How Not to Make the Joint Extended Kalman Filter Fail with Unstructured Mechanistic Models
Abstract
1. Introduction
- We provide proof of JEKF failure when acting as an unshared parameter estimator under specific biomanufacturing conditions that represent a failure case. To our knowledge, this is the first work to formally report this failure case regarding the JEKF.
- An approach to avoid the JEKF failure that enables using JEKF with UMM for real-time bioprocess monitoring. This is helpful in the macro-scale modeling of a phenomenon with UMM where the underlying process mechanism is not fully understood.
2. Related Work
3. Background
3.1. Unstructured Mechanistic Model (UMM)
3.2. Continuous-Discrete Extended Kalman Filter
3.3. JEKF
4. Theoretical Analysis
4.1. JEKF Failure
- Unshared parameters: They are parameters used only in one term of an ODE and not used by other ODEs of the same UMM. See the example in Section S3.1 of the Supplementary Material.
- Weak and Strong term of an ODE: A weak term is a term of an ODE with a low percentage of variables of the state variable vector, and a "strong term" is one with a high percentage of variables of the state variable vector. See the example in Section S3.2 of the Supplementary Material.
- Weak and Strong variable of an ODE: A weak variable is a variable used only in the first member of an ODE in UMM, and a strong variable is a variable used in the first member and different terms of the second member of an ODE. Furthermore, it is used in the second member of other ODEs of the same UMM. See the example in Section S3.3 of the Supplementary Material.
4.1.1. Failure Case: Biomanufacturing Conditions
- ODEs of UMM with unshared parameters. This parameter type is commonly used in ODE to model the dynamic of product formation in biomanufacturing [53,54,55]. See the example in Section S3.1 of the Supplementary Material.
- P and Q with uncorrelated elements. In case of the limited amount of data, it is very common to assume P and Q with uncorrelated elements in EKF applications [19,20,21,47]. This assumption means that the error covariance matrices P and Q are diagonal, with the diagonal elements being the noise variances (P and Q) and off-diagonal elements equal to zero (P and Q). The Q constant and with uncorrelated elements is used only to build the MRDE, and the P with uncorrelated elements can be used to build an MRDE and as an initial condition of MRDE (the initial predicted state error covariance P(t = 0)).This assumption raises two scenarios:
- The use of P with uncorrelated elements to build the MRDE (Equation (6)) and P(t = 0) with uncorrelated elements as the initial condition. When P with uncorrelated elements is used to build the MRDE, the ODEs of MRDE are based only on noise variance of P and Q and elements of Jacobian . See the example in Section S3.4 of the Supplementary Material. It is important to point out that depending on the partial derivative, the ODE to predict a state error covariance can be time-invariant . See Section S3.2 of the Supplementary Material.
- The use of P with correlated elements to build the MRDE (Equation (6)) and P(t = 0) with uncorrelated elements as the initial condition. This means that the ODE of MRDE can be composed of off-diagonal elements of P, and it can reduce the number of the time-invariant ODE to predict a state error covariance between two state variables.
- ODEs of UMM with weak terms. A strong term contributes more than a weak term to compute the predicted state error covariance . Many elements of Jacobian result from the partial derivation of a strong term. See the example in Section S3.2 of the Supplementary Material.
- ODEs of UMM with weak variables. In the Jacobian , the first-order partial derivatives of all functions with respect to a weak variable are equal to zero. Consequently, this variable type does not contribute to the calculations of predicted error covariance since it will not be part of any element of MRDE to predict the state error covariance matrix . On the other hand, a strong variable contributes to the calculations of predicted error covariance . See the example in Section S3.3 of Supplementary Material.
- Only one measured state variable. In some cases (JEKF application), measuring only one state variable is possible. This measured state variable determines which column of the predicted state error covariance is used to compute the Kalman gain through in Equation (10). If this column has a row with a value equal to zero (no covariance between the measured variable and state variable represented by the row), the Kalman gain cannot be computed to the state variable defined by the row. See the example in Section S3.5 of the Supplementary Material.
4.1.2. Lemma: Inability to Update Kalman Gain for Unshared Parameters based P(t = 0) and Q with Uncorrelated Elements
- A general UMM with an unshared parameter in a weak term represented by a system of nonlinear differential equations of the form:where and are the variables of the system, are the functions defining the system, and are the parameters of the system, and is an unshared parameter.
- A joint state variables vector defined as
- A process model defined as
- as the unique measured state variable (MSV) and = [1 0 … 0 0].
- R as measurement noise variance of .
- as the unshared parameter (UP) to be evolved (estimated) and presented in only one weak term.
4.1.3. Theorem: JEKF Failure
- H = [1 0 … 0 0] and as obtained in the proof of Lemma 1 in Section 4.1.2, where = 0.
- as a measured value of .
- Predicted mean of the state variable vector with regard to the general UMM used in the proof of Lemma 1 in Section 4.1.2.
4.2. SANTO: Specific Initial Condition for MRDE ( )
- A positive quantity .
- as the unique measured state variable (MSV) and = [1 0 … 0 0].
- R as measurement noise variance of .
- as the unshared parameter (UP) to be evolved (estimated) and presented in only one weak term.
- A specific initial predicted state error covariance matrix with uncorrelated elements and as following
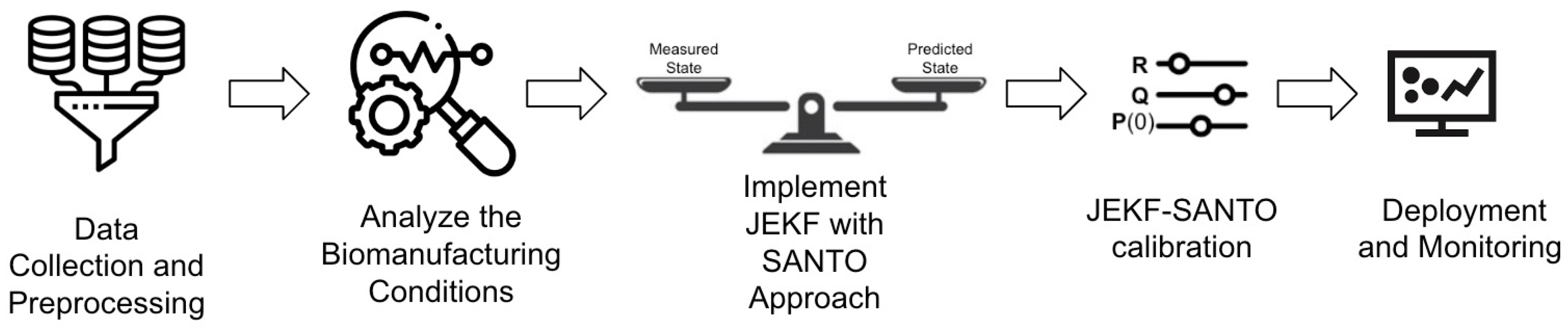
- Step 1: Data Collection and Preprocessing. The first step in developing a soft sensor for bioprocess monitoring using the JEKF-SANTO approach involves comprehensive data collection and preprocessing. Once collected, these data must be meticulously cleaned and preprocessed to remove outliers and address any missing values. This preprocessing is crucial to ensure the quality and reliability of the data, which forms the foundation for accurate modeling and estimation in subsequent steps.
- Step 2: Analyze the Biomanufacturing Conditions. This step involves a comprehensive analysis of the biomanufacturing conditions where JEKF fails to estimate an unshared parameter that is part of a state variable vector and part of a weak term in a UMM if the initial state error covariance matrix P(t = 0) and Q are composed of uncorrelated elements, and there is only one state variable measured.
- Step 3: Implement JEKF with the SANTO approach. Implement the JEKF algorithm, defining the process model and the measurement model. Modify the initial state error covariance matrix as per the SANTO approach, adding a specific positive quantity to the covariance between the measured state variable and the unshared parameter.
- Step 4: JEKF-SANTO calibration. Tune the R and Q of JEKF-SANTO based on consistency tests, and adjust the parameter model based on the estimates obtained from JEKF-SANTO related to the unshared parameter and the associated weak variable.
- Step 5: Deployment and Monitoring. Integrate the JEKF-SANTO as a soft sensor into the biomanufacturing process control system to monitor critical quality attributes (CQAs) and critical process parameters (CPPs) in real time.
5. Empirical Evaluation
- (G1) Experimentally test Theorem 1 (JEKF Failure ) in Section 4.1.3;
- (G2) Test whether SANTO can avoid the JEKF failure and compare its performance with KPH2.
- (RQ1-G1) Is there any variation in the unshared parameter estimation completed by JEKF-Classic with the biomanufacturing conditions (failure case), or are the estimations constant in the entire process?
- (RQ2-G2) Is there any variation in the unshared parameter estimation completed by SANTO and KPH2 with the biomanufacturing conditions (failure case), and which one has the best estimations (performance)?
- (RQ3-G2) Can the SANTO simultaneously estimate more than one unshared parameter, performing better than KPH2?
5.1. Experimental Setup
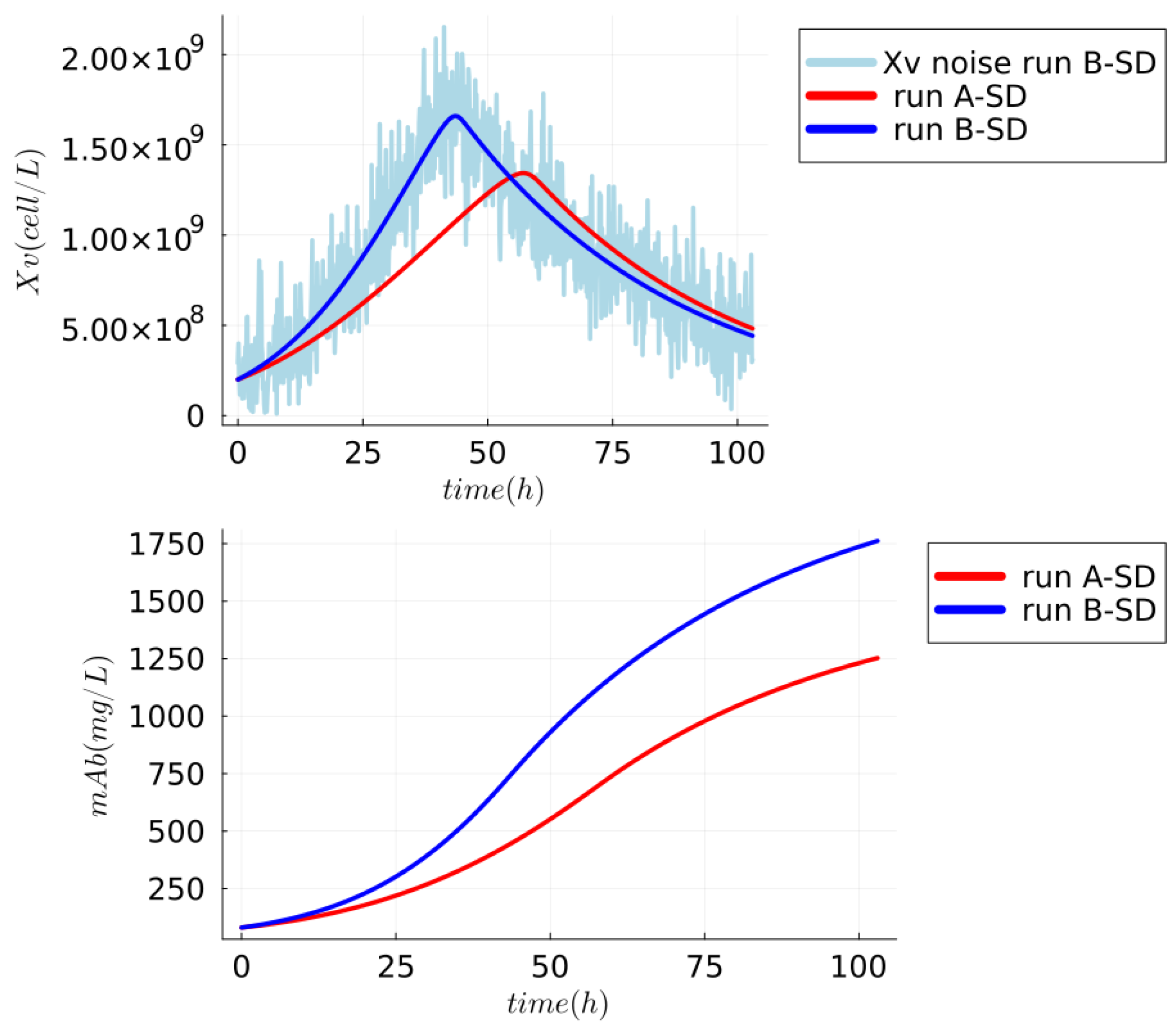
5.1.1. Synthetic Dataset—mAb Production
5.1.2. Real Dataset: AAV Production
5.1.3. NSEs Assessment with Synthetic Dataset to Address RQ1-G1 and RQ2-G2
5.1.4. NSEs Assessment with Real Dataset to Address RQ3-G2
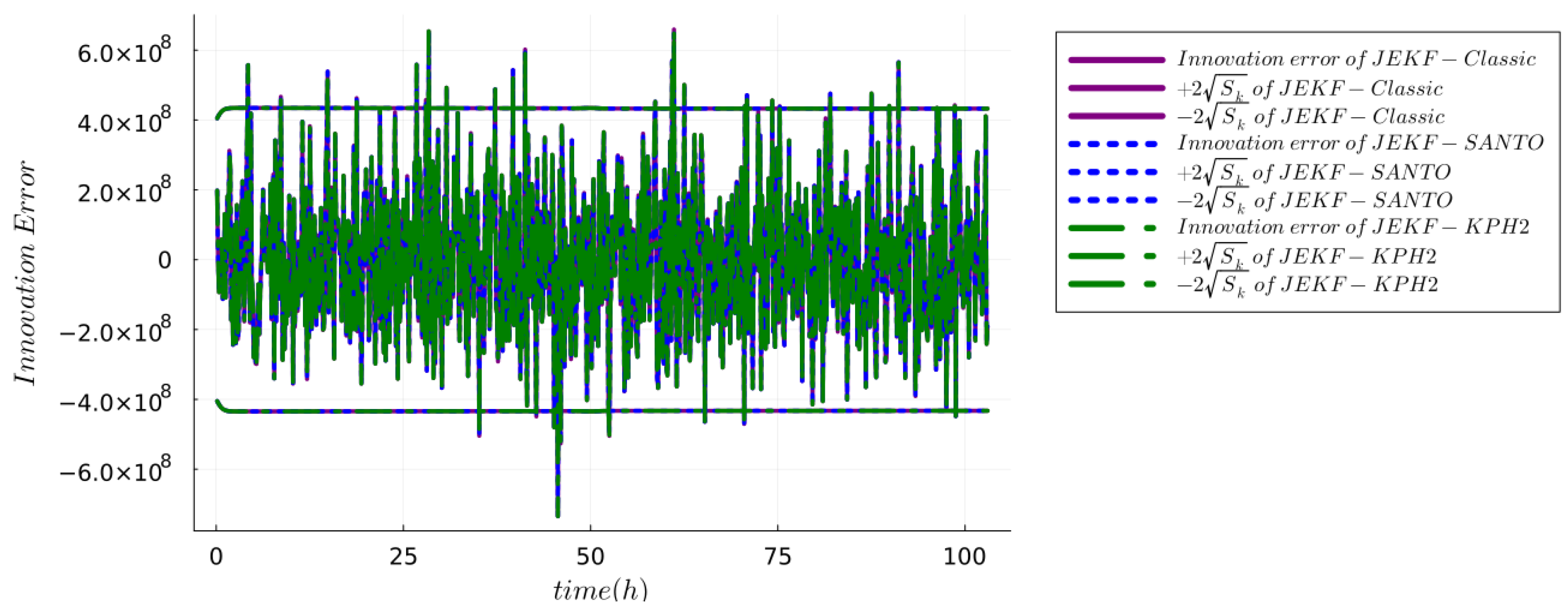
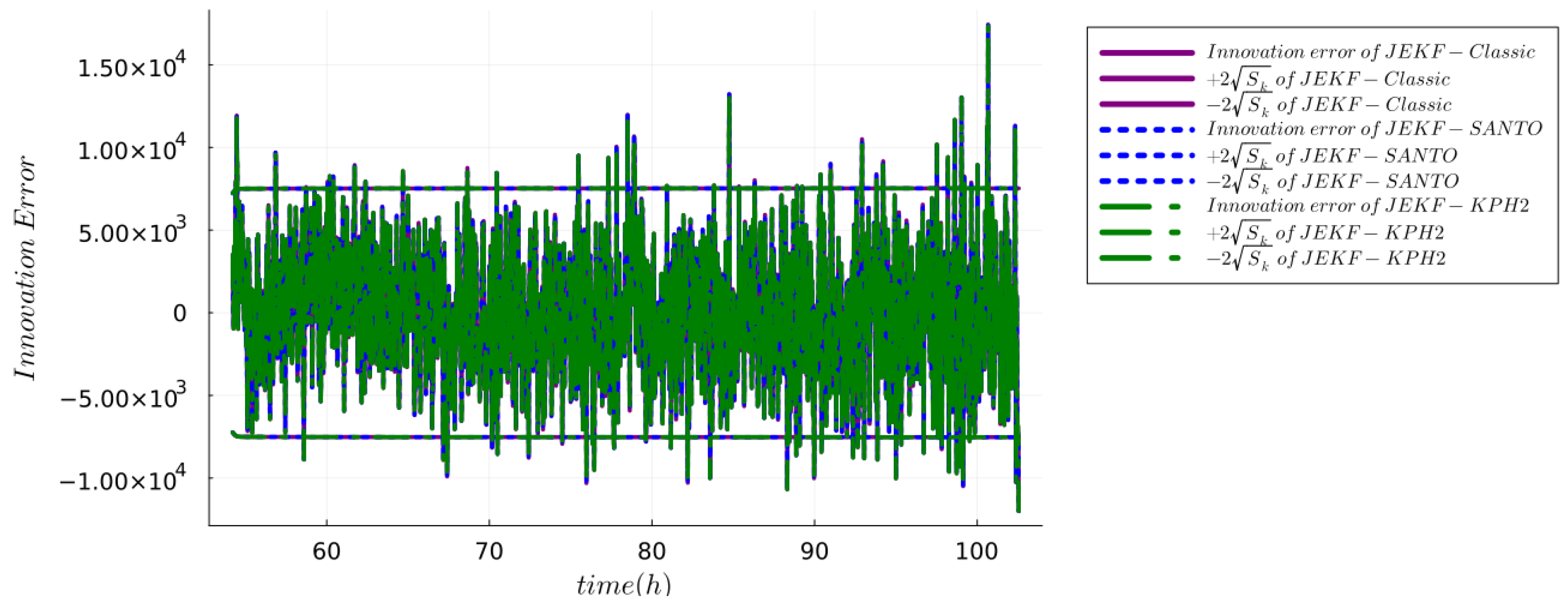
5.1.5. Checking Consistency and Efficiency
6. Results
7. Discussion
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| JEKF | Joint estimation of states and parameters with Extended Kalman Filter |
| NSE | Nonlinear State Estimator |
| UMM | Unstructured Mechanistic Model |
| SMM | Structured Mechanistic Model |
| MRDE | Matrix Ricatti Differential Equation |
| MSV | Measured State Variable |
| UP | Unshared parameter |
| SANTO | Specific initiAl coNdiTiOn |
| CD-EKF | Continuous-Discrete EKF |
| RD | Real dataset |
| SD | Synthetic dataset |
| rAAV | Recombinant Adeno-Associated Virus |
| mAb | Monoclonal Antibody |
References
- Jin, X.B.; Robert Jeremiah, R.J.; Su, T.L.; Bai, Y.T.; Kong, J.L. The new trend of state estimation: From model-driven to hybrid-driven methods. Sensors 2021, 21, 2085. [Google Scholar] [CrossRef] [PubMed]
- Alexander, R.; Campani, G.; Dinh, S.; Lima, F.V. Challenges and opportunities on nonlinear state estimation of chemical and biochemical processes. Processes 2020, 8, 1462. [Google Scholar] [CrossRef]
- Kalman, R.E. A new approach to linear filtering and prediction problems. J. Basic Eng. Mar 1960, 82, 35–45. [Google Scholar] [CrossRef]
- Jazwinski, A. Stochastic Processes and Filtering Theory; Mathematics in Science and Engineering; Academic Press: Cambridge, MA, USA, 1970; Available online: https://books.google.ch/books?id=nGlSNvKyY2MC (accessed on 20 November 2023).
- Aswal, N.; Bhattacharya, B.; Sen, S. Joint and Dual Estimation of States and Parameters with Extended and Unscented Kalman Filters. In Recent Developments in Structural Health Monitoring and Assessment–Opportunities and Challenges: Bridges, Buildings and Other Infrastructures; World Scientific: Singapore, 2022; pp. 223–252. [Google Scholar]
- Ljung, L. Asymptotic behavior of the extended Kalman filter as a parameter estimator for linear systems. IEEE Trans. Autom. Control 1979, 24, 36–50. [Google Scholar] [CrossRef]
- Kopp, R.E.; Orford, R.J. Linear regression applied to system identification for adaptive control systems. Aiaa J. 1963, 1, 2300–2306. [Google Scholar] [CrossRef]
- Haykin, S.S.; Haykin, S.S. Kalman Filtering and Neural Networks; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2001; Volume 284. [Google Scholar] [CrossRef]
- Cox, H. On the estimation of state variables and parameters for noisy dynamic systems. IEEE Trans. Autom. Control 1964, 9, 5–12. [Google Scholar] [CrossRef]
- Urrea, C.; Agramonte, R. Kalman filter: Historical overview and review of its use in robotics 60 years after its creation. J. Sensors 2021, 2021, 1–21. [Google Scholar] [CrossRef]
- Aswal, N.; Sen, S.; Mevel, L. Switching Kalman filter for damage estimation in the presence of sensor faults. Mech. Syst. Signal Process. 2022, 175, 109116. [Google Scholar] [CrossRef]
- Stojanovic, V.; He, S.; Zhang, B. State and parameter joint estimation of linear stochastic systems in presence of faults and non-Gaussian noises. Int. J. Robust Nonlinear Control 2020, 30, 6683–6700. [Google Scholar] [CrossRef]
- Beelen, H.; Bergveld, H.J.; Donkers, M. Joint estimation of battery parameters and state of charge using an extended Kalman filter: A single-parameter tuning approach. IEEE Trans. Control Syst. Technol. 2020, 29, 1087–1101. [Google Scholar] [CrossRef]
- Dhanalakshmi, R.; Bhavani, N.; Raju, S.S.; Shaker Reddy, P.C.; Marvaluru, D.; Singh, D.P.; Batu, A. Onboard Pointing Error Detection and Estimation of Observation Satellite Data Using Extended Kalman Filter. Comput. Intell. Neurosci. 2022, 2022, 4340897. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Yuen, K.V.; Wang, L. Real-time simultaneous input-state-parameter estimation with modulated colored noise excitation. Mech. Syst. Signal Process. 2022, 165, 108378. [Google Scholar] [CrossRef]
- Huang, K.; Yuen, K.V. Online dual-rate decentralized structural identification for wireless sensor networks. Struct. Control Health Monit. 2019, 26, e2453. [Google Scholar] [CrossRef]
- Yuen, K.V.; Huang, K. Real-time substructural identification by boundary force modeling. Struct. Control Health Monit. 2018, 25, e2151. [Google Scholar] [CrossRef]
- Kleyman, V.; Schaller, M.; Mordmuller, M.; Wilson, M.; Brinkmann, R.; Worthmann, K.; Muller, M.A. State and parameter estimation for retinal laser treatment. arXiv 2022, arXiv:2203.12452. [Google Scholar] [CrossRef]
- Iglesias, C.F., Jr.; Xu, X.; Mehta, V.; Akassou, M.; Venereo-Sanchez, A.; Belacel, N.; Kamen, A.; Bolic, M. Monitoring the Recombinant Adeno-Associated Virus Production using Extended Kalman Filter. Processes 2022, 10, 2180. [Google Scholar] [CrossRef]
- Yousefi-Darani, A.; Paquet-Durand, O.; Hitzmann, B. The Kalman filter for the supervision of cultivation processes. Digital Twins 2020, 177, 95–125. [Google Scholar]
- Paquet-Durand, O.; Zettel, V.; Yousefi-Darani, A.; Hitzmann, B. The supervision of dough fermentation using image analysis complemented by a continuous discrete extended Kalman filter. Processes 2020, 8, 1669. [Google Scholar] [CrossRef]
- Song, H.; Hu, S. Open Problems in Applications of the Kalman Filtering Algorithm. In Proceedings of the 2019 International Conference on Mathematics, Big Data Analysis and Simulation and Modelling (MBDASM 2019), Changsha, China, 30–31 August 2019; Atlantis Press: Amsterdam, The Netherlands, 2019; pp. 185–190. [Google Scholar] [CrossRef]
- Khodarahmi, M.; Maihami, V. A Review on Kalman Filter Models. Arch. Comput. Methods Eng. 2022, 30, 727–747. [Google Scholar] [CrossRef]
- Nelson, L.; Stear, E. The simultaneous on-line estimation of parameters and states in linear systems. IEEE Trans. Autom. Control 1976, 21, 94–98. [Google Scholar] [CrossRef]
- Herwig, C.; Pörtner, R.; Möller, J. Digital Twins: Tools and Concepts for Smart Biomanufacturing; Springer Nature: Berlin/Heidelberg, Germany, 2021; Volume 176. [Google Scholar] [CrossRef]
- Herwig, C.; Pörtner, R.; Möller, J. Digital Twins: Applications to the Design and Optimization of Bioprocesses; Springer Nature: Berlin/Heidelberg, Germany, 2021; Volume 177. [Google Scholar] [CrossRef]
- Sinner, P.; Daume, S.; Herwig, C.; Kager, J. Usage of Digital Twins Along a Typical Process Development Cycle. In Digital Twins: Tools and Concepts for Smart Biomanufacturing; Herwig, C., Pörtner, R., Möller, J., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 71–96. [Google Scholar] [CrossRef]
- Moser, A.; Appl, C.; Brüning, S.; Hass, V.C. Mechanistic mathematical models as a basis for digital twins. Digit. Twins 2020, 176, 133–180. [Google Scholar]
- Narayanan, H.; Sokolov, M.; Morbidelli, M.; Butté, A. A new generation of predictive models: The added value of hybrid models for manufacturing processes of therapeutic proteins. Biotechnol. Bioeng. 2019, 116, 2540–2549. [Google Scholar] [CrossRef] [PubMed]
- Fernandes-Platzgummer, A.; Badenes, S.M.; da Silva, C.L.; Cabral, J.M. Bioreactors for Stem Cell and Mammalian Cell Cultivation. Bioprocess. Technol. Prod. Biopharm. Bioprod. 2018, 4, 131–173. [Google Scholar]
- Iglesias, C.F., Jr.; Ristovski, M.; Bolic, M.; Cuperlovic-Culf, M. rAAV Manufacturing: The Challenges of Soft Sensing during Upstream Processing. Bioengineering 2023, 10, 229. [Google Scholar] [CrossRef] [PubMed]
- Gargalo, C.L.; Udugama, I.; Pontius, K.; Lopez, P.C.; Nielsen, R.F.; Hasanzadeh, A.; Mansouri, S.S.; Bayer, C.; Junicke, H.; Gernaey, K.V. Towards smart biomanufacturing: A perspective on recent developments in industrial measurement and monitoring technologies for bio-based production processes. J. Ind. Microbiol. Biotechnol. Off. J. Soc. Ind. Microbiol. Biotechnol. 2020, 47, 947–964. [Google Scholar] [CrossRef] [PubMed]
- Udugama, A.; Öner, M.; Lopez, P.C.; Beenfeldt, C.; Bayer, C.; Huusom, J.K.; Gernaey, K.V.; Sin, G. Towards Digitalization in Bio-Manufacturing Operations: A Survey on Application of Big Data and Digital Twin Concepts in Denmark. Front. Chem. Eng. 2021, 3, 727152. [Google Scholar] [CrossRef]
- Luo, Y.; Kurian, V.; Ogunnaike, B.A. Bioprocess systems analysis, modeling, estimation, and control. Curr. Opin. Chem. Eng. 2021, 33, 100705. [Google Scholar] [CrossRef]
- Wang, K.; Li, Y.; Rizos, C. Practical approaches to Kalman filtering with time-correlated measurement errors. IEEE Trans. Aerosp. Electron. Syst. 2012, 48, 1669–1681. [Google Scholar] [CrossRef]
- Ji, Z.; Brown, M. Joint state and parameter estimation for biochemical dynamic pathways with iterative extended Kalman filter: Comparison with dual state and parameter estimation. Open Autom. Control Syst. J. 2009, 2, 69–77. [Google Scholar] [CrossRef]
- Mariani, S.; Corigliano, A. Impact induced composite delamination: State and parameter identification via joint and dual extended Kalman filters. Comput. Methods Appl. Mech. Eng. 2005, 194, 5242–5272. [Google Scholar] [CrossRef]
- Ljung, L.; Söderström, T. Theory and Practice of Recursive Identification; MIT Press: Cambridge, MA, USA, 1983; Volume 4, pp. 136–249. [Google Scholar]
- Kyriakopoulos, S.; Ang, K.S.; Lakshmanan, M.; Huang, Z.; Yoon, S.; Gunawan, R.; Lee, D.Y. Kinetic modeling of mammalian cell culture bioprocessing: The quest to advance biomanufacturing. Biotechnol. J. 2018, 13, 1700229. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Park, C.H.; Choi, D.H.; Hong, J.K.; Lee, D.Y. Bioprocess digital twins of mammalian cell culture for advanced biomanufacturing. Curr. Opin. Chem. Eng. 2021, 33, 100702. [Google Scholar] [CrossRef]
- Tsopanoglou, A.; del Val, I.J. Moving towards an era of hybrid modelling: Advantages and challenges of coupling mechanistic and data-driven models for upstream pharmaceutical bioprocesses. Curr. Opin. Chem. Eng. 2021, 32, 100691. [Google Scholar] [CrossRef]
- Mears, L.; Stocks, S.M.; Albaek, M.O.; Sin, G.; Gernaey, K.V. Mechanistic fermentation models for process design, monitoring, and control. Trends Biotechnol. 2017, 35, 914–924. [Google Scholar] [CrossRef] [PubMed]
- Reyes, S.J.; Durocher, Y.; Pham, P.L.; Henry, O. Modern Sensor Tools and Techniques for Monitoring, Controlling, and Improving Cell Culture Processes. Processes 2022, 10, 189. [Google Scholar] [CrossRef]
- Zhang, D.; Del Rio-Chanona, E.A.; Petsagkourakis, P.; Wagner, J. Hybrid physics-based and data-driven modeling for bioprocess online simulation and optimization. Biotechnol. Bioeng. 2019, 116, 2919–2930. [Google Scholar] [CrossRef]
- Kourti, T. Multivariate Statistical Process Control and Process Control, Using Latent Variables; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar] [CrossRef]
- Brockwell, P. Time series analysis. Encycl. Stat. Behav. Sci. 2005. [Google Scholar] [CrossRef]
- Ohadi, K.; Legge, R.L.; Budman, H.M. Development of a soft-sensor based on multi-wavelength fluorescence spectroscopy and a dynamic metabolic model for monitoring mammalian cell cultures. Biotechnol. Bioeng. 2015, 112, 197–208. [Google Scholar] [CrossRef]
- Assimakis, N.; Adam, M. Kalman filter Riccati equation for the prediction, estimation, and smoothing error covariance matrices. Int. Sch. Res. Not. 2013, 2013, 249594. [Google Scholar] [CrossRef]
- Kulikova, M.V.; Kulikov, G.Y. Adaptive ODE solvers in extended Kalman filtering algorithms. J. Comput. Appl. Math. 2014, 262, 205–216. [Google Scholar] [CrossRef]
- Särkkä, S.; Svensson, L. Bayesian Filtering and Smoothing; Cambridge University Press: Cambridge, UK, 2023; Volume 17. [Google Scholar] [CrossRef]
- Murphy, K.P. Machine Learning: A Probabilistic Perspective; MIT Press: Cambridge, MA, USA, 2012; Volume 1, pp. 631–660. [Google Scholar]
- Haldar, A.; Al-hussein, A.A.A. Recent Developments in Structural Health Monitoring and Assessment-Opportunities and Challenges: Bridges, Buildings and Other Infrastructures; World Scientific: Singapore, 2022. [Google Scholar] [CrossRef]
- Goudar, C.T. Computer programs for modeling mammalian cell batch and fed-batch cultures using logistic equations. Cytotechnology 2012, 64, 465–475. [Google Scholar] [CrossRef]
- Kornecki, M.; Strube, J. Accelerating biologics manufacturing by upstream process modelling. Processes 2019, 7, 166. [Google Scholar] [CrossRef]
- Narayanan, H.; Behle, L.; Luna, M.F.; Sokolov, M.; Guillén-Gosálbez, G.; Morbidelli, M.; Butté, A. Hybrid-EKF: Hybrid Model coupled with Extended Kalman Filter for real-time monitoring and control of mammalian cell culture. Biotechnol. Bioeng. 2020, 117, 2703–2714. [Google Scholar] [CrossRef] [PubMed]
- Ueno, G.; Nakamura, N. Bayesian estimation of the observation-error covariance matrix in ensemble-based filters. Q. J. R. Meteorol. Soc. 2016, 142, 2055–2080. [Google Scholar] [CrossRef]
- Michel, V.; Gramfort, A.; Varoquaux, G.; Eger, E.; Thirion, B. Total variation regularization for fMRI-based prediction of behavior. IEEE Trans. Med Imaging 2011, 30, 1328–1340. [Google Scholar] [CrossRef] [PubMed]
- Jyothilekshmi, I.; Jayaprakash, N. Trends in monoclonal antibody production using various bioreactor systems. J. Microbiol. Biotechnol. 2021, 31, 349–357. [Google Scholar] [CrossRef]
- Liu, Y.; Gunawan, R. Bioprocess optimization under uncertainty using ensemble modeling. J. Biotechnol. 2017, 244, 34–44. [Google Scholar] [CrossRef]
- Bulcha, J.T.; Wang, Y.; Ma, H.; Tai, P.W.; Gao, G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 2021, 6, 53. [Google Scholar] [CrossRef]
- Evangelidis, A.; Parker, D. Quantitative verification of Kalman filters. Form. Asp. Comput. 2021, 33, 669–693. [Google Scholar] [CrossRef]
- Bar-Shalom, Y.; Li, X.R.; Kirubarajan, T. Estimation with Applications to Tracking and Navigation: Theory Algorithms and Software; John Wiley & Sons: Hoboken, NJ, USA, 2001; Volume 5, pp. 199–265. [Google Scholar]
- Simon, D. Optimal State Estimation: Kalman, H Infinity, and Nonlinear Approaches; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar] [CrossRef]
- Maybeck, P.S. Stochastic Models, Estimation, and Control; Academic Press: Cambridge, MA, USA, 1982; Volume 3, pp. 126–132. [Google Scholar]
- Boulkroune, B.; Geebelen, K.; Wan, J.; van Nunen, E. Auto-tuning extended Kalman filters to improve state estimation. In Proceedings of the 2023 IEEE Intelligent Vehicles Symposium (IV), Anchorage, AK, USA, 4–7 June 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 1–6. [Google Scholar] [CrossRef]
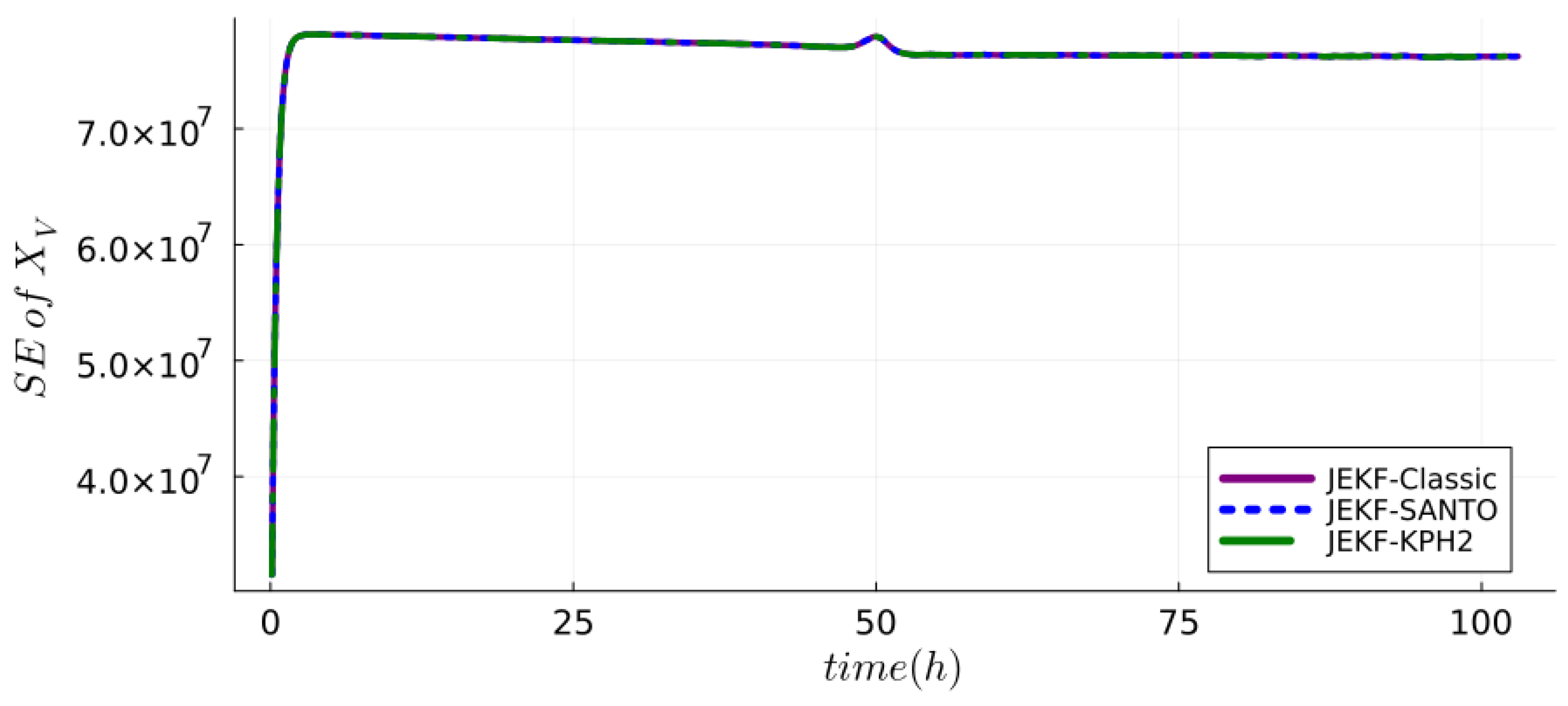

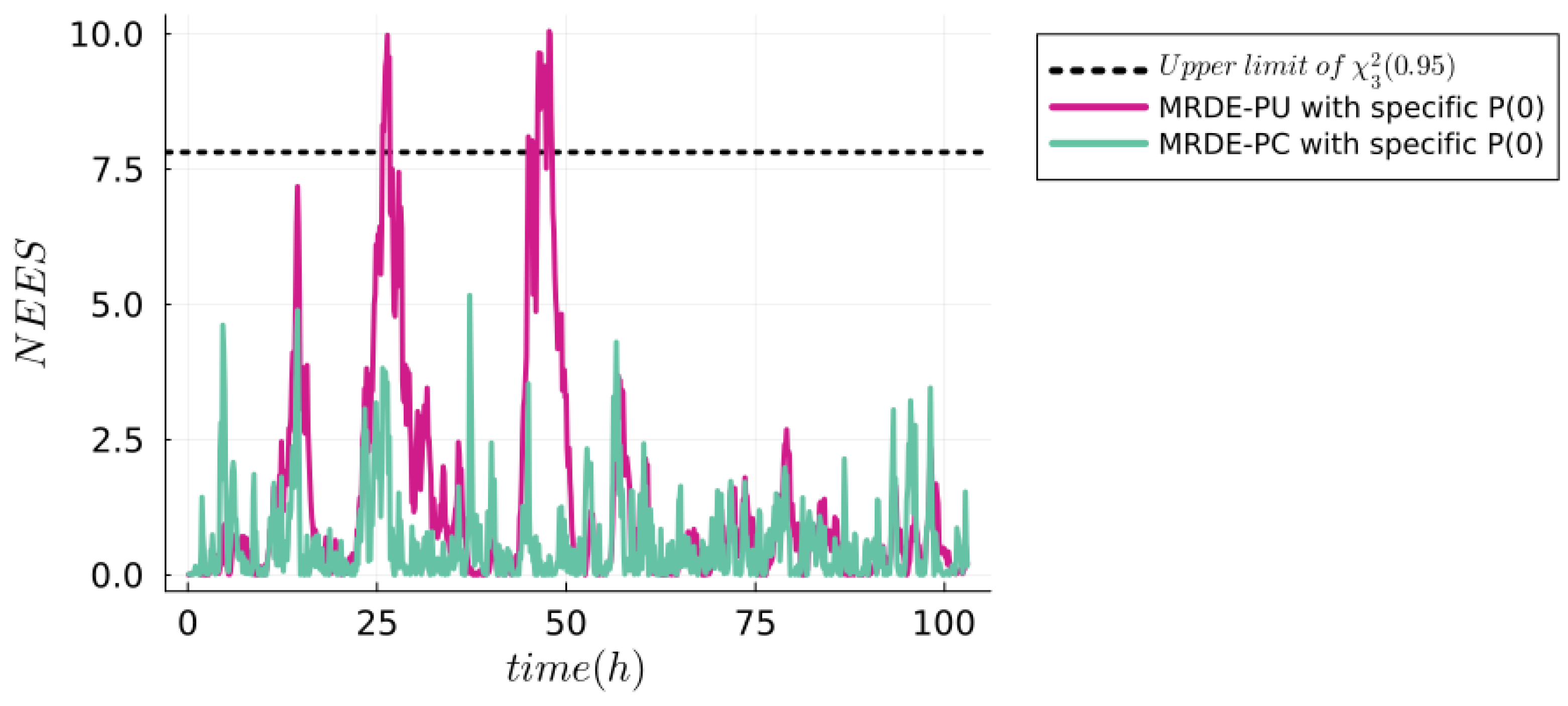
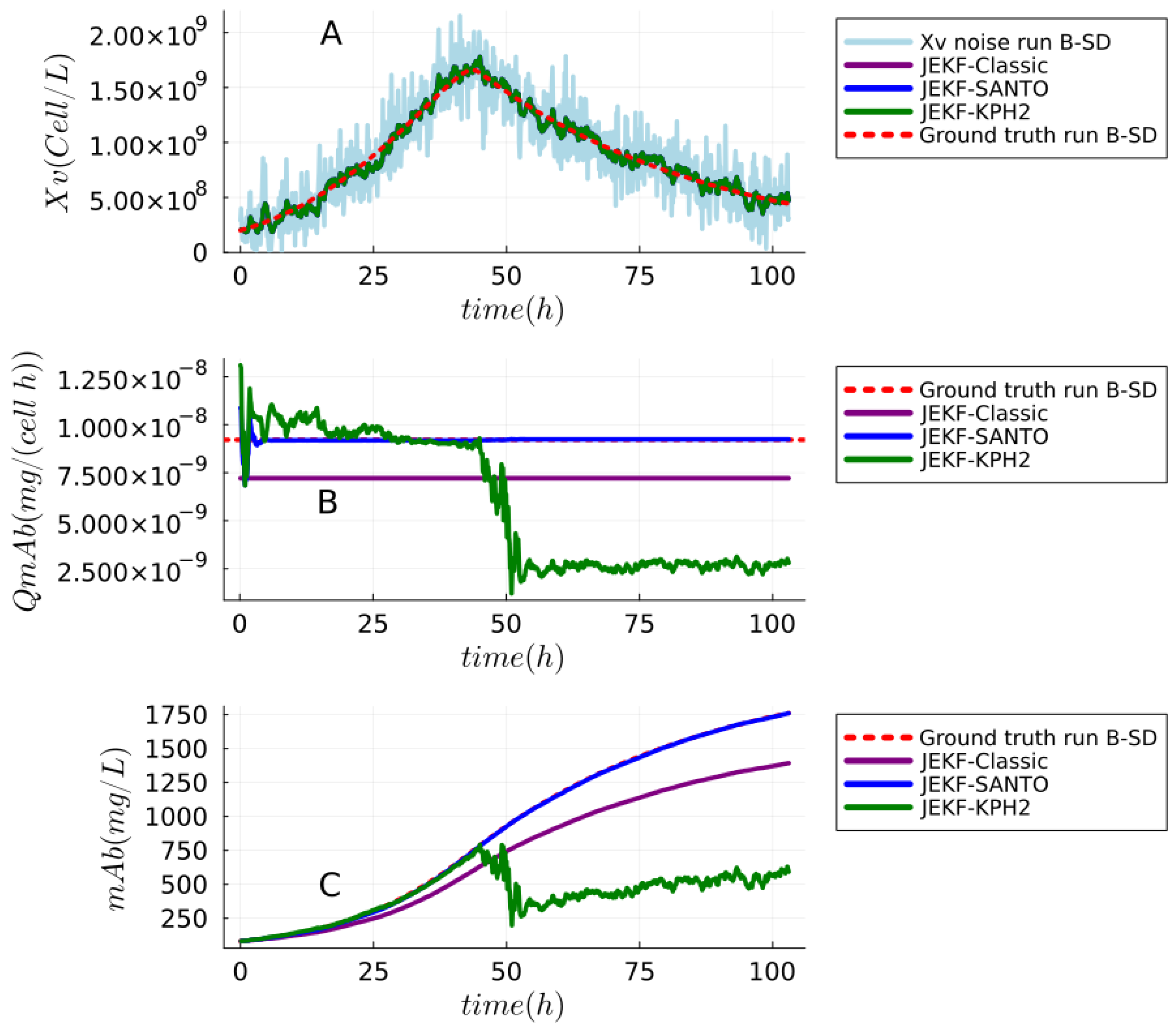
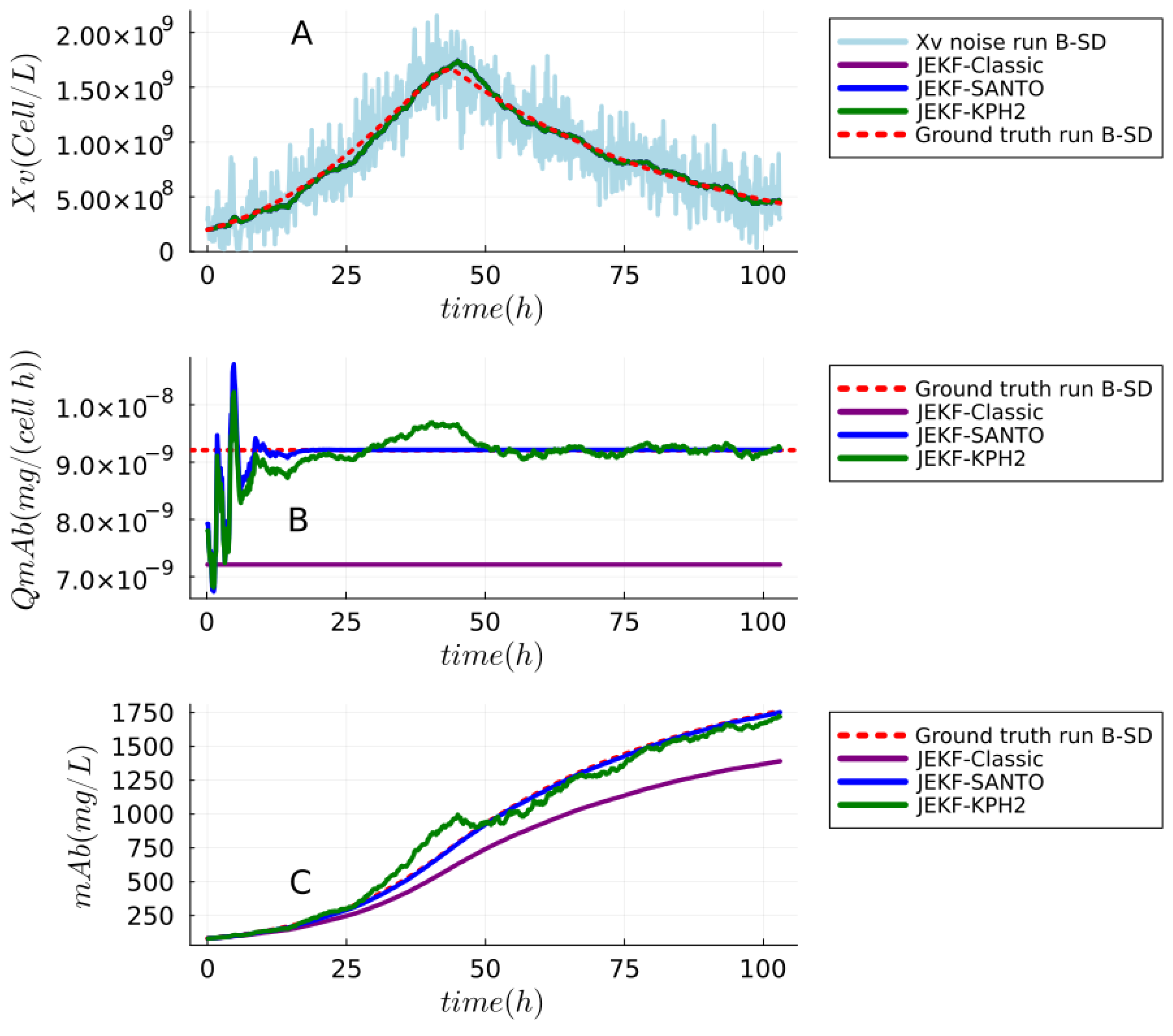
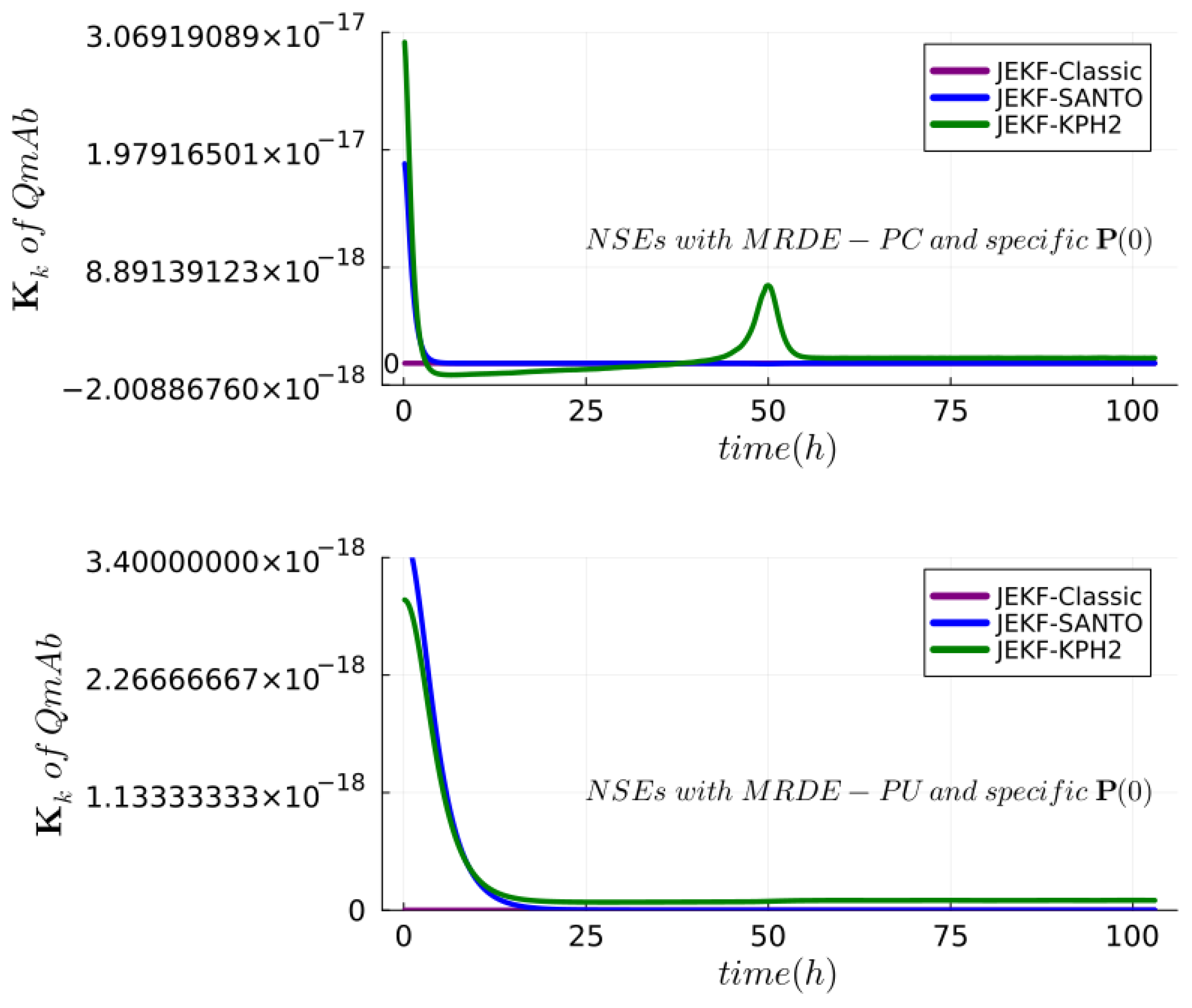

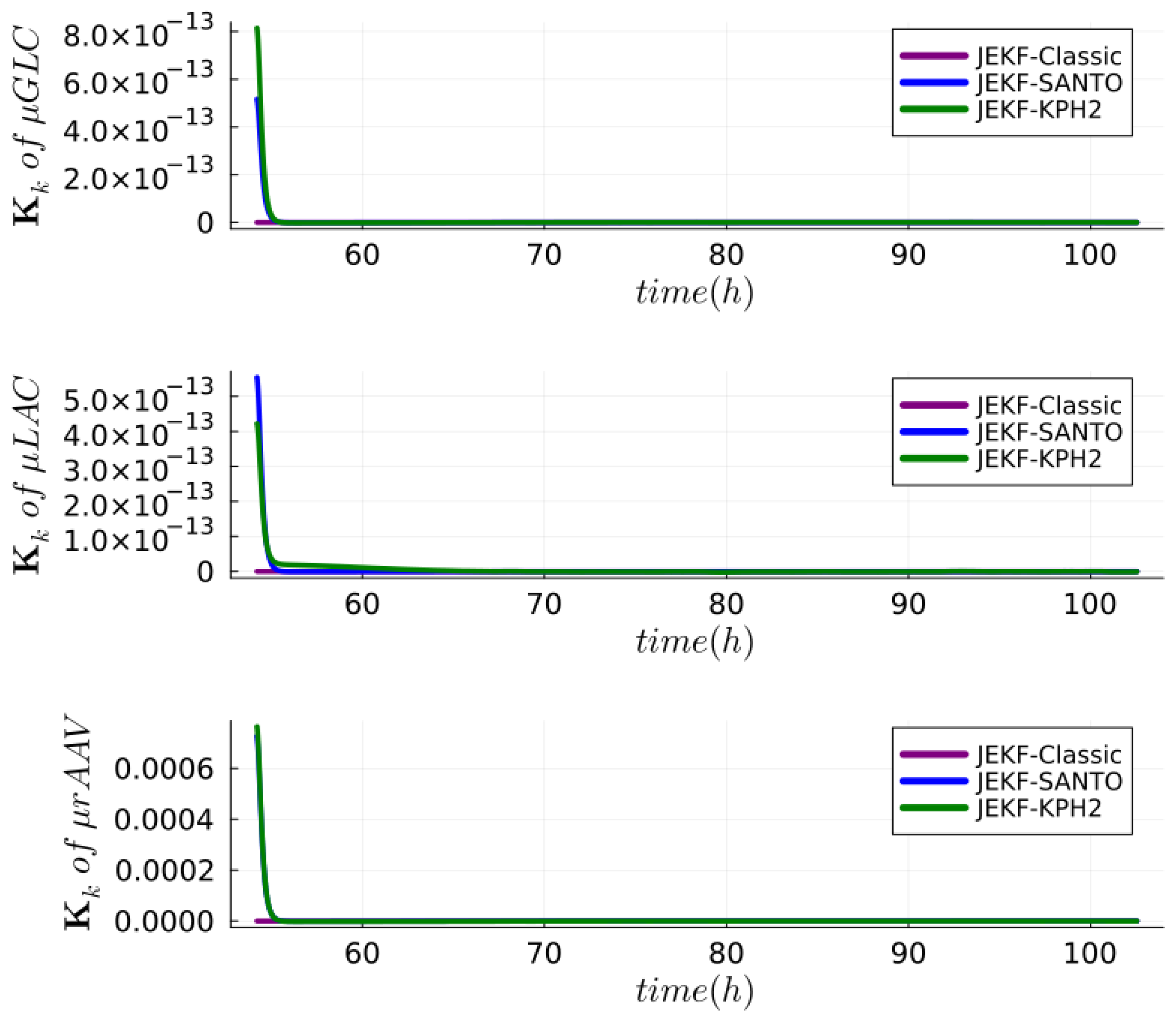
| NSE | RMSPE (MRDE-PU) | RMSPE (MRDE-PC) |
|---|---|---|
| JEKF-SANTO | 2.06% | 1.30% |
| JEKF-KPH2 | 10.11% | 48.44% |
| JEKF-Classic | 18.80% | 18.65% |
| Ground Truth | JEKF-Classic | JEKF-SANTO | JEKF-KPH2 |
|---|---|---|---|
| GLC | 11.6% | 3.48% | 3.58% |
| LAC | 16.66% | 1.01% | 1.61% |
| rAAV (titer) | 41.41% | 2.87% | 3.28% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iglesias, C.F., Jr.; Bolic, M. How Not to Make the Joint Extended Kalman Filter Fail with Unstructured Mechanistic Models. Sensors 2024, 24, 653. https://doi.org/10.3390/s24020653
Iglesias CF Jr., Bolic M. How Not to Make the Joint Extended Kalman Filter Fail with Unstructured Mechanistic Models. Sensors. 2024; 24(2):653. https://doi.org/10.3390/s24020653
Chicago/Turabian StyleIglesias, Cristovão Freitas, Jr., and Miodrag Bolic. 2024. "How Not to Make the Joint Extended Kalman Filter Fail with Unstructured Mechanistic Models" Sensors 24, no. 2: 653. https://doi.org/10.3390/s24020653
APA StyleIglesias, C. F., Jr., & Bolic, M. (2024). How Not to Make the Joint Extended Kalman Filter Fail with Unstructured Mechanistic Models. Sensors, 24(2), 653. https://doi.org/10.3390/s24020653







