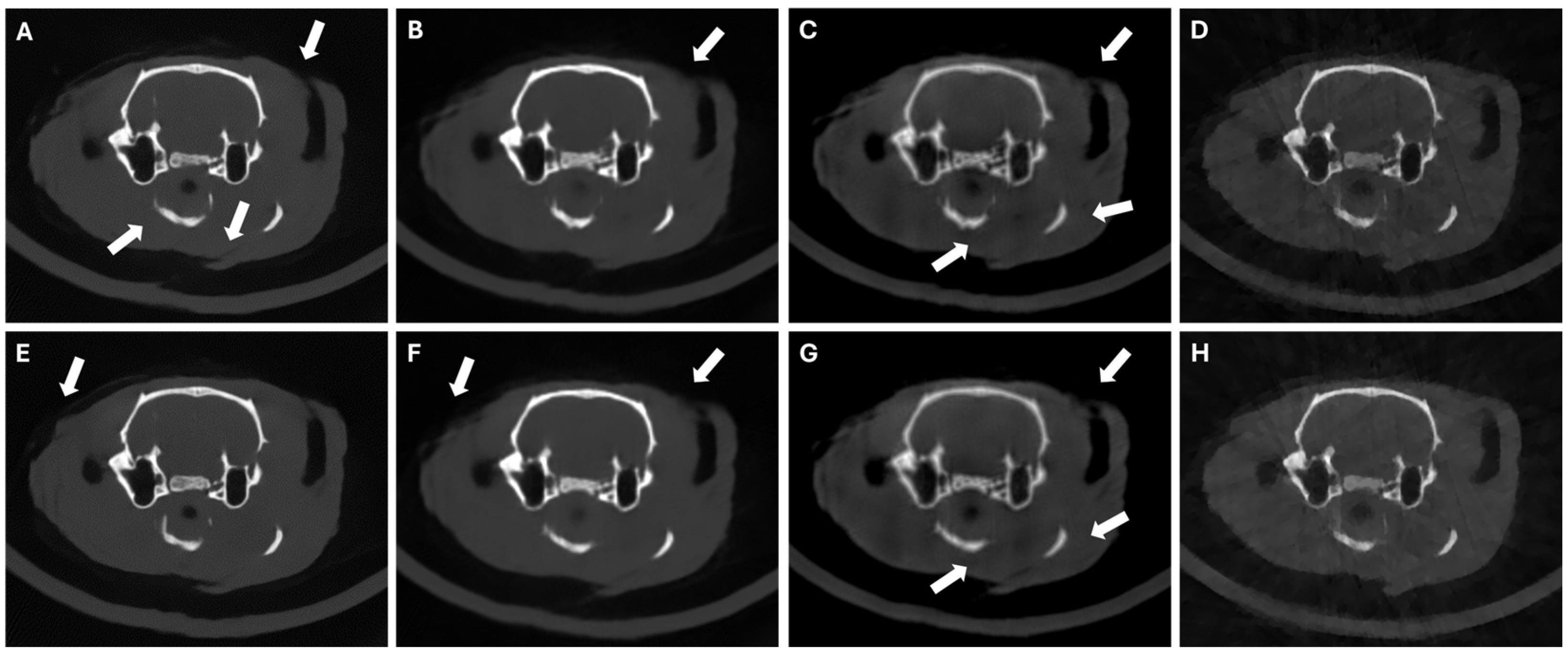
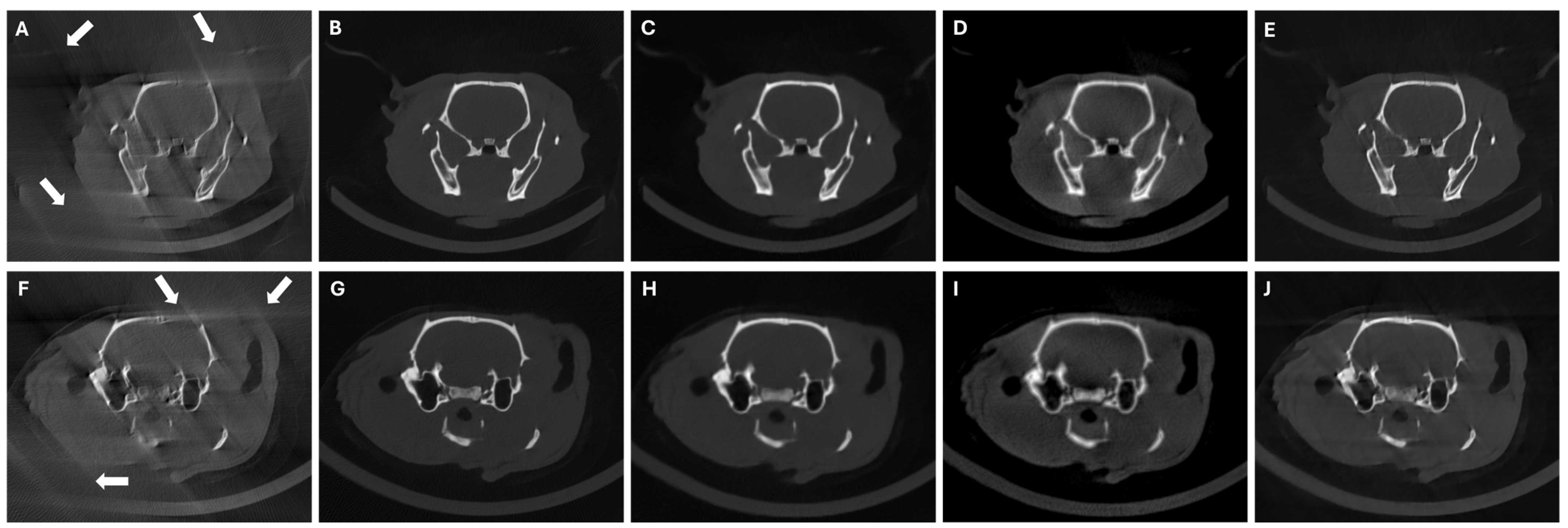
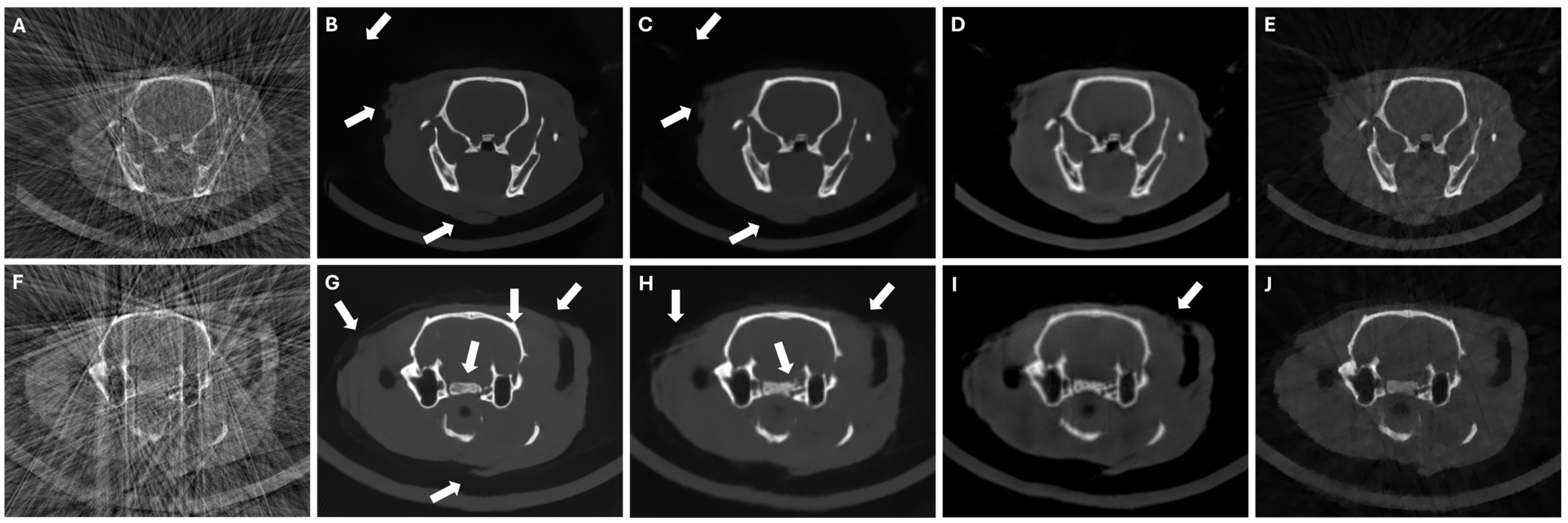
3. Experiments and Results
3.1. Datasets
- Standard dose (SD) scenario. Complete data, that is, 360 projections with a span angle of 360 degrees, resulting in seven studies for training, two for validation, and two for test.
- Low-dose (LD) scenario. We selected every second projection from each study (180 projections covering a 360-degree span), resulting in seven studies for training, two for validation, and two for test.
- Limited span angle (LSA) scenario. This scenario entailed the random selection of three span angles between 90 and 160 degrees for each of the 11 rodent studies. Consequently, we obtained a total of 21 studies for training, 6 for validation, and an additional 6 for testing.
- Limited number of random projections (LNP) scenario. We conducted three rounds of selection, randomly choosing a varying number of projections between 30 and 60 within a 360-degree span for each of the 11 datasets. This process yielded a total of 21 studies allocated for training, 6 for validation, and another 6 for testing.
3.2. Parameters of Iterative Methods and Evaluation Metrics
3.3. Results in Conventional Scenarios
3.4. Results in Highly Limited-Data Scenarios
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barrett, J.F.; Keat, N. Artifacts in CT: Recognition and Avoidance. RadioGraphics 2004, 24, 1679–1691. [Google Scholar] [CrossRef] [PubMed]
- Brooks, R.A.; Di Chiro, G. Beam hardening in x-ray reconstruction tomography. Phys. Med. Biol. 1976, 21, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Nalcioglu, O.; Lou, R.Y. Post-reconstruction Method for Beam Hardening in Computerised Tomography. Phys. Med. Biol. 1979, 24, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Joseph, P.M.; Spital, R.D. A method for correcting bone induced artifacts in computed tomography scanners. J. Comput. Assist. Tomogr. 1978, 2, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Martinez, C.; Fessler, J.A.; Desco, M.; Abella, M. Simple beam-hardening correction method (2DCalBH) based on 2D linearization. Phys. Med. Biol. 2022, 67, 115005. [Google Scholar] [CrossRef]
- Kyriakou, Y.; Meyer, E.; Prell, D.; Kachelrieß, M. Empirical beam hardening correction (EBHC) for CT. Med. Phys. 2010, 37, 5179–5187. [Google Scholar] [CrossRef]
- Schüller, S.; Sawall, S.; Stannigel, K.; Hülsbusch, M.; Ulrici, J.; Hell, E.; Kachelrieß, M. Segmentation-free empirical beam hardening correction for CT. Med. Phys. 2015, 42, 794–803. [Google Scholar] [CrossRef]
- Elbakri, I.A.; Fessler, J.A. Statistical Image Reconstruction for Polyenergetic X-Ray Computed Tomography. IEEE Trans. Med. Imaging 2002, 21, 89–99. [Google Scholar] [CrossRef]
- Martinez, C.; Fessler, J.A.; Desco, M.; Abella, M. Segmentation-free statistical method for polyenergetic X-ray computed tomography with a calibration step. In Proceedings of the 6th International Conference on Image Formation in X-Ray Computed Tomography, Regensburg, Germany, 3–7 August 2020; pp. 118–121. [Google Scholar]
- Sanderson, D.; Martinez, C.; Fessler, J.A.; Desco, M.; Abella, M. Statistical image reconstruction with beam-hardening com-pensation for X-ray CT by a calibration step (2DIterBH). Med. Phys. 2024, 51, 5204–5213. [Google Scholar] [CrossRef]
- Li-ping, Z.; Yi, S.; Kai, C.; Jian-qiao, Y. Deep learning based beam hardening artifact reduction in industrial X-ray CT. CT Theory Appl. 2018, 27, 227–240. [Google Scholar]
- Ji, X.; Gao, D.; Gan, Y.; Zhang, Y.; Xi, Y.; Quan, G.; Lu, Z.; Chen, Y. A Deep-Learning-Based Method for Correction of Bone-Induced CT Beam-Hardening Artifacts. IEEE Trans. Instrum. Meas. 2023, 72, 4504012. [Google Scholar] [CrossRef]
- Kalare, K.; Bajpai, M.; Sarkar, S.; Munshi, P. Deep neural network for beam hardening artifacts removal in image reconstruction. Appl. Intell. 2022, 52, 6037–6056. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Adishesha, A.S.; Vanselow, D.J.; La Riviere, P.; Cheng, K.C.; Huang, S.X. Sinogram domain angular upsampling of sparse-view micro-CT with dense residual hierarchical transformer and attention-weighted loss. Comput. Methods Programs Biomed. 2023, 242, 107802. [Google Scholar]
- Qiao, Z.; Du, C. Rad-unet: A residual; attention-based, dense unet for CT sparse reconstruction. J. Digit. Imaging 2022, 35, 1748–1758. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, H. Convolutional neural network based metal artifact reduction in x-ray computed tomography. IEEE Trans. Med. Imaging 2018, 37, 1370–1381. [Google Scholar] [CrossRef]
- Zhang, P.; Li, K. A dual-domain neural network based on sinogram synthesis for sparse-view CT reconstruction. Comput. Methods Programs Biomed. 2022, 226, 107168. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, S.; Guan, Y.; Maier, A. Limited angle tomography for transmission X-ray microscopy using deep learning. J. Synchrotron Radiat. 2020, 27, 477–485. [Google Scholar] [CrossRef]
- Zhang, H.; Li, L.; Qiao, K.; Wang, L.; Yan, B.; Li, L.; Hu, G. Image prediction for limited-angle tomography via deep learning with convolutional neural network. arXiv 2016, arXiv:1607.08707. [Google Scholar]
- Gu, J.; Ye, J.C. Multi-scale wavelet domain residual learning for limited-angle CT reconstruction. arXiv 2017, arXiv:1703.01382. [Google Scholar]
- Zhang, Q.; Hu, Z.; Jiang, C.; Zheng, H.; Ge, Y.; Liang, D. Artifact removal using a hybrid-domain convolutional neural network for limited-angle computed tomography imaging. Phys. Med. Biol. 2020, 65, 155010. [Google Scholar] [CrossRef]
- Bhadra, S.; Kelkar, V.A.; Brooks, F.J.; Anastasio, M.A. On hallucinations in tomographic image reconstruction. IEEE Trans. Med. Imaging 2021, 40, 3249–3260. [Google Scholar] [CrossRef] [PubMed]
- Cascarano, P.; Piccolomini, E.L.; Morotti, E.; Sebastiani, A. Plug-and-Play gradient-based denoisers applied to CT image enhancement. Appl. Math. Comput. 2022, 422, 126967. [Google Scholar] [CrossRef]
- Hu, D.; Zhang, Y.; Liu, J.; Luo, S.; Chen, Y. DIOR: Deep iterative optimization-based residual-learning for limited-angle CT reconstruction. IEEE Trans. Med. Imaging 2022, 41, 1778–1790. [Google Scholar] [CrossRef] [PubMed]
- Adler, J.; Öktem, O. Learned primal-dual reconstruction. IEEE Trans. Med. Imaging 2018, 37, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Preuhs, A.; Lauritsch, G.; Manhart, M.; Huang, X.; Maier, A. Data consistent artifact reduction for limited angle tomography with deep learning prior. In Proceedings of the International Workshop on Machine Learning for Medical Image Reconstruction, Shenzhen, China, 17 October 2019; Springer: Cham, Switzerland, 2019; pp. 101–112. [Google Scholar]
- Zhang, C.; Li, Y.; Chen, G.H. Accurate and robust sparse-view angle CT image reconstruction using deep learning and prior image constrained compressed sensing (DL-PICCS). Med. Phys. 2021, 48, 5765–5781. [Google Scholar] [CrossRef]
- Chen, G.H.; Thériault-Lauzier, P.; Tang, J.; Nett, B.; Leng, S.; Zambelli, J.; Qi, Z.; Bevins, N.; Raval, A.; Reeder, S.; et al. Time-resolved interventional cardiac C-arm cone-beam CT: An application of the PICCS algorithm. IEEE Trans. Med. Imaging 2011, 31, 907–923. [Google Scholar] [CrossRef]
- Chen, G.H.; Tang, J.; Leng, S. Prior image constrained compressed sensing (PICCS): A method to accurately reconstruct dynamic CT images from highly undersampled projection data sets. Med. Phys. 2008, 35, 660–663. [Google Scholar] [CrossRef]
- Abascal, J.F.; Abella, M.; Sisniega, A.; Vaquero, J.J.; Desco, M. Investigation of different sparsity transforms for the PICCS algorithm in small-animal respiratory gated CT. PLoS ONE 2015, 10, e0120140. [Google Scholar]
- Goldstein, T.; Osher, S. The Split Bregman Method for L1 Regularized Problems. SIAM J. Imaging Sci. 2009, 2, 323–343. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 770–778. [Google Scholar]
- Aitken, A.; Ledig, C.; Theis, L.; Caballero, J.; Wang, Z.; Shi, W. Checkerboard artifact free sub-pixel convolution: A note on sub-pixel convolution, resize convolution and convolution resize. arXiv 2017, arXiv:1707.02937. [Google Scholar]
- Kingma, D.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 1026–1034. [Google Scholar]
- Smith, L.N. Cyclical Learning Rates for Training Neural Networks. In Proceedings of the IEEE Winter Conference on Applications of Computer Vision (WACV), Santa Rosa, CA, USA, 24–31 March 2017. [Google Scholar]
- Hu, Z.; Zheng, H. Improved total variation minimization method for few-view computed tomography image reconstruction. BioMedical Eng. OnLine 2014, 13, 70. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vaquero, J.J.; Redondo, S.; Lage, E.; Abella, M.; Sisniega, A.; Tapias, G.; Montenegro, M.L.S.; Desco, M. Assessment of a New High-Performance Small- Animal X-ray Tomograph. IEEE Trans. Nucl. Sci 2008, 55, 898–905. [Google Scholar] [CrossRef]
- Abella, M.; Serrano, E.; Garcia-Blas, J.; García, I.; De Molina, C.; Carretero, J.; Desco, M. FUX-Sim: An implementation of a fast universal simulation/reconstruction framework for X-ray systems. PLoS ONE 2017, 12, e0180363. [Google Scholar] [CrossRef] [PubMed]
- Necasová, T.; Burgos, N.; Svoboda, D. Chapter 25-Validation and evaluation metrics for medical and biomedical image synthesis. In Biomedical Image Synthesis and Simulation; The MICCAI Society book Series; Burgos, N., Svoboda, D., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 573–600. [Google Scholar]
- Teukolsky, S.A.; Flannery, B.P.; Press, W.H.; Vetterling, W. Numerical Recipes in C; Cambridge University Press: New York, NY, USA, 1992. [Google Scholar]
- Monga, V.; Li, Y.; Eldar, Y. Algorithm unrolling: Interpretable, efficient deep learning for signal and image processing. IEEE Signal Process. Mag. 2021, 38, 18–44. [Google Scholar] [CrossRef]
- Wu, D.; Kim, K.; Li, Q. Computationally efficient deep neural network for computed tomography image reconstruction. Med. Phys. 2019, 46, 4763–4776. [Google Scholar] [CrossRef]




| Scenario | SD | LD | LSA | LNP |
|---|---|---|---|---|
| Training | 3361 | 3361 | 10,083 | 10,083 |
| Validation | 992 | 992 | 2976 | 2976 |
| Test | 992 | 992 | 2976 | 2976 |
| Scenario | Rodent 1 | Rodent 2 | ||||
|---|---|---|---|---|---|---|
| Μ | λ | α | μ | λ | α | |
| LNP | 1.6 | 0.12 | 0.5 | 1.4 | 0.1 | 0.9 |
| LSA | 1.4 | 0.12 | 3 | 1.4 | 0.12 | 3 |
| Random Seed Value | PSNR (dB) | SSIM | CC | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Prior | SART | PICDL | Prior | SART | PICDL | Prior | SART | PICDL | |
| 42 | 25.82 | 23.99 | 32.07 | 0.50 | 0.61 | 0.83 | 0.93 | 0.91 | 0.96 |
| 33 | 25.47 | 24.41 | 31.85 | 0.52 | 0.63 | 0.84 | 0.93 | 0.91 | 0.96 |
| Metric | Reconstruction Method | Rodent 1 | Rodent 2 | ||
|---|---|---|---|---|---|
| LSA | LNP | LSA | LNPs | ||
| PSNR (dB) | FDK | 19.81 | 15.81 | 15.09 | 15.52 |
| Prior | 26.45 | 23.63 | 27.97 | 25.82 | |
| SART | 23.32 | 24.66 | 25.10 | 23.99 | |
| PICDL | 31.37 | 29.37 | 30.24 | 32.07 | |
| SSIM | FDK | 0.680 | 0.357 | 0.512 | 0.327 |
| Prior | 0.674 | 0.454 | 0.738 | 0.500 | |
| SART | 0.733 | 0.619 | 0.732 | 0.610 | |
| PICDL | 0.840 | 0.789 | 0.856 | 0.831 | |
| CC | FDK | 0.793 | 0.782 | 0.824 | 0.722 |
| Prior | 0.968 | 0.964 | 0.935 | 0.925 | |
| SART | 0.912 | 0.933 | 0.907 | 0.913 | |
| PICDL | 0.961 | 0.955 | 0.953 | 0.960 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piol, A.; Sanderson, D.; del Cerro, C.F.; Lorente-Mur, A.; Desco, M.; Abella, M. Hybrid Reconstruction Approach for Polychromatic Computed Tomography in Highly Limited-Data Scenarios. Sensors 2024, 24, 6782. https://doi.org/10.3390/s24216782
Piol A, Sanderson D, del Cerro CF, Lorente-Mur A, Desco M, Abella M. Hybrid Reconstruction Approach for Polychromatic Computed Tomography in Highly Limited-Data Scenarios. Sensors. 2024; 24(21):6782. https://doi.org/10.3390/s24216782
Chicago/Turabian StylePiol, Alessandro, Daniel Sanderson, Carlos F. del Cerro, Antonio Lorente-Mur, Manuel Desco, and Mónica Abella. 2024. "Hybrid Reconstruction Approach for Polychromatic Computed Tomography in Highly Limited-Data Scenarios" Sensors 24, no. 21: 6782. https://doi.org/10.3390/s24216782
APA StylePiol, A., Sanderson, D., del Cerro, C. F., Lorente-Mur, A., Desco, M., & Abella, M. (2024). Hybrid Reconstruction Approach for Polychromatic Computed Tomography in Highly Limited-Data Scenarios. Sensors, 24(21), 6782. https://doi.org/10.3390/s24216782






