A Microbial Cocaine Bioreporter
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Bacterial Strain
2.3. Plasmid Design and Construction
- Cocaine sensor plasmids
- I.
- Plasmid pCocE-benR contained the genes encoding the cocaine esterase enzyme (cocE) and the P. putida transcription factor gene benR. The benR sequence was amplified from plasmid pBEAST-BenR (a gift from Dr. Jerome Bonnet, CNRS, France; Addgene plasmid #114597) and was inserted into plasmid pSB4C5_J23101-CocE (a gift from Dr. Jean-Loup Faulon, INRAE, France; Addgene plasmid #128129).
- II.
- The second plasmid, pBen::luxPleio, harbored a fusion of the P. leiognathi luxCDABEG gene cassette to the BenR-inducible P. putida promotor PBen. It is based on the C55_luxPleio plasmid described by Shemer et al. (2020), the yqjF gene promotor in which was replaced by double restriction enzyme digest with the PBen gene promoter using Gibson assembly.
- Benzoate sensor plasmid
- III.
- A third plasmid (pBen::luxPleio, Figure 1C,D) was constructed for the purpose of improving Pben responses to benzoate by accelerated evolution. The plasmid included the benR gene sequence, along with the E. coli arabinose-inducible expression system, composed of the PBAD promoter and its regulatory gene araC [19]. The benR sequence was amplified from plasmid pCocE-benR (this work), and those of araC and PBAD were amplified from plasmid PBAD-mTagBFP2 (Addgene cat. 34632). The vector backbone was digested with SalI, and the PCR products were assembled in the digested vector pBen::luxPleio using Gibson assembly.
| Component | Origin | Description |
|---|---|---|
| cocE | Rhococcus sp. | E. coli-optimized cocaine esterase; cleaves cocaine into ecgonine methyl ester and benzoate. |
| benR | P. putida | Regulatory gene of the ben operon, activated by benzoate [20]. |
| PBen | P. putida | The ben operon promoter, induced by a BenR/benzoate complex. |
| luxCDABEG | P. leiognathi | Bioluminescence gene cassette of Photobacterium leiognathi; luxA and luxB encode the heterodimeric luciferase, luxCDE a fatty acid reductase complex, and luxG a flavin reductase [21]. |
| araC | E. coli | Regulatory gene of the arabinose operon [19]. |
| PBAD | E. coli | Promotor of the arabinose operon, activated by arabinose [19]. |
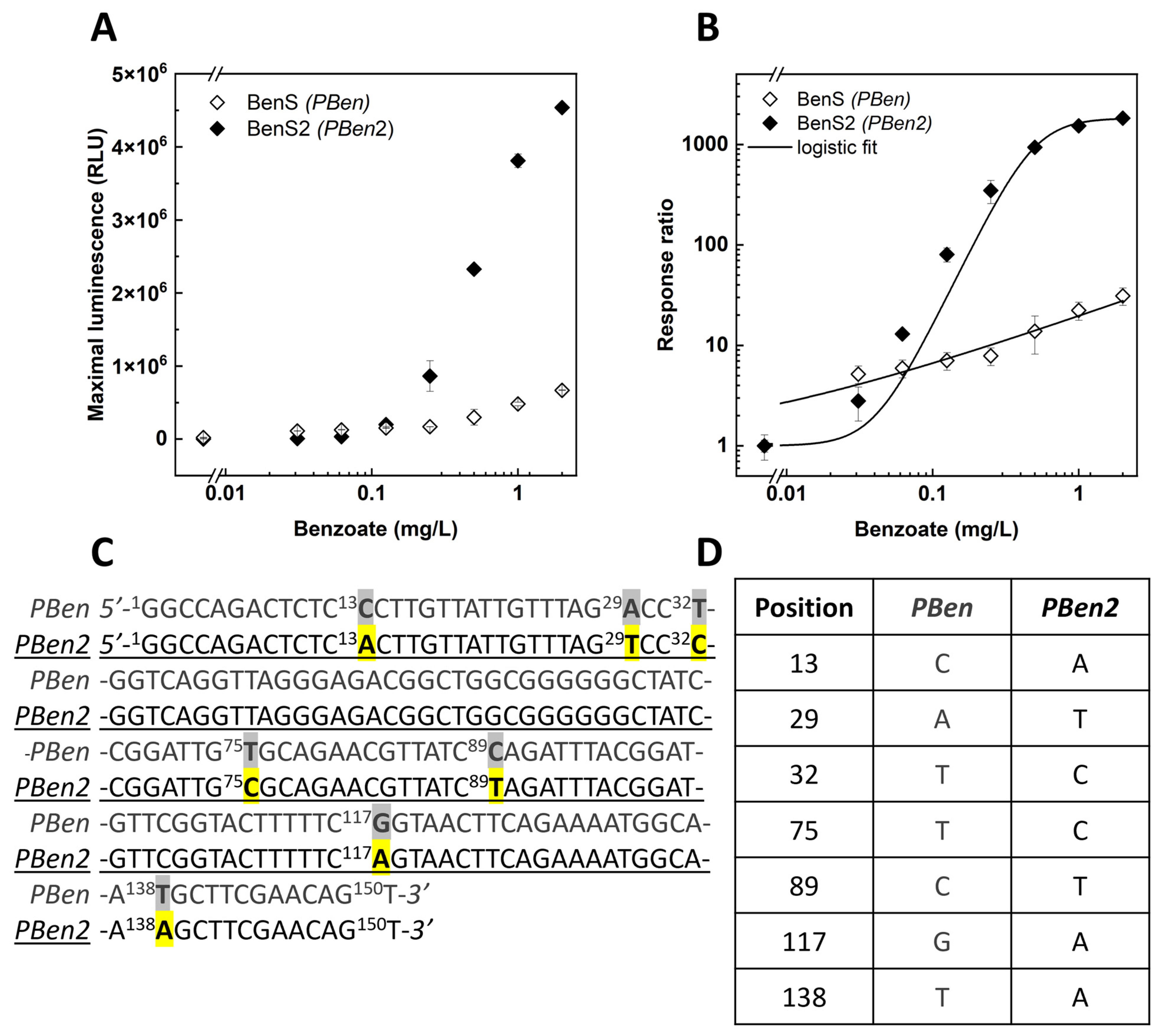
| PBen2 | P. putida | Mutated PBen (this work). |
2.4. Bacterial Sensor Strains
2.5. Luminescence Assay
2.6. Cocaine Detection by CocS Paper Strips
2.7. Random Mutagenesis of the PBen Gene Promoter
2.8. Calculations
3. Results
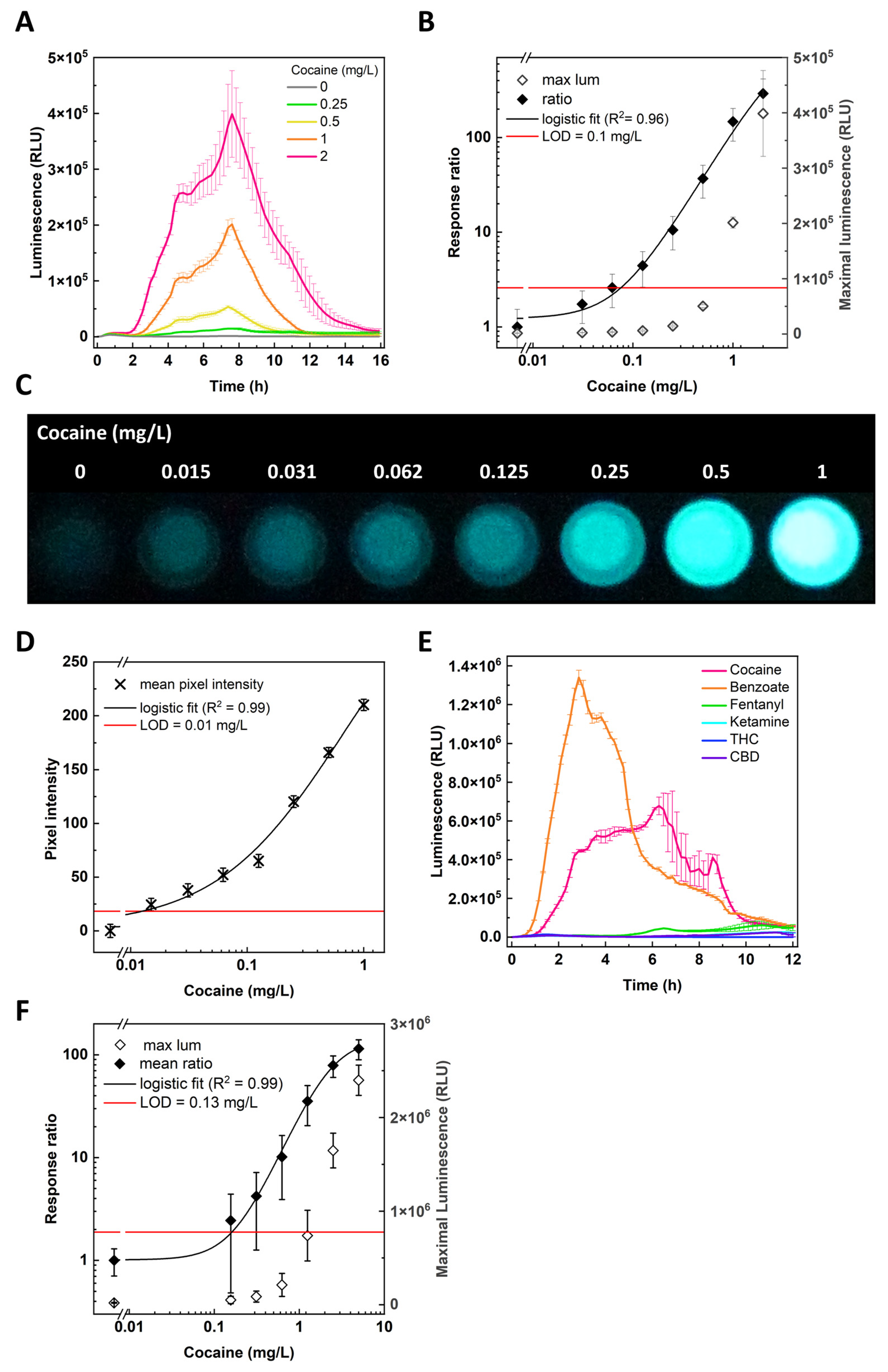
3.1. Cocaine Detection by CocS
3.2. Cocaine Detection by CocS Paper Strips
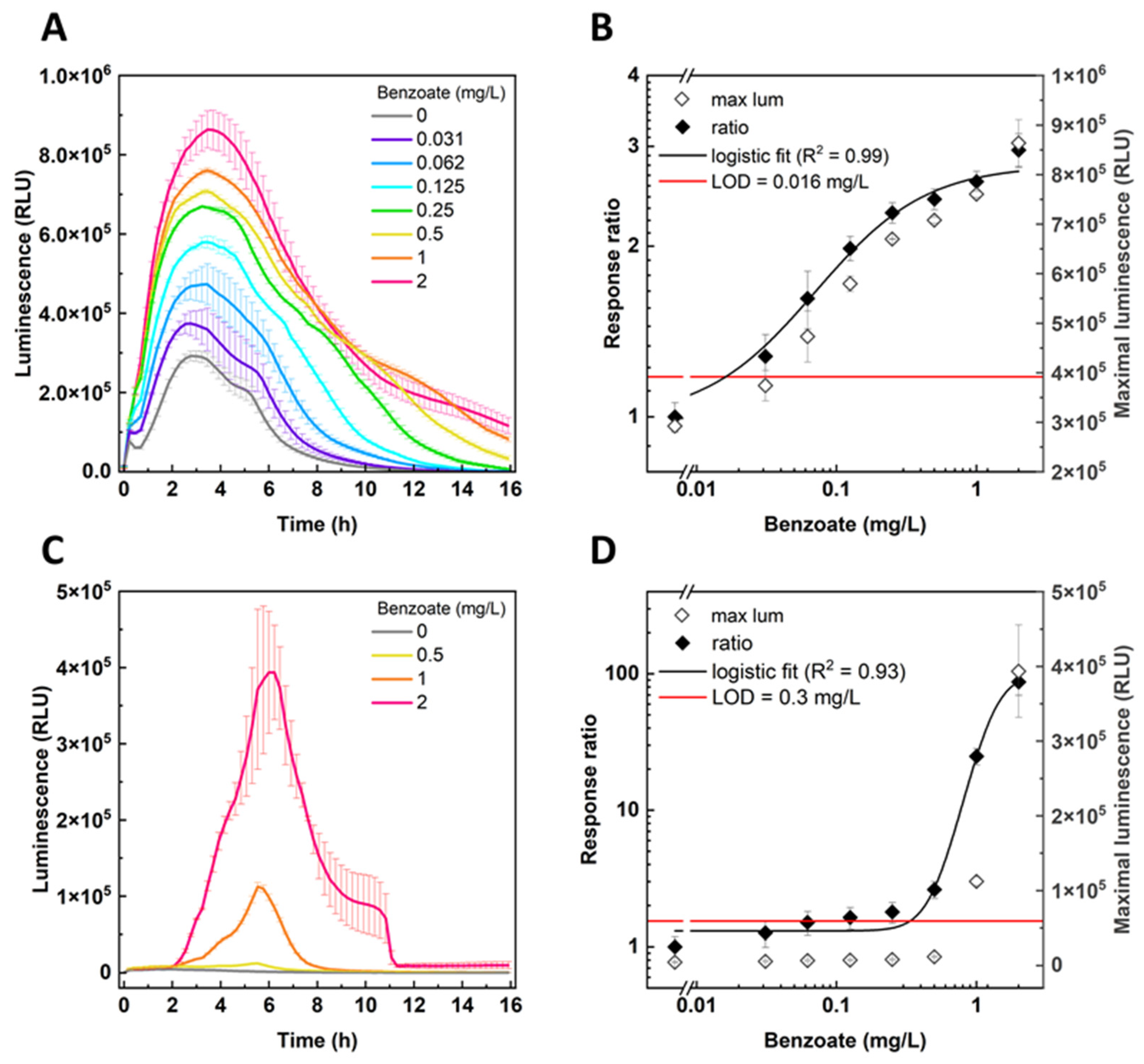
3.3. Benzoate Detection by BenS
3.4. Directed Evolution of the PBen Promotor
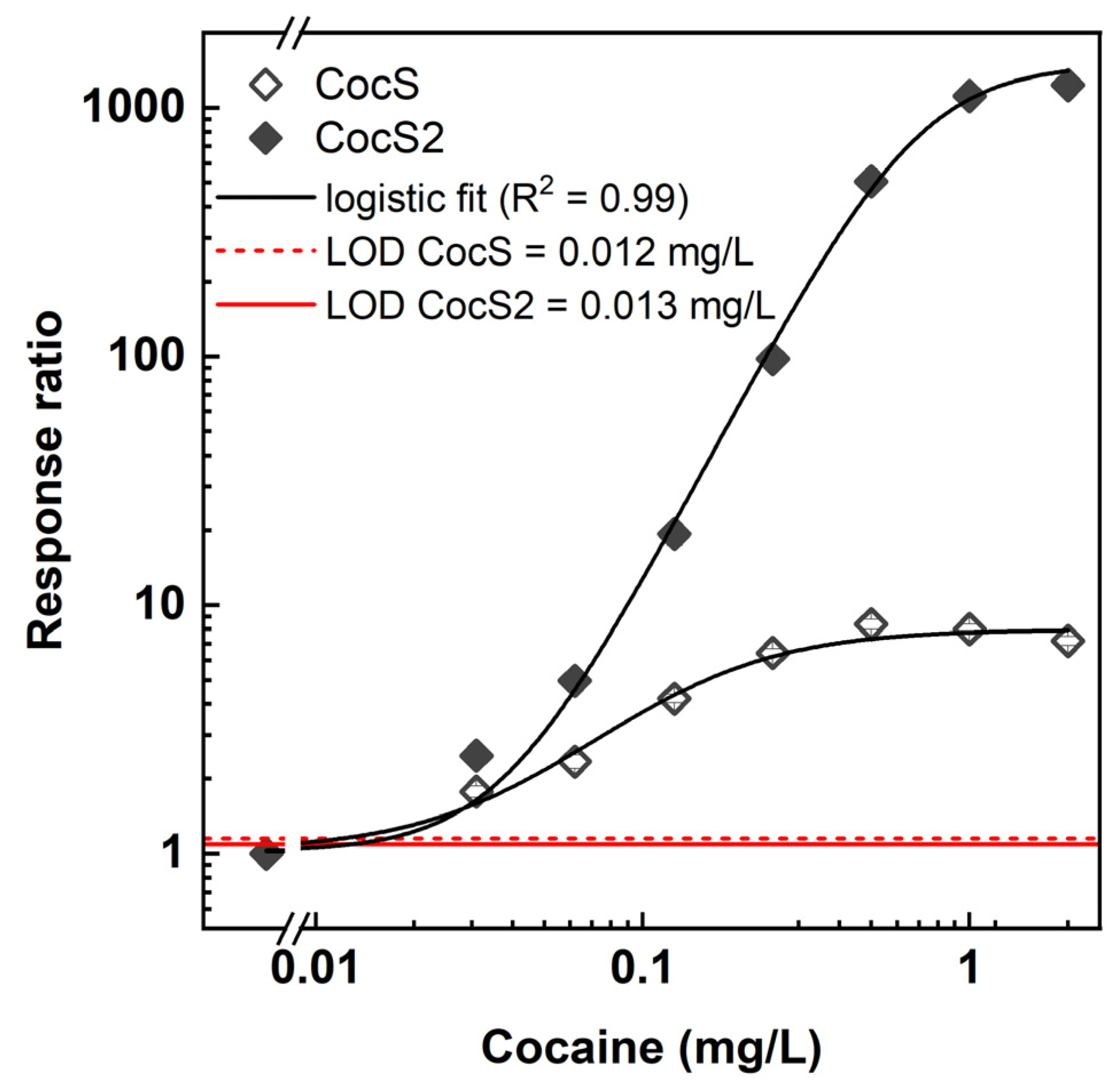
3.5. Cocaine Detection with PBen2
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van der Meer, J.R.; Belkin, S. Where microbiology meets microengineering: Design and applications of reporter bacteria. Nat. Rev. Microbiol. 2010, 8, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Yagi, K. Applications of whole-cell bacterial sensors in biotechnology and environmental science. Appl. Microbiol. Biotechnol. 2007, 3, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Elad, T.; Almog, R.; Yagur-Kroll, S.; Levkov, K.; Melamed, S.; Shacham-Diamand, Y.; Belkin, S. Online monitoring of water toxicity by use of bioluminescent reporter bacterial biochips. Environ. Sci. Technol. 2011, 45, 8536–8544. [Google Scholar] [CrossRef] [PubMed]
- Reifferscheid, G.; Buchinger, S. Cell-based genotoxicity testing: Genetically modified and genetically engineered bacteria in environmental genotoxicology. Adv. Biochem. Eng. Biotechnol. 2010, 118, 85–111. [Google Scholar]
- Belkin, S.; Yagur-Kroll, S.; Kabessa, Y.; Korouma, V.; Septon, T.; Anati, Y.; Zohar-Perez, C.; Rabinovitz, Z.; Nussinovitch, A.; Agranat, A.J. Remote detection of buried landmines using a bacterial sensor. Nat. Biotechnol. 2017, 35, 308–310. [Google Scholar] [CrossRef]
- Finckh, S.; Buchinger, S.; Escher, B.I.; Hollert, H.; König, M.; Krauss, M.; Leekitratanapisan, W.; Schiwy, S.; Schlichting, R.; Shuliakevich, A.; et al. Endocrine disrupting chemicals entering European rivers: Occurrence and adverse mixture effects in treated wastewater. Environ. Int. 2022, 170, 107608. [Google Scholar] [CrossRef]
- National Survey on Drug Use and Health, 2022 NSDUH National Releases. Available online: https://www.samhsa.gov/data/data-we-collect/nsduh-national-survey-drug-use-and-health (accessed on 28 August 2024).
- National Center for Drug Abuse Statistics, Drug Abuse Statistics 2023. Available online: https://drugabusestatistics.org (accessed on 28 August 2024).
- Britt, A.J.; Bruce, N.C.; Lowe, C.R. Identification of a cocaine esterase in a strain of Pseudomonas maltophilia. J. Bacteriol. 1992, 174, 2087–2094. [Google Scholar] [CrossRef]
- Shimomura, E.T.; Jackson, G.F.; Paul, B.D. Cocaine, Crack Cocaine, and Ethanol. In Critical Issues in Alcohol and Drugs of Abuse Testing; Dasgupta, A., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 215–224. [Google Scholar]
- Isenschmid, D.S.; Levine, B.S.; Caplan, Y.H. A comprehensive study of the stability of cocaine and its metabolites. J. Anal. Toxicol. 1989, 13, 250–256. [Google Scholar] [CrossRef]
- Gao, D.; Narasimhan, D.L.; Macdonald, J.; Brim, R.; Ko, M.-C.; Landry, D.W.; Woods, J.H.; Sunahara, R.K.; Zhan, C.-G. Thermostable variants of cocaine esterase for long-time protection against cocaine toxicity. Mol. Pharmacol. 2009, 75, 318–323. [Google Scholar] [CrossRef]
- Zhang, H.-Y.; Bi, G.-H.; Yang, H.-J.; He, Y.; Xue, G.; Cao, J.; Tanda, G.; Gardner, E.L.; Newman, A.H.; Xi, Z.-X. The Novel Modafinil Analog, JJC8-016, as a Potential Cocaine Abuse Pharmacotherapeutic. Neuropsychopharmacology 2017, 42, 1871–1883. [Google Scholar] [CrossRef]
- Brim, R.L.; Nance, M.R.; Youngstrom, D.W.; Narasimhan, D.; Zhan, C.-J.; Tesmer, J.J.G.; Sunahara, R.K.; Woods, J.H. A thermally stable form of bacterial cocaine esterase: A potential therapeutic agent for treatment of cocaine abuse. Mol. Pharmacol. 2010, 77, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, D.; Woods, J.H.; Sunahara, R.K. Bacterial cocaine esterase: A protein-based therapy for cocaine overdose and addiction. Future Med. Chem. 2012, 4, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Voyvodic, P.L.; Pandi, A.; Koch, M.; Conejero, I.; Valjent, E.; Courtet, P.; Renard, E.; Faulon, J.-L.; Bonnet, J. Plug-and-play metabolic transducers expand the chemical detection space of cell-free biosensors. Nat. Commun. 2019, 10, 1697. [Google Scholar] [CrossRef] [PubMed]
- Shemer, B.; Shpigel, E.; Hazan, C.; Kabessa, Y.; Agranat, A.J.; Belkin, S. Detection of buried explosives with immobilized bacterial bioreporters. Microb. Biotechnol. 2021, 14, 251–261. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1996, 177, 4121–4130. [Google Scholar] [CrossRef]
- Silva-Rocha, R.; de Lorenzo, V. A GFP-lacZ bicistronic reporter system for promoter analysis in environmental gram-negative bacteria. PLoS ONE 2012, 7, e34675. [Google Scholar] [CrossRef]
- Brodl, E.; Winkler, A.; Macheroux, P. Molecular Mechanisms of Bacterial Bioluminescence. Comput. Struct. Biotechnol. J. 2018, 16, 551–564. [Google Scholar] [CrossRef]
- Stocker, J.; Balluch, D.; Gsell, M.; Harms, H.; Feliciano, J.; Daunert, S.; Malik, K.A.; van der Meer, J.R. Development of a set of simple bacterial biosensors for quantitative and rapid measurements of arsenite and arsenate in potable water. Environ. Sci. Technol. 2003, 37, 4743–4750. [Google Scholar] [CrossRef]
- Dymond, J.S. PCR-based random mutagenesis. Methods Enzymol. 2013, 529, 249–258. [Google Scholar]
- Banerjee, S.; Salunkhe, S.S.; Apte-Deshpande, A.D.; Mandi, N.S.; Mandal, G.; Padmanabhan, S. Over-expression of proteins using a modified pBAD24 vector in E. coli expression system. Biotechnol. Lett. 2009, 31, 1031–1036. [Google Scholar] [CrossRef] [PubMed]
- Lozano Terol, G.; Gallego-Jara, J.; Sola Martínez, R.A.; Martínez Vivancos, A.; Cánovas Díaz, M.; de Diego Puente, T. Impact of the expression system on recombinant protein production in Escherichia coli BL21. Front. Microbiol. 2021, 12, 682001. [Google Scholar] [CrossRef] [PubMed]
- Rossi, L.; Pierigè, F.; Agostini, M.; Bigini, N.; Termopoli, V.; Cai, Y.; Zheng, F.; Zhan, C.-G.; Landry, D.W.; Magnani, M. Efficient Cocaine Degradation by Cocaine Esterase-Loaded Red Blood Cells. Front. Physiol. 2020, 11, 573492. [Google Scholar] [CrossRef]
- Schulz, M.; Iwersen-Bergmann, S.; Andresen, H.; Schmoldt, A. Therapeutic and toxic blood concentrations of nearly 1000 drugs and other xenobiotics. Crit. Care 2012, 16, R136. [Google Scholar] [CrossRef] [PubMed]
- Musshoff, F.; Hokamp, E.G.; Bott, U.; Madea, B. Performance evaluation of on-site oral fluid drug screening devices in normal police procedure in Germany. Forensic Sci. Int. 2014, 238, 120–124. [Google Scholar] [CrossRef]
- Vidal, J.C.; Bertolín, J.R.; Bonel, L.; Asturias, L.; Arcos-Martínez, M.J.; Castillo, J.R. Rapid determination of recent cocaine use with magnetic particles-based enzyme immunoassays in serum, saliva, and urine fluids. J. Pharm. Biomed. Anal. 2016, 125, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Roque Bravo, R.; Faria, A.C.; Brito-da-Costa, A.M.; Carmo, H.; Mladěnka, P.; Da Dias Silva, D.; Remião, F. Cocaine: An updated overview on chemistry, detection, biokinetics, and pharmacotoxicological aspects including abuse pattern. Toxins 2022, 14, 278. [Google Scholar] [CrossRef]
- United States Nuclear Regulatory Commission. Available online: https://www.nrc.gov/about-nrc.html (accessed on 30 September 2024).
- Yagur-Kroll, S.; Amiel, E.; Rosen, R.; Belkin, S. Detection of 2,4-dinitrotoluene and 2,4,6-trinitrotoluene by an Escherichia coli bioreporter: Performance enhancement by directed evolution. Appl. Microbiol. Biotechnol. 2015, 99, 7177–7188. [Google Scholar] [CrossRef]
- Pandi, A.; Koch, M.; Voyvodic, P.L.; Soudier, P.; Bonnet, J.; Kushwaha, M.; Faulon, J.-L. Metabolic perceptrons for neural computing in biological systems. Nat. Commun. 2019, 10, 3880. [Google Scholar] [CrossRef]
- Shemer, B.; Shpigel, E.; Glozman, A.; Yagur-Kroll, S.; Kabessa, Y.; Agranat, A.J.; Belkin, S. Genome-wide gene-deletion screening identifies mutations that significantly enhance explosives vapor detection by a microbial sensor. N. Biotechnol. 2020, 59, 65–73. [Google Scholar] [CrossRef]
- Subach, O.M.; Cranfill, P.J.; Davidson, M.W.; Verkhusha, V.V. An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS ONE 2011, 6, e28674. [Google Scholar] [CrossRef]


| Sensor Strain | Plasmid | Functional Genes | Function |
|---|---|---|---|
| CocS | I. pCocE-benR | cocE, benR | Cleavage of cocaine into benzoate and activation of BenR |
| II. pBen::luxPleio | PBen, luxCDABEG | Monitoring PBen activation by the benzoate–BenR complex | |
| CocS2 | I: pCocE-benR | CocE, benR | Cleavage of cocaine into benzoate and activation of BenR |
| II: pBen2::luxPleio | PBen2, luxCDABEG | Monitoring PBen2 activation | |
| BenS | pBR-araBAD:benR-pBen::luxPleio | PBAD, araC, benR, PBen, luxCDABEG | benR expression in the presence of arabinose and activation of BenR in the presence of benzoate. Monitoring PBen activation. |
| BenS2 | pBR-araBAD:benR-pBen2::luxPleio | PBAD, araC, benR, PBen2, luxCDABEG | benR expression in the presence of arabinose and activation of BenR in the presence of benzoate. Monitoring of PBen2 activation. |
| Sensor | Medium | Maximal Luminescence (RLU, 2 mg/mL Cocaine) | Maximal Response Ratio | LOD (mg/L) |
|---|---|---|---|---|
| CocS | Buffer | 4.6 × 105 ± 1.8 × 104 | 7.3 ± 0.3 | 0.017 ± 0.008 ** (plate reader) 0.01 ± 0.02 (camera images) |
| CocS * | Urine | 7 × 105 ± 3 × 105 (1.23 mg/L cocaine) | 40 ± 15 | 0.13 ± 0.05 |
| CocS2 | Buffer | 1.39 × 107 ± 8 × 105 | 1230 ± 80 | 0.13 ± 0.009 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grimm, A.-K.; Rozanes, D.; Shpigel, E.; Moscovici, L.; Belkin, S. A Microbial Cocaine Bioreporter. Sensors 2024, 24, 6549. https://doi.org/10.3390/s24206549
Grimm A-K, Rozanes D, Shpigel E, Moscovici L, Belkin S. A Microbial Cocaine Bioreporter. Sensors. 2024; 24(20):6549. https://doi.org/10.3390/s24206549
Chicago/Turabian StyleGrimm, Anne-Kathrin, Dor Rozanes, Etai Shpigel, Liat Moscovici, and Shimshon Belkin. 2024. "A Microbial Cocaine Bioreporter" Sensors 24, no. 20: 6549. https://doi.org/10.3390/s24206549
APA StyleGrimm, A.-K., Rozanes, D., Shpigel, E., Moscovici, L., & Belkin, S. (2024). A Microbial Cocaine Bioreporter. Sensors, 24(20), 6549. https://doi.org/10.3390/s24206549









