Oxygen Sensor-Based Respirometry and the Landscape of Microbial Testing Methods as Applicable to Food and Beverage Matrices
Abstract
1. Introduction
2. Culture-Based Methods of Microbiological Testing
2.1. The TEMPO System
2.2. The Soleris System
3. Bioluminescent Detection of Cellular ATP
4. Molecular Methods
4.1. Immunoassays
4.1.1. Modifications of ELISA Tests
4.1.2. Lateral Flow Assays
4.1.3. Immuno-Magnetic Separation Systems (IMS)
4.2. Polymerase Chain Reaction (PCR) Methods
4.3. Loop-Mediated Isothermal Amplification (LAMP)
4.4. Next-Generationation Sequencing Methods
5. Instrumental Methods
5.1. Flow Cytometry and Cell Counting
5.2. Raman Spectroscopy
5.3. Mass Spectrometry Methods
5.4. Calorimetry
6. Oxygen Sensor-Based Respirometry in Microbial Testing
6.1. Measurement Principles
6.2. O2 Sensing Materials
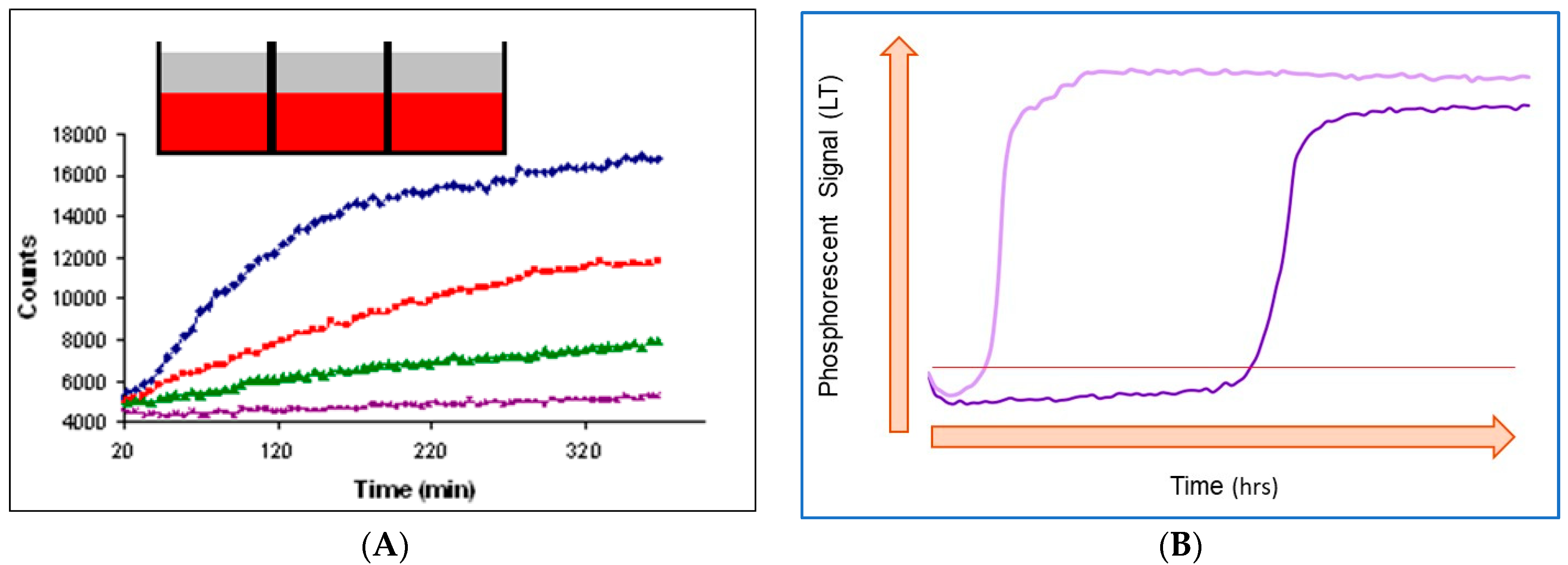
6.3. Typical Profiles in Optical O2 Respirometry and Their Analysis
6.3.1. Mammalian Cell Respiration
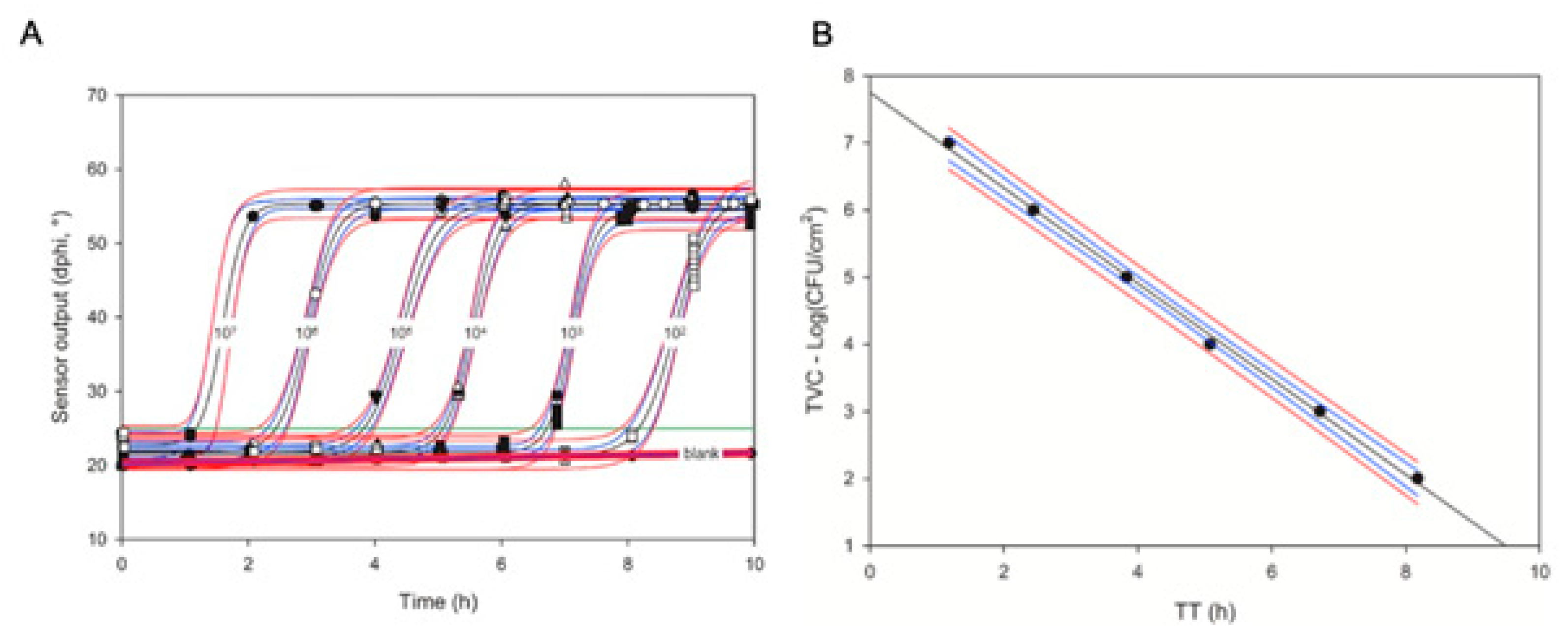
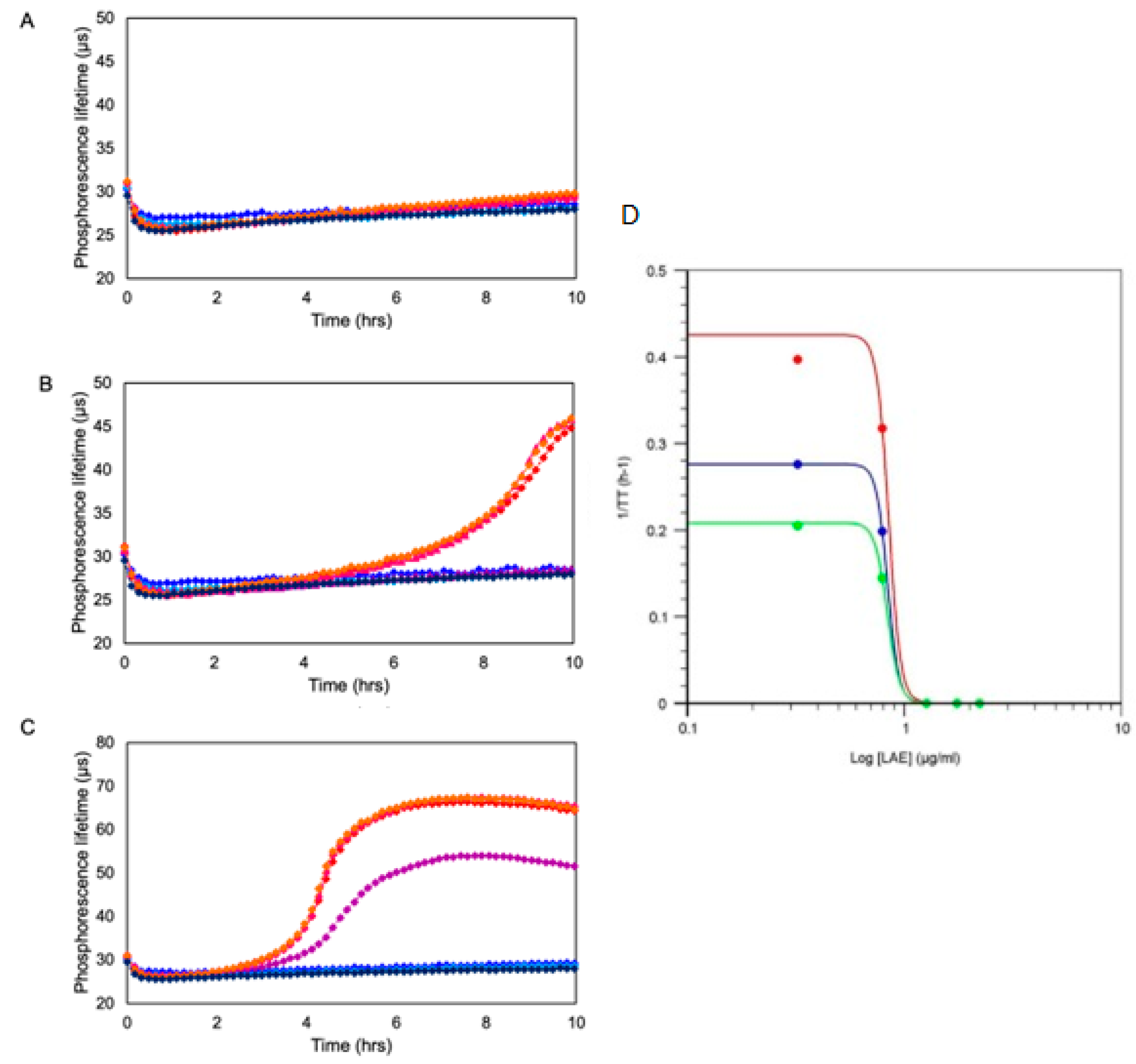
6.3.2. Respiration Profiles of Bacterial Cells and their Characteristic Features
- Take sample specimen and put it in assay medium that promotes microbial cell growth; homogenize, if required;
- Transfer the sample to a measurement chamber and record its sensor signals over time under standard (optimal) assay conditions;
- Determine (once-off) the optimal threshold sensor signal for these conditions and the recorded respiration profile(s), which accounts for sensor, sample and assay variability and avoids any false-positive or false-negative results;
- Apply the threshold and determine the corresponding TT value for each profile and sample;
- Apply the mathematical equation (pre-determined, see above) to convert measured TT values into corresponding cell counts (log (CFU/mL) or log (CFU/g) for each tested sample.
6.4. Existing Formats and Applications of Bacterial Respirometry
- (i)
- A soluble, dispensable O2-sensing probe MitoXpress-Xtra (Agilent) based on a Pt-porphyrin dye instead of the pre-made, solid-state sensor coatings;
- (ii)
- Standard 96/384-well plates, uncoated, tissue-culture treated and even customized assay substrates [74];
- (iii)
- Sealing the samples on the plate with mineral oil;
- (iv)
- Measuring the plate on a multilabel plater reader in the TRF/RLD mode [77], thus implementing LT-based O2 sensing.
- Measurement vessels type and size: 2 mL vials for Platform 6, 30–50 mL vials for Platform 7, 15 mL vials with swab brushes for Platform 8 and flexible plastic pouches for Platform 9;
- Sensor integration method: permanent coatings (dots at vial bottom) for Platforms 6, 7 or on vial side for Platform 8 or small inserts membrane type in Platform 9;
- Detector type: automated benchtop reader with incubator and carousel for sensor vials (GreenLight-930 in Platform 6) or autonomous handheld reader for in-field operation (Platforms 7–9).
6.5. Comparison of O2 Respirometry with Established Microbial Testing Methods
- Detection and enumeration of only viable cells via monitoring of their growth.
- Ultimate single cell sensitivity, broad concentrations range and minimal number of steps.
- Rapid mix-and-measure assay in liquid media with real-time signal output and time-to-result 1–10 h. Quantitative, accurate and automated.
- Assays require only disposable sensor vials, a simple sensor reader and an incubator. No special facilities, equipment or skills are needed.
- Various types of samples can be analyzed: swabs, crude homogenates, food and environmental samples, etc. Sample preparation is the same as in the ISO method.
- Choice of different sensor materials, assay substrates (microplates, vials, pouches) and detectors.
- Portable, autonomous, low-cost commercial sensor readers that provide LT-based sensing and low start-up costs for the whole system (from $2000 upwards).
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Franz, C.M.A.P.; den Besten, H.M.W.; Böhnlein, C.; Gareis, M.; Zwietering, M.H.; Fusco, V. Microbial food safety in the 21st century: Emerging challenges and foodborne pathogenic bacteria. Trends Food Sci. Technol. 2018, 81, 155–158. [Google Scholar] [CrossRef]
- Stellato, G.; La Storia, A.; De Filippis, F.; Borriello, G.; Villani, F.; Ercolini, D. Overlap of Spoilage-Associated Microbiota between Meat and the Meat Processing Environment in Small-Scale and Large-Scale Retail Distributions. Appl. Environ. Microbiol. 2016, 82, 4045–4054. [Google Scholar] [CrossRef]
- Nisa, M.; Dar, R.A.; Fomda, B.A.; Nazir, R. Combating food spoilage and pathogenic microbes via bacteriocins: A natural and eco-friendly substitute to antibiotics. Food Control. 2023, 149, 109710. [Google Scholar] [CrossRef]
- Capita, R.; Prieto, M.; Alonso-Calleja, C. Sampling Methods for Microbiological Analysis of Red Meat and Poultry Carcasses. J. Food Prot. 2004, 67, 1303–1308. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.L.; Ricke, S.C.; Roper, D.K.; Gibson, K.E. Swabbing the surface: Critical factors in environmental monitoring and a path towards standardization and improvement. Crit. Rev. Food Sci. Nutr. 2018, 60, 225–243. [Google Scholar] [CrossRef] [PubMed]
- ISO 4833-1:2013; Microbiology of the Food Chain—Horizontal Method for the Enumeration of Microorganisms. Part 1: Colony Count at 30 °C by the Pour Plate Technique. International Standards Organisation: Geneva, Switzerland, 2013.
- ISO 18593:2018; Microbiology of the Food Chain—Horizontal Methods for Surface Sampling. International Standards Organisation: Geneva, Switzerland, 2018.
- Jasson, V.; Jacxsens, L.; Luning, P.; Rajkovic, A.; Uyttendaele, M. Alternative microbial methods: An overview and selection criteria. Food Microbiol. 2010, 27, 710–730. [Google Scholar] [CrossRef]
- Bonnet, M.; Lagier, J.; Raoult, D.; Khelaifia, S. Bacterial culture through selective and non-selective conditions: The evolution of culture media in clinical microbiology. New Microbes New Infect. 2019, 34, 100622. [Google Scholar] [CrossRef]
- Cirolini, A.; Baseggio, A.M.; Miotto, M.; Ramos, R.J.; Cattani, C.S.D.O.; Vieira, C.R.W. Evaluation of the PetrifilmTM and TEMPO® systems and the conventional method for counting microorganisms in pasteurized milk. Food Sci. Technol. 2013, 33, 784–789. [Google Scholar] [CrossRef]
- Cayer, M.-P.; Dussault, N.; De Grandmont, M.J.; Cloutier, M.; Lewin, A.; Brouard, D. Evaluation of the Tempo® System: Improving the Microbiological Quality Monitoring of Human Milk. Front. Pediatr. 2020, 8, 494. [Google Scholar] [CrossRef]
- Gómez-Govea, M.; Solís-Soto, L.; Heredia, N.; García, S.; Moreno, G.; Tovar, O.; Isunza, G. Analysis of microbial contamination levels of fruits and vegetables at retail in Monterrey, Mexico. J. Food Agric. Environ. 2012, 10, 152–156. [Google Scholar]
- Foti, D.; Romano, L.; Alles, S.; A Mozola, M. Validation of the Soleris®E. coli Method for Detection and Semi-Quantitative Determination of Escherichia coli in Foods. J. AOAC Int. 2012, 95, 786–794. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tolan, J.; Lavigne, N.; Montei, C.; Donofrio, R.; Biswas, P. Soleris® Automated System for the Rapid Detection of Burkholderia cepacia Complex in Cosmetic Products. J. AOAC Int. 2022, 106, 171–178. [Google Scholar] [CrossRef]
- Bottari, B.; Santarelli, M.; Neviani, E. Determination of microbial load for different beverages and foodstuff by assessment of intracellular ATP. Trends Food Sci. Technol. 2015, 44, 36–48. [Google Scholar] [CrossRef]
- Adan, A.; Kiraz, Y.; Baran, Y. Cell proliferation and cytotoxicity assays. Curr. Pharm. Biotechnol. 2016, 17, 1213–1221. [Google Scholar] [CrossRef] [PubMed]
- Ríos-Castillo, A.G.; Ripolles-Avila, C.; Rodríguez-Jerez, J.J. Evaluation of bacterial population using multiple sampling methods and the identification of bacteria detected on supermarket food contact surfaces. Food Control. 2020, 119, 107471. [Google Scholar] [CrossRef]
- Monica, S.; Bancalari, E.; Castellone, V.; Rijkx, J.; Wirth, S.; Jahns, A.; Bottari, B. ATP Bioluminescence for Rapid and Selective Detection of Bacteria and Yeasts in Wine. Appl. Sci. 2021, 11, 4953. [Google Scholar] [CrossRef]
- Møretrø, T.; Normann, M.A.; Sæbø, H.R.; Langsrud, S. Evaluation of ATP bioluminescence-based methods for hygienic assessment in fish industry. J. Appl. Microbiol. 2019, 127, 186–195. [Google Scholar] [CrossRef]
- Nagaraj, S.; Ramlal, S.; Kingston, J.; Batra, H.V. Development of IgY based sandwich ELISA for the detection of staphylococcal enterotoxin G (SEG), an egc toxin. Int. J. Food Microbiol. 2016, 237, 136–141. [Google Scholar] [CrossRef]
- Xiao, X.; Hu, S.; Lai, X.; Peng, J.; Lai, W. Developmental trend of immunoassays for monitoring hazards in food samples: A review. Trends Food Sci. Technol. 2021, 111, 68–88. [Google Scholar] [CrossRef]
- Seddaoui, N.; Amine, A. Smartphone-based competitive immunoassay for quantitative on-site detection of meat adulteration. Talanta 2021, 230, 122346. [Google Scholar] [CrossRef]
- Mandli, J.; EL Fatimi, I.; Seddaoui, N.; Amine, A. Enzyme immunoassay (ELISA/immunosensor) for a sensitive detection of pork adulteration in meat. Food Chem. 2018, 255, 380–389. [Google Scholar] [CrossRef]
- Pang, B.; Zhao, C.; Li, L.; Song, X.; Xu, K.; Wang, J.; Liu, Y.; Fu, K.; Bao, H.; Song, D.; et al. Development of a low-cost paper-based ELISA method for rapid Escherichia coli O157:H7 detection. Anal. Biochem. 2018, 542, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Alamer, S.; Eissa, S.; Chinnappan, R.; Zourob, M. A rapid colorimetric immunoassay for the detection of pathogenic bacteria on poultry processing plants using cotton swabs and nanobeads. Microchim. Acta 2018, 185, 164. [Google Scholar] [CrossRef]
- Shan, S.; Lai, W.; Xiong, Y.; Wei, H.; Xu, H. Novel Strategies To Enhance Lateral Flow Immunoassay Sensitivity for Detecting Foodborne Pathogens. J. Agric. Food Chem. 2015, 63, 745–753. [Google Scholar] [CrossRef]
- Di Nardo, F.; Chiarello, M.; Cavalera, S.; Baggiani, C.; Anfossi, L. Ten Years of Lateral Flow Immunoassay Technique Applications: Trends, Challenges and Future Perspectives. Sensors 2021, 21, 5185. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cai, R.; Gao, Z.; Yuan, Y.; Yue, T. Immunomagnetic separation: An effective pretreatment technology for isolation and enrichment in food microorganisms detection. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3802–3824. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.-C.; Park, J.Y.; Park, K.; Ok, G.; Jang, H.-J.; Choi, S.-W. An automated system for separation and concentration of food-borne pathogens using immunomagnetic separation. Food Control. 2017, 73, 1541–1547. [Google Scholar] [CrossRef]
- Mansfield, L.P.; Forsythe, S. The detection of Salmonella serovars from animal feed and raw chicken using a combined immunomagnetic separation and ELISA method. Food Microbiol. 2001, 18, 361–366. [Google Scholar] [CrossRef]
- LeJeune, J.T.; Hancock, D.D.; Besser, T.E. Sensitivity of Escherichia coli O157 Detection in Bovine Feces Assessed by Broth Enrichment followed by Immunomagnetic Separation and Direct Plating Methodologies. J. Clin. Microbiol. 2006, 44, 872–875. [Google Scholar] [CrossRef]
- Hameed, S.; Xie, L.; Ying, Y. Conventional and emerging detection techniques for pathogenic bacteria in food science: A review. Trends Food Sci. Technol. 2018, 81, 61–73. [Google Scholar] [CrossRef]
- Kasturi, K.N.; Drgon, T. Real-Time PCR Method for Detection of Salmonella spp. in Environmental Samples. Appl. Environ. Microbiol. 2017, 83, e00644-17. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, A.; Gordillo, R.; Andrade, M.J.; Córdoba, J.J.; Rodríguez, M. Development of an efficient real-time PCR assay to quantify enterotoxin-producing staphylococci in meat products. Food Control 2016, 60, 302–308. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic. Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zou, D.; Dong, D.; Yang, Z.; Ao, D.; Liu, W.; Huang, L. Development of a multiplex loop-mediated isothermal amplification method for the simultaneous detection of Salmonella spp. and Vibrio parahaemolyticus. Sci. Rep. 2017, 7, srep45601. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, S.; Zhao, Y.; Yang, X.; Fu, S.; McKillip, J.L.; Fox, E.M.; Man, C. Multiplex loop-mediated isothermal amplification-based lateral flow dipstick for simultaneous detection of 3 food-borne pathogens in powdered infant formula. J. Dairy Sci. 2020, 103, 4002–4012. [Google Scholar] [CrossRef]
- Jagadeesan, B.; Gerner-Smidt, P.; Allard, M.W.; Leuillet, S.; Winkler, A.; Xiao, Y.; Chaffron, S.; Van Der Vossen, J.; Tang, S.; Katase, M.; et al. The use of next generation sequencing for improving food safety: Translation into practice. Food Microbiol. 2018, 79, 96–115. [Google Scholar] [CrossRef]
- Li, S.; Mann, D.A.; Zhang, S.; Qi, Y.; Meinersmann, R.J.; Deng, X. Microbiome-Informed Food Safety and Quality: Longitudinal Consistency and Cross-Sectional Distinctiveness of Retail Chicken Breast Microbiomes. mSystems 2020, 5(5), e00589-20. [Google Scholar] [CrossRef]
- Doster, E.; Thomas, K.M.; Weinroth, M.D.; Parker, J.K.; Crone, K.K.; Arthur, T.M.; Schmidt, J.W.; Wheeler, T.L.; Belk, K.E.; Morley, P.S. Metagenomic Characterization of the Microbiome and Resistome of Retail Ground Beef Products. Front. Microbiol. 2020, 11, 541972. [Google Scholar] [CrossRef]
- Wang, Y.; Salazar, J.K. Culture-Independent Rapid Detection Methods for Bacterial Pathogens and Toxins in Food Matrices. Compr. Rev. Food Sci. Food Saf. 2016, 15, 183–205. [Google Scholar] [CrossRef]
- Liu, S.; Wang, B.; Sui, Z.; Wang, Z.; Li, L.; Zhen, X.; Zhao, W.; Zhou, G. Faster Detection of Staphylococcus aureus in Milk and Milk Powder by Flow Cytometry. Foodborne Pathog. Dis. 2021, 18, 346–353. [Google Scholar] [CrossRef]
- Genovese, M.; Poulain, E.; Doppler, F.; Toussaint, R.; Boyer, M. Bacillus spore enumeration using flow cytometry: A proof of concept for probiotic application. J. Microbiol. Methods 2021, 190, 106336. [Google Scholar] [CrossRef] [PubMed]
- Afari, G.K.; Hung, Y.-C. Detection and Verification of the Viable but Nonculturable (VBNC) State of Escherichia coli O157:H7 and Listeria monocytogenes Using Flow Cytometry and Standard Plating. J. Food Sci. 2018, 83, 1913–1920. [Google Scholar] [CrossRef] [PubMed]
- Pahlow, S.; Meisel, S.; Cialla-May, D.; Weber, K.; Rösch, P.; Popp, J. Isolation and identification of bacteria by means of Raman spectroscopy. Adv. Drug Deliv. Rev. 2015, 89, 105–120. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Wang, S.; Qiu, J.; Li, M.; Li, D.; Xu, D.; Li, D.; Liu, Q. Raman spectroscopy combined with machine learning for rapid detection of food-borne pathogens at the single-cell level. Talanta 2021, 226, 122195. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Taylor, J.N.; Koseki, S.; Koyama, K. Classification of food spoilage bacterial species and their sodium chloride, sodium acetate and glycine tolerance using chemometrics analysis and Raman spectroscopy. J. Microbiol. Methods 2021, 190, 106326. [Google Scholar] [CrossRef]
- Meisel, S.; Stöckel, S.; Rösch, P.; Popp, J. Identification of meat-associated pathogens via Raman microspectroscopy. Food Microbiol. 2014, 38, 36–43. [Google Scholar] [CrossRef]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef]
- Domínguez, I.; Frenich, A.G.; Romero-González, R. Mass spectrometry approaches to ensure food safety. Anal. Methods 2020, 12, 1148–1162. [Google Scholar] [CrossRef]
- Jadhav, S.; Sevior, D.; Bhave, M.; Palombo, E.A. Detection of Listeria monocytogenes from selective enrichment broth using MALDI–TOF Mass Spectrometry. J. Proteom. 2014, 97, 100–106. [Google Scholar] [CrossRef]
- Vithanage, N.R.; Yeager, T.R.; Jadhav, S.R.; Palombo, E.A.; Datta, N. Comparison of identification systems for psychrotrophic bacteria isolated from raw bovine milk. Int. J. Food Microbiol. 2014, 189, 26–38. [Google Scholar] [CrossRef]
- Peruzy, M.; Murru, N.; Yu, Z.; Kerkhof, P.-J.; Neola, B.; Joossens, M.; Proroga, Y.; Houf, K. Assessment of microbial communities on freshly killed wild boar meat by MALDI-TOF MS and 16S rRNA amplicon sequencing. Int. J. Food Microbiol. 2019, 301, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Fricke, C.; Harms, H.; Maskow, T. How to speed up the detection of aerobic microbial contaminations by using isothermal microcalorimetry. J. Therm. Anal. Calorim. 2020, 142, 1933–1949. [Google Scholar] [CrossRef]
- von Ah, U.; Shani, N.; Chollet, M.; Solokhina, A.; Braissant, O. Measuring antibiotic resistance in mixed cultures: Isothermal microcalorimetry as a novel analytical tool. Int. Dairy J. 2018, 77, 73–79. [Google Scholar] [CrossRef]
- Khalef, N.; Campanella, O.; Bakri, A. Isothermal calorimetry: Methods and applications in food and pharmaceutical fields. Curr. Opin. Food Sci. 2016, 9, 70–76. [Google Scholar] [CrossRef]
- Papkovsky, D.B.; Dmitriev, R.I. Biological detection by optical oxygen sensing. Chem. Soc. Rev. 2013, 42, 8700–8732. [Google Scholar] [CrossRef] [PubMed]
- Borisov, S.M. CHAPTER 1 Fundamentals of Quenched Phosphorescence O2 Sensing and Rational Design of Sensor Materials, Quenched-Phosphorescence Detection of Molecular Oxy-Gen: Applications in Life Sciences; The Royal Society of Chemistry: London, UK, 2018; pp. 1–18. [Google Scholar] [CrossRef]
- Wang, X.-D.; Wolfbeis, O.S. Optical methods for sensing and imaging oxygen: Materials, spectroscopies and applications. Chem. Soc. Rev. 2014, 43, 3666–3761. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, NY, USA, 2006. [Google Scholar]
- Demas, J.N.; DeGraff, B.A.; Xu, W. Modeling of Luminescence Quenching-Based Sensors: Comparison of Multisite and Nonlinear Gas Solubility Models. Anal. Chem. 1995, 67, 1377–1380. [Google Scholar] [CrossRef]
- Banerjee, S.; Kelly, C.; Kerry, J.P.; Papkovsky, D.B. High throughput non-destructive assessment of quality and safety of packaged food products using phosphorescent oxygen sensors. Trends Food Sci. Technol. 2016, 50, 85–102. [Google Scholar] [CrossRef]
- Papkovsky, D.B.; Hynes, J.; Will, Y. Respirometric Screening Technology for ADME-Tox studies. Expert Opin. Drug Metab. Toxicol. 2006, 2, 313–323. [Google Scholar] [CrossRef]
- Ballew, R.M.; Demas, J.N. An error analysis of the rapid lifetime determination method for the evaluation of single exponential decays. Anal. Chem. 1989, 61, 30–33. [Google Scholar] [CrossRef]
- Ogurtsov, V.I.; Papkovsky, D.B. Selection of modulation frequency of excitation for luminescence lifetime-based oxygen sensors. Sens. Actuators B Chem. 1998, 51, 377–381. [Google Scholar] [CrossRef]
- Schmälzlin, E.; van Dongen, J.T.; Klimant, I.; Marmodée, B.; Steup, M.; Fisahn, J.; Geigenberger, P.; Löhmannsröben, H.-G. An Optical Multifrequency Phase-Modulation Method Using Microbeads for Measuring Intracellular Oxygen Concentrations in Plants. Biophys. J. 2005, 89, 1339–1345. [Google Scholar] [CrossRef] [PubMed]
- Koren, K.; Dmitriev, R.I.; Borisov, S.M.; Papkovsky, D.B.; Klimant, I. Complexes of IrIII-Octaethylporphyrin with Peptides as Probes for Sensing Cellular O2. ChemBioChem 2012, 13, 1184–1190. [Google Scholar] [CrossRef] [PubMed]
- Hempel, A.; Borchert, N.; Walsh, H.; Choudhury, K.R.; Kerry, J.; Papkovsky, D. Analysis of Total Aerobic Viable Counts in Raw Fish by High-Throughput Optical Oxygen Respirometry. J. Food Prot. 2011, 74, 776–782. [Google Scholar] [CrossRef]
- Elisseeva, S.; Santovito, E.; Linehan, E.; Kerry, J.P.; Papkovsky, D.B. Performance assessment of the two oxygen sensor based respirometric platforms with complex media and in selective bacterial assays. Sens. Actuators B Chem. 2023, 383, 133582. [Google Scholar] [CrossRef]
- Zhdanov, A.V.; Ogurtsov, V.I.; Taylor, C.T.; Papkovsky, D.B. Monitoring of cell oxygenation and responses to metabolic stimulation by intracellular oxygen sensing technique. Integr. Biol. 2010, 2, 443–451. [Google Scholar] [CrossRef]
- Zitova, A.; O’Mahony, F.C.; Cross, M.; Davenport, J.; Papkovsky, D.B. Toxicological profiling of chemical and environmental samples using panels of test organisms and optical oxygen respirometry. Environ. Toxicol. 2009, 24, 116–127. [Google Scholar] [CrossRef]
- Hynes, J.; Floyd, S.; Soini, A.E.; O’Connor, R.; Papkovsky, D.B. Fluorescence-Based Cell Viability Screening Assays Using Water-Soluble Oxygen Probes. SLAS Discov. Adv. Sci. Drug Discov. 2003, 8, 264–272. [Google Scholar] [CrossRef]
- Ferrick, D.A.; Neilson, A.; Beeson, C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov. Today 2008, 13, 268–274. [Google Scholar] [CrossRef]
- O’Donovan, C.; Twomey, E.; Alderman, J.; Moore, T.; Papkovsky, D. Development of a respirometric biochip for embryo assessment. Lab A Chip 2006, 6, 1438–1444. [Google Scholar] [CrossRef]
- Müller, B.; Sulzer, P.; Walch, M.; Zirath, H.; Buryška, T.; Rothbauer, M.; Ertl, P.; Mayr, T. Measurement of respiration and acidification rates of mammalian cells in thermoplastic microfluidic devices. Sens. Actuators B Chem. 2021, 334, 29664. [Google Scholar] [CrossRef]
- Gerencser, A.A.; Neilson, A.; Choi, S.W.; Edman, U.; Yadava, N.; Oh, R.J.; Ferrick, D.A.; Nicholls, D.G.; Brand, M.D. Quantitative Microplate-Based Respirometry with Correction for Oxygen Diffusion. Anal. Chem. 2009, 81, 6868–6878. [Google Scholar] [CrossRef] [PubMed]
- Will, Y.; Hynes, J.; I Ogurtsov, V.; Papkovsky, D. Analysis of mitochondrial function using phosphorescent oxygen-sensitive probes. Nat. Protoc. 2006, 1, 2563–2572. [Google Scholar] [CrossRef] [PubMed]
- O’Mahony, F.C.; Papkovsky, D.B. Rapid High-Throughput Assessment of Aerobic Bacteria in Complex Samples by Fluorescence-Based Oxygen Respirometry. Appl. Environ. Microbiol. 2006, 72, 1279–1287. [Google Scholar] [CrossRef]
- Santovito, E.; Elisseeva, S.; Cruz-Romero, M.C.; Duffy, G.; Kerry, J.P.; Papkovsky, D.B. A Simple Sensor System for Onsite Monitoring of O2 in Vacuum-Packed Meats during the Shelf Life. Sensors 2021, 21, 4256. [Google Scholar] [CrossRef]
- Santovito, E.; Elisseeva, S.; Bukulin, A.; Kerry, J.P.; Papkovsky, D.B. Facile biosensor-based system for on-site quantification of total viable counts in food and environmental swabs. Biosens. Bioelectron. 2020, 176, 112938. [Google Scholar] [CrossRef]
- O’Mahony, F.; Green, R.A.; Baylis, C.; Fernandes, R.; Papkovsky, D.B. Analysis of total aerobic viable counts in samples of raw meat using fluorescence-based probe and oxygen consumption assay. Food Control. 2009, 20, 129–135. [Google Scholar] [CrossRef]
- Fernandes, R.; Carey, C.; Hynes, J.; Papkovsky, D. GreenLight™ Model 960. J. AOAC Int. 2019, 96, 369–385. [Google Scholar] [CrossRef]
- Borchert, N.; Hempel, A.; Walsh, H.; Kerry, J.P.; Papkovsky, D.B. High throughput quality and safety assessment of packaged green produce using two optical oxygen sensor based systems. Food Control. 2012, 28, 87–93. [Google Scholar] [CrossRef]
- Jasionek, G.; Ogurtsov, V.; Papkovsky, D. Rapid detection and respirometric profiling of aerobic bacteria on panels of selective media. J. Appl. Microbiol. 2012, 114, 423–432. [Google Scholar] [CrossRef]
- Elisseeva, S.; Kelly, C.; Cruz-Romero, M.; Zhdanov, A.V.; Kerry, J.P.; Papkovsky, D.B. The use of optical oxygen sensing and respirometry to quantify the effects of antimicrobials on common food spoilage bacteria and food samples. Sensors Actuators B Chem. 2020, 322, 128572. [Google Scholar] [CrossRef]
- Li, L.; Zhdanov, A.V.; Papkovsky, D.B. Advanced multimodal solid-state optochemical pH and dual pH/O2 sensors for cell analysis. Sens. Actuators B Chem. 2022, 371, 132486. [Google Scholar] [CrossRef]
- Flamholz, A.I.; Saccomano, S.; Cash, K.; Newman, D.K. Optical O2 Sensors Also Respond to Redox Active Molecules Commonly Secreted by Bacteria. mBio 2022, 13, e02076-22. [Google Scholar] [CrossRef]
- Ardito, F.; Posteraro, B.; Sanguinetti, M.; Zanetti, S.; Fadda, G. Evaluation of BACTEC Mycobacteria Growth Indicator Tube (MGIT 960) Automated System for Drug Susceptibility Testing of Mycobacterium tuberculosis. J. Clin. Microbiol. 2001, 39, 4440–4444. [Google Scholar] [CrossRef] [PubMed]
- Guarino, R.D.; Dike, L.E.; Haq, T.A.; Rowley, J.A.; Pitner, J.B.; Timmins, M.R. Method for determining oxygen consumption rates of static cultures from microplate measurements of pericellular dissolved oxygen concentration. Biotechnol. Bioeng. 2004, 86, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Hutter, B.; John, G.T. Evaluation of OxoPlate for real-time assessment of antibacterial activities. Curr. Microbiol. 2004, 48, 57–61. [Google Scholar] [CrossRef]
- Santovito, E.E.S.; Kerry, J.P.; Papkovsky, D.B. Rapid detection of bacterial load in food samples using disposable respirometric sensor sachets. Sens. Actuat. B, 2023; in press. [Google Scholar]
- Hsu, W.-T.; Aulakh, R.P.S.; Traul, D.L.; Yuk, I.H. Advanced microscale bioreactor system: A representative scale-down model for bench-top bioreactors. Cytotechnology 2012, 64, 667–678. [Google Scholar] [CrossRef]
- Ungerböck, B.; Mayr, T. CHAPTER 14 Microfluidic Systems and Optical Oxygen Sensors: A Perfect Match for Advancing Bioprocessing and Microbiology, Quenched-Phosphorescence Detection of Molecular Oxygen: Applications in Life Sciences; The Royal Society of Chemistry: London, UK, 2018; pp. 278–297. [Google Scholar] [CrossRef]
- IKlimant, I.; Meyer, V.; Kühl, M. Fiber-optic oxygen microsensors, a new tool in aquatic biology. Limnol. Oceanogr. 1995, 40, 1159–1165. [Google Scholar] [CrossRef]
- Sandner, V.; Pybus, L.; McCreath, G.; Glassey, J. Scale-Down Model Development in ambr systems: An Industrial Perspective. Biotechnol. J. 2018, 14, e1700766. [Google Scholar] [CrossRef]

| Parameter | Mammalian Cell Respirometry (1) | Bacterial Cell Respirometry (2) | Comments |
|---|---|---|---|
| O2 Sensor/probe type | Various, soluble or solid-state | Various, soluble or solid-state | Interchangeable, different dyes |
| Assay substrate | Microwell plates, special substrates | Standard plates and plastic tubes, sachets | More critical for (1) than (2) |
| Sample sealing options | Oil seal, sealable microchambers | Oil seal or no seal (liquid barrier) | Critical for (1) Less critical for (2) |
| Assay volume and sample type | 0.05–1 mL, simple and uniform | 0.1–50 mL, complex and variable | Larger for (2) |
| Cell number range | 104–2 × 105 per well | Single-cell—107 cells/mL | May vary for the different apps |
| Assay T, °C | 37 °C | 7 °C, 30 °C, 37 °C | Cell-specific |
| Instrumentation | Benchtop reader with T-control | Handheld reader + Incubator | Stationary vs transportable |
| Measurement time | 5 min–2 h | 1–16 h | Longer for (2) |
| Frequency of reads | 1–5 min Or end-point reads | 10–60 min Or end-point reads | Slower for manual readers |
| Repeated measurements | Desirable and common | No | Samples discarded |
| Signal monitored | Int, LT, [O2] | Int, LT, [O2] | LT is preferred |
| Profile shape | Linear or extended sigmoid | Steep sigmoid | May be different in some apps |
| What parts are analyzed and how | Linear part–signal slope or [O2] slope | Signal onset—time to threshold, TT | |
| Calibration used | Sensor LT vs [O2] | TT vs log(CFU/mL) | Once-off calibrations |
| Assay readout | Absolute or relative OCR values | Cell counts—CFU/g or CFU/mL | May vary in different apps |
| Assay Volume | LOD1 for 1 CFU/Sample | LOD10 for 10 CFU/Sample | LOD10 plus 1:10 Sample Dilution | Application |
|---|---|---|---|---|
| 0.1 mL | 10 CFU/mL | 100 CFU/mL | 1000 CFU/ml | Screening |
| 1.0 mL | 1 CFU/mL | 10 CFU/mL | 100 CFU/ml | Research, food |
| 10 mL | 0.1 CFU/mL | 1 CFU/mL | 10 CFU/ml | Food safety |
| 50 mL | 0.02 CFU/mL | 0.2 CFU/mL | 1 CFU/ml | Sterility tests |
| No. | Platform | Sensor Dye and Format | Assay Substrate and Settings | Detector, Optical Readout | Application | Refs. |
|---|---|---|---|---|---|---|
| 1. | Bactec System 1 | Ru(dpp)3 (470/615 nm), solid-state coating | Coated glass vials with caps | Customized detector with incubator, intensity-based | Detection of microbial infections in blood samples | [88] |
| 2. | BD Biosensor System 1 | Ru-dye (470/615 nm), solid-state coating | Bottom-coated 96-WP | Standard plate reader with T control, Intensity-based | Microbial respiration (general), antimicrobials | [89] |
| 3. | Sensor Plates 2.3 e.g., OxoDish | PtPFPP2 (525/650 nm) or PtBP3 (615/760 nm), solid-state coating | Bottom-coated 24-WP | Multichannel phase detector, phase shift | Detection of microbial respiration (general) | [90] |
| 4. | Seahorse XF Analyser 4 | PtPFPP dye, solid- state coating on lid pins | Special 24/96-WP, plus lid with pins and sensors. Seal able microwells | Customized detector with incubator, intensity-based | Mammalian cell respiration, OCRs, multiparametric analysis of cell bio- energetics. Research apps | [73] |
| 5. | MitoXpress probe 5 | PtCP dye (390/650 nm), liquid dispensable probe | Standard 96/384- WP, other sub- strates, biochips. Oil seal. | Standard TR-F reader with T control, LT based | Detection of bacterial growth, enumeration—TVCs Analysis of food samples, antimicrobials, etc. | [78,81,82,83,84] |
| 6. | GreenLight 930 System 6 | PtBP dye (615/760 nm), solid-state coating | 2 mL plastic vials with sensor dots | Benchtop carousel reader and incubator, LT based | Enumeration of bacteria, bacterial growth and inhibition assays | [82] |
| 7. | Sensor vial system | PtBP dye (615/760 nm), solid-state coating | 15/50 mL plastic vials with sensor dots | Handheld reader, LT or phase shift | Detection of bacteria in cultures, complex samples (homogenates), enumeration—TVCs | [79] |
| 8. | Swab vial system | PtBP dye (615/760 nm), solid-state coating | 15 mL vials with swab brushes and sensor dots | Handheld reader, LT or phase shift measurements | Detection of bacteria in surface, carcass, envi- ronmental swabs, enumeration—TVCs | [80] |
| 9. | Sensor pouches | PtBP dye (615/760 nm), membrane insert | Sealable plastic sachets with sensor inserts | Handheld reader, LT or phase shift measurements | Detection of bacteria in various foods, enumeration —TVCs | [91] |
| 10. | ambr® micro bioreactor system 7 | PtPFPP dye (525/650 nm), solid-state O2 and pH-sensitive coatings | Disposable 15 mL microbioreactors with O2 and pH sensors | Multichannel phase detector, phase shift | High throughput optimiza- tion of cell culturing conditions and media— bioprocessing | [92] |
| 11. | Fluidic biochips | PtBP dye (615/760 nm), soluble NP probe 5 | Microfluidic biochips, probe in the media | Portable benchtop reader, phase shift measurements | Mammalian cell respiration, OCRs, research apps | [75,93] |
| 12. | Micro-sensors 2,3 | PtPFPP or PtBP dye in sol–gel matrix | Sensor on tip of optical fiber probe | Portable reader, LT- based sensing— phase shift | Dipstick O2 and OCR probe, microbial communities, biofilms | [94] |
| Parameter | Sensor Respi-Rometry | Agar Plating | Tempo BioMerieux | Soleris | ATP BL | PCR, LAMP and Alike | ELISA, Lateral Flow | IMS | Flow Cytometry | Raman, SCRS | MS |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Time to Result, h | 1–8 h | 24–72 h | 6–24 h | 6-24 h | <1 h | 6–12 h | 3 h | 4 h | <1 h | <1 h | 1 h |
| Detection of viable cells only | +++ | +++ | +++ | +++ | ++ | + | + | ++ | ++ | + | - |
| Sensitivity/LOD, CFU | 1 CFU | 1 CFU | 1 CFU | 1 CFU | 10–100 | 10–100 | 102–105 | 10–100 | >1 cell | >1 cell | 102–105 |
| Dynamic range | 7–8 Logs | several Logs4 | several Logs | several Logs | 4–6 Logs | 3–4 Logs | 2–4 Logs | 3–4 Logs | 3–4 Logs | unknown | unknown |
| Total/Selective cell counts possible | yes/yes 1 | yes/yes | yes/yes 3 | yes/yes | yes/yes 3 | no/yes | no/yes | no/yes | yes/yes 2 | no/yes 2 | no/yes |
| Sample/assay volume, mL | 0.1–50 | 10 | 1 | 10 | 0.1–1 | 1–2 | 0.1 mL | 10 mL | 0.1–1 | 0.1 mL | 0.1 mL |
| No of samples processed (batch size) | 1–50 | 1–20 | 1–100 | 1–20 | 10–100 | 1–10 | 1–10 | 1–10 | 10–20 | 1–10 | 1–20 |
| Sample preparation time, h | 0.3 | 1 | 0.5 | 0.3 | 0.3 | 3 h | 1 h | 1 h | 0.5 h | 0.5 h | 1 h |
| Cell lysis/enrichment/ clarification required | no | no | partial | partial | yes | yes | yes/no | yes/no | no | yes | yes |
| Matrix effects, false +/--ves | +/- | - | + | + | ++ | ++ | ++ | + | ++ | ++ | - |
| Waste and risk of contamination | + | +++ | + | + | + | ++ | ++ | ++ | + | + | + |
| Detector costs, $ | 1–30 k | none | >30 k | 1 k | 5 k | 5 0k | 0–5 k | >5 k | >50 k | >50 k | >200 k |
| Additional equipment and facilities required | + | ++ | +++ | + | + | +++ | + | ++ | ++ | ++ | +++ |
| Assay costs, $ | 1–5 $ | 2–0 | >20 | 30 | 10 | >10 | 3–10 | 10–20 | >5 | >10 | >3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papkovsky, D.B.; Kerry, J.P. Oxygen Sensor-Based Respirometry and the Landscape of Microbial Testing Methods as Applicable to Food and Beverage Matrices. Sensors 2023, 23, 4519. https://doi.org/10.3390/s23094519
Papkovsky DB, Kerry JP. Oxygen Sensor-Based Respirometry and the Landscape of Microbial Testing Methods as Applicable to Food and Beverage Matrices. Sensors. 2023; 23(9):4519. https://doi.org/10.3390/s23094519
Chicago/Turabian StylePapkovsky, Dmitri B., and Joseph P. Kerry. 2023. "Oxygen Sensor-Based Respirometry and the Landscape of Microbial Testing Methods as Applicable to Food and Beverage Matrices" Sensors 23, no. 9: 4519. https://doi.org/10.3390/s23094519
APA StylePapkovsky, D. B., & Kerry, J. P. (2023). Oxygen Sensor-Based Respirometry and the Landscape of Microbial Testing Methods as Applicable to Food and Beverage Matrices. Sensors, 23(9), 4519. https://doi.org/10.3390/s23094519





