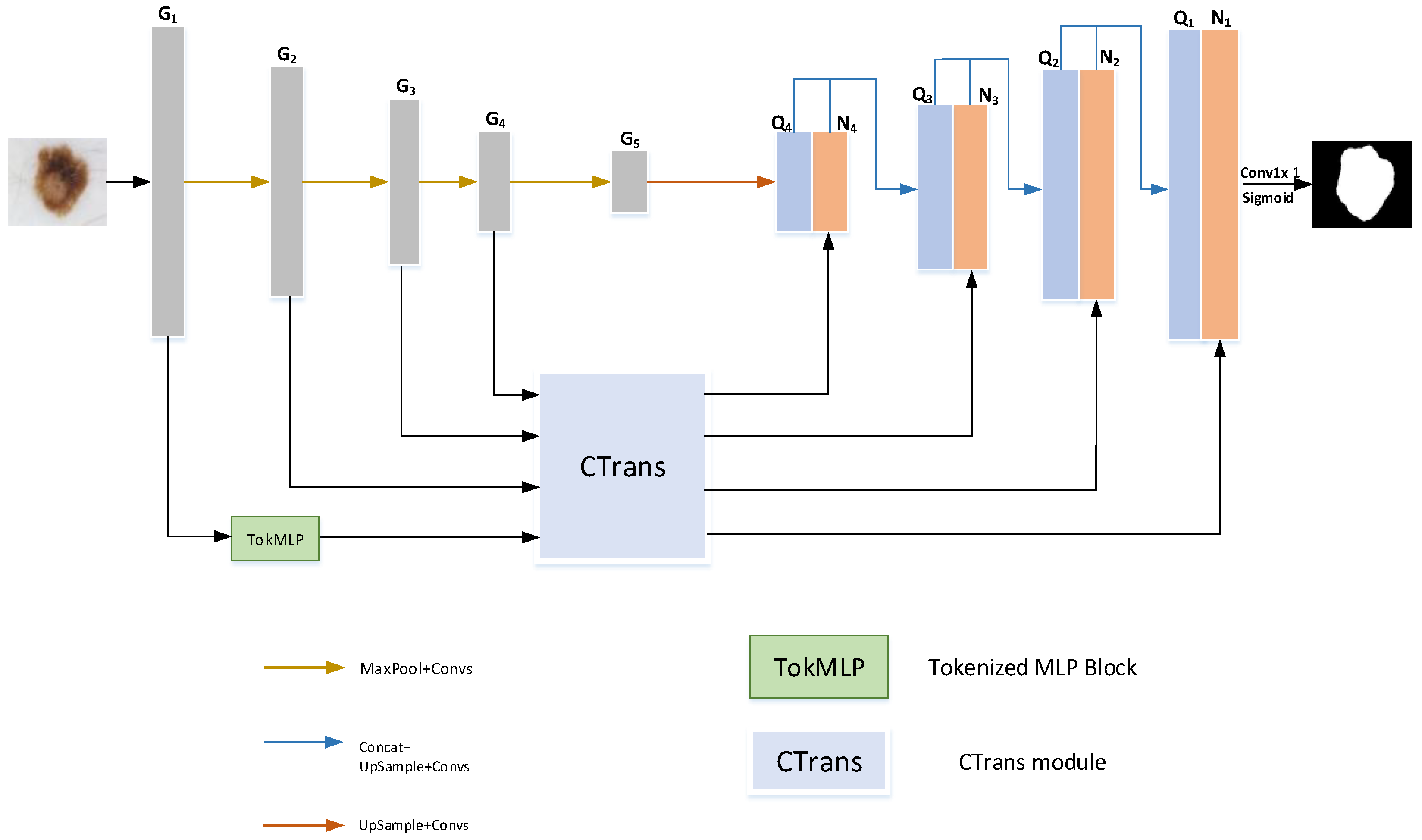
This chapter begins with a brief introduction to the overall framework of HMT-Net. Then, the composition, structure, and implementation details of the TokMLP block are introduced. Finally, we introduce the CTrans module in detail.
3.2. TokMLP Block
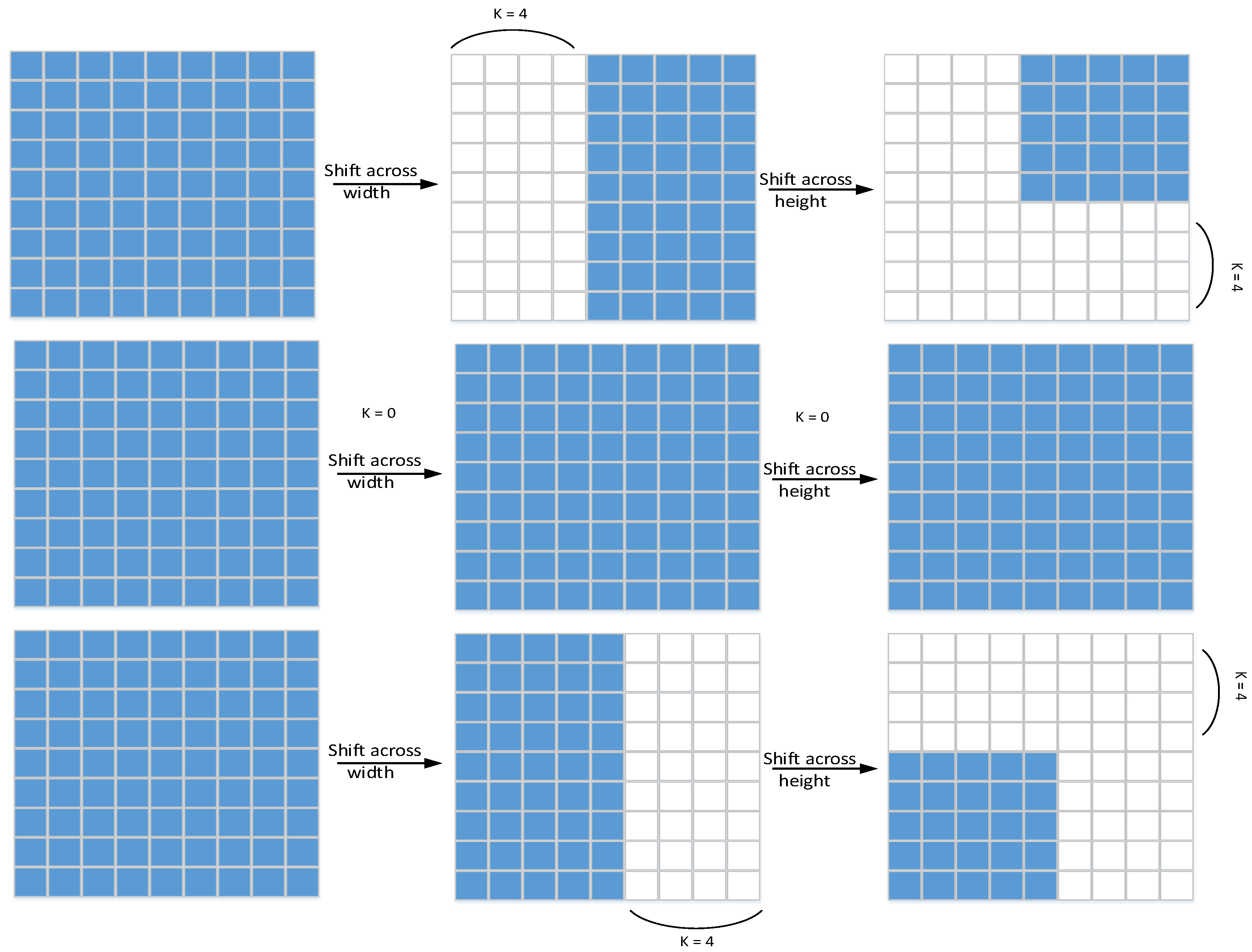
We first shift the convolutional features in an orderly manner along the channel axis. Since this operation can enhance MLP’s attention to certain local feature information, the MLP block produces locality. We first move the feature map horizontally in the width and height axes, respectively (the shift operation principle is shown in
Figure 2), and its principle is very similar to axial attention [
31]. First, the feature map is divided into m partitions, and then they are sequentially moved by k positions according to the specified different axes.
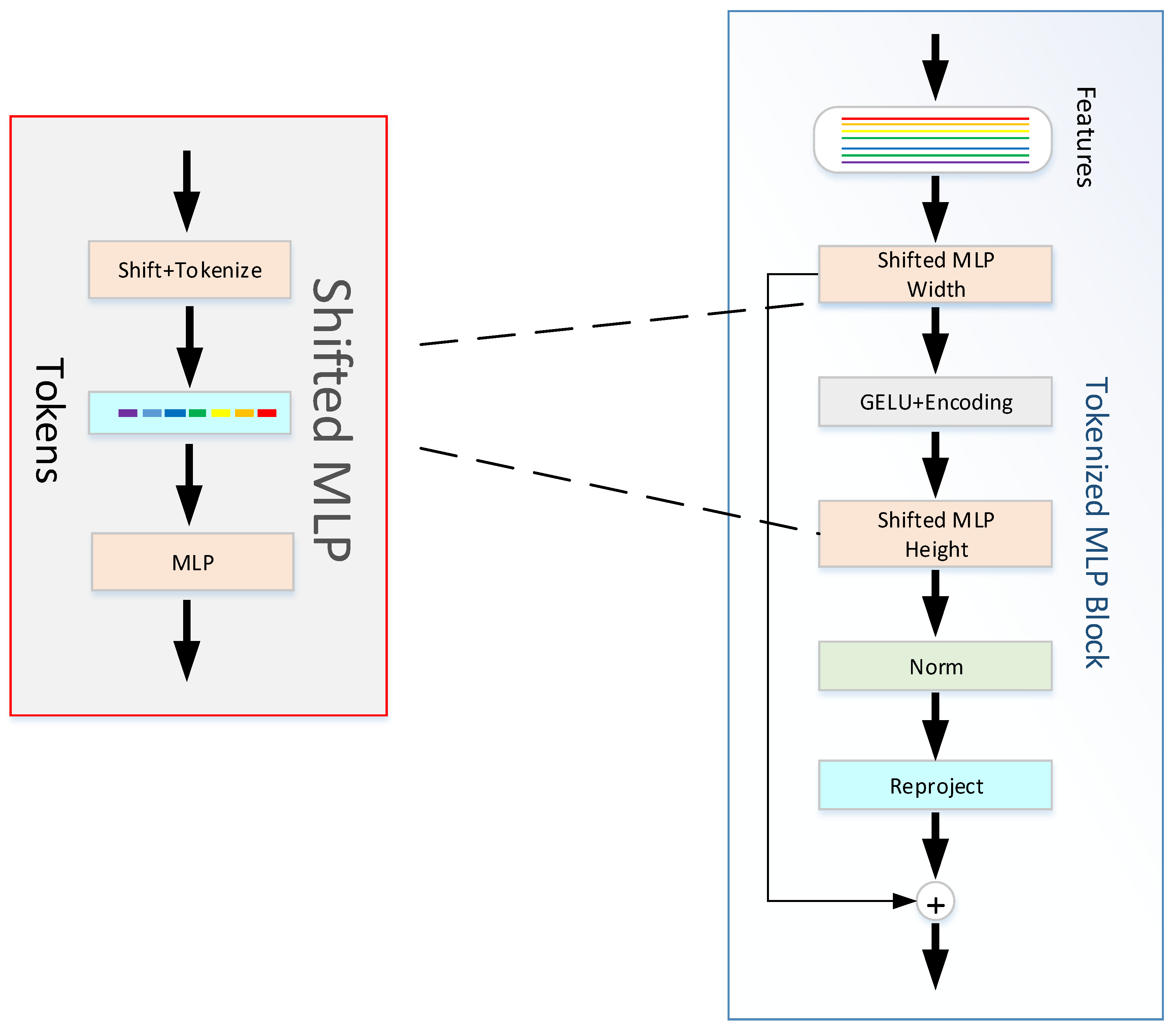
In the TokMLP block (
Figure 3), we first map the shifted feature information into tokens. Two MLP modules make up the TokMLP block we use (as shown in
Figure 3). These tokens are first passed by us to the shifted MLP (span width). Then, this feature information needs to go through the depth direction convolution layer (DWConv) operation. It should be noted that the operation of the GELU [
31] activation layer is included in the DWConv operation. GELU is a common activation function, the full name of which is “Gaussian error linear unit”. This activation function has random regular operation. Even state-of-the-art architectures, such as ViT [
32] and BERT [
33], achieve good results due to the use of GELU. Our approach of using GELU instead of RELU is a smoother alternative with better performance. Next, we need to pass the feature information into another moving MLP (across height). Finally, we feed the layer normalized (LN) features into the next module. LN is a normalization operation on a single neuron in an intermediate layer, and we use LN because in the TokMLP block, layer normalization along tokens makes more sense than batch normalization (BN). The calculation process in the TokMLP block is as follows:
where
is for height,
is for width,
is for deep convolution,
stands for Shift operation in width direction,
stands for shift operation in the height direction,
stands for feature mapping operation,
stands for tokens gained by feature mapping operation,
stands for deep convolution, and
for layer normalization. As shown in the above formula, firstly, the feature information obtained by convolution is obtained by shift operation and feature mapping operation along the width direction to obtain
, and then the output
Y is obtained by
MLP and
DWConv operations on
. We carry out a displacement operation and a feature mapping operation along the height direction for the obtained output
Y to get
. We carry out an MLP and GELU operations on
. After adding the output obtained with
, we carry out an
LN operation. Our network enhances the recognition of local features of skin lesions by using TokMLP blocks to strengthen the association of elements with surrounding elements.
3.3. CTrans Module
The CTrans module we used is composed of two modules, CCT and CCA. CCT is a channel cross-fusion transformer, which is composed of three parts: multiscale feature embedding, multihead channel cross-attention, and multilayer perceptrons (
MLP) (see
Figure 4). It has the effect of fusing features from multiscale encoders. CCA is channel cross-attention, which has the effect of fusing features from multiscale encoders.
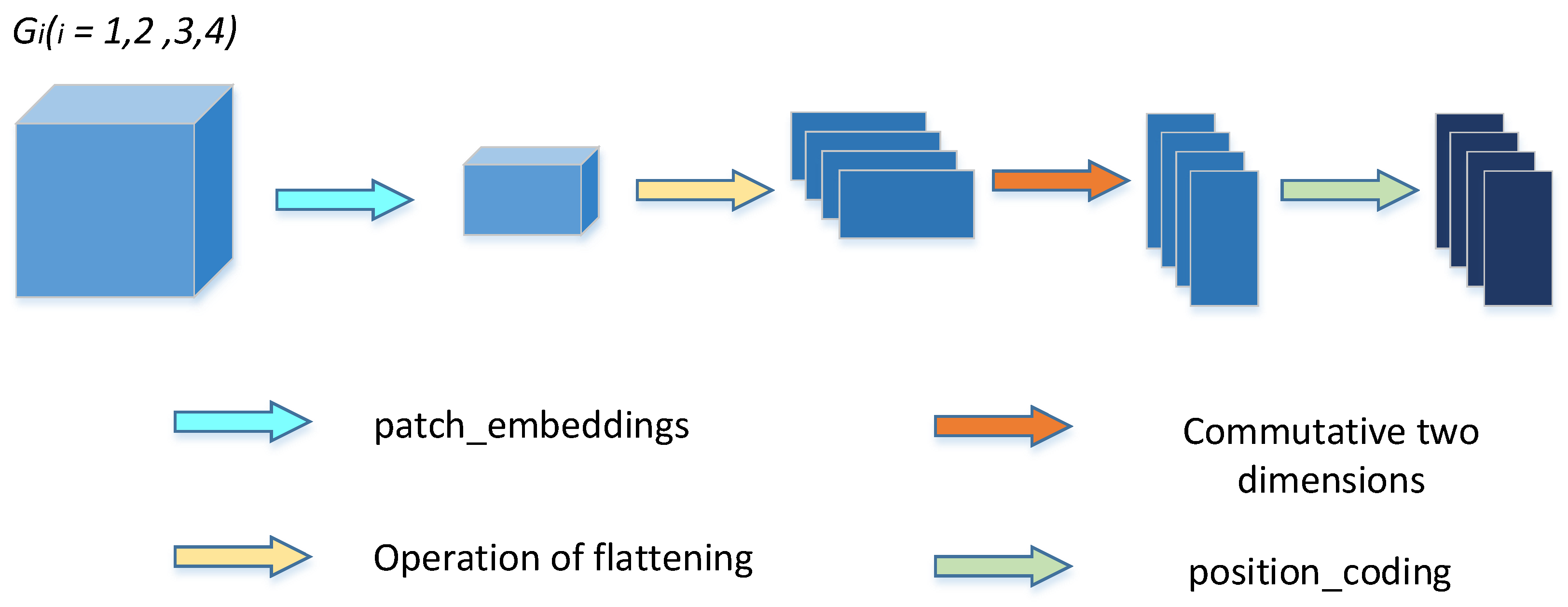
Firstly, the feature graph () of its four skip connection layers (including those operated by the TokMLP block) is reconstructed by a multiscale feature embedding operation.
We first embed the feature graph
(as shown in
Figure 4) and
and then perform the patch_embeddings operation (that is, after a convolution operation with kernel size of (16, 16) and step size of 16). The output
is obtained, and then the output
is flattened. We perform a dimension swap operation on its length and width to obtain the output
. Finally, we perform position coding operations on it to obtain the output
. Similarly, for
,
, and
and after the operation to obtain new features (
) in the same way. The four resulting results are entered into the encoder in sequence and then spliced according to their channel dimensions. Then, the output
obtained after concatenation.
We first input
and
into the CCT module for transformer operation, and its working principle is shown in
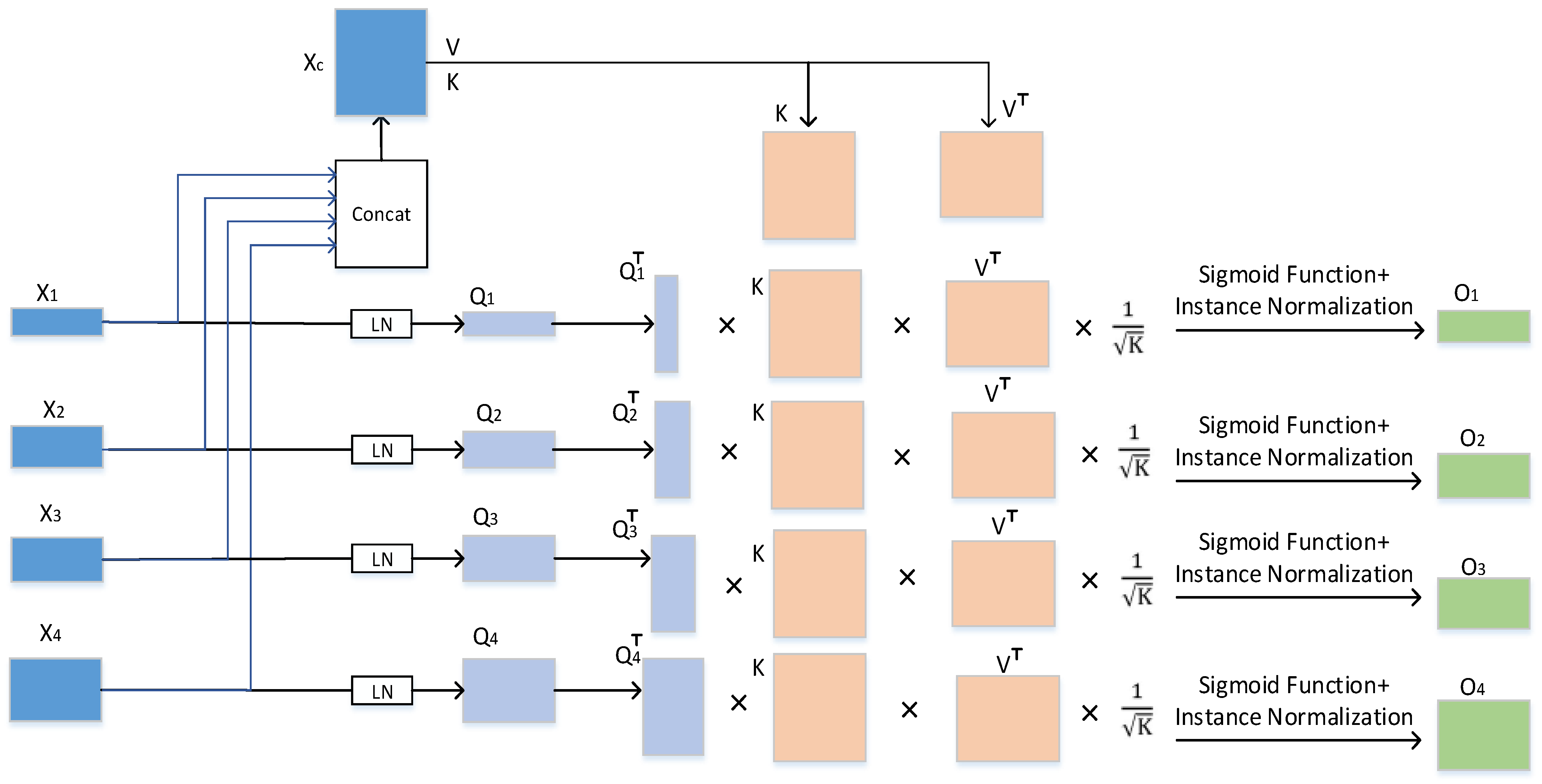
Figure 5.
In Formula (5), , , and represent the weight of each input, respectively, , , and . b represents the sequence length (i.e., number of patches) and ( = 1, 2, 3, 4) represents the channel size of each jump connection layer. The channel sizes of the four jump connection layers in our experiment were 64, 128, 256, and 512, respectively.
The matrix
is generated using the cross-concern (CA) mechanism in the multihead cross-self-attention. We use the matrix
to weight the value
V:
where
represents the instance normalization operation, sigma is the softmax function.
The
is computed for each
, and four
are generated for each of the four inputs. In the case of “
S many heads of attention”, its output can be calculated as follows:
In Formula (7), represents the output of multiheaded cross-self-attention. In Formula (8), MLP represents the multilayer perceptron in the CCT module, represents the layer normalization, and represents the four outputs operated by the CCT module.
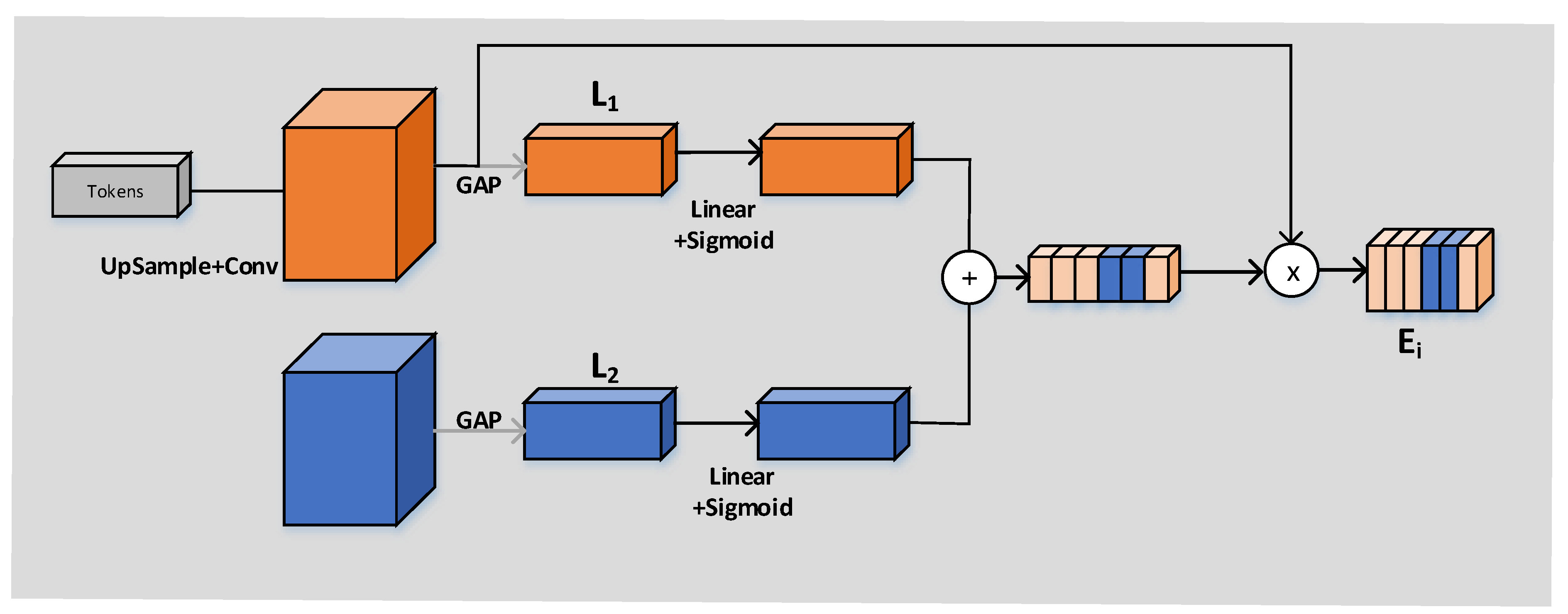
The working principle of the CCA module is shown in
Figure 6. The input of the CCA module is, respectively, every output
obtained through the operation of the CCT module and every feature figure
in the decoder. The vector
can be obtained by compressing the GAP layer’s space. The full connection layer should not only expand the convolutional layer into vectors but also classify each feature map. The idea of GAP is to combine the above two processes into one and carry them out together. The purpose of GAP we use is to solve the overfitting risk caused by fewer parameters of the full connection but also to achieve the same conversion function of the full connection. Then, linear and sigmoid operations were carried out, respectively, and finally (as shown in
Figure 3), pixel point addition and pixel point multiplication were successfully carried out.
We obtain four outputs after CCA module operation (), and we need to concatenate these four outputs, respectively, with the output () of the decoder obtained by the up-sampling operation in accordance with the channel dimension. After a convolution (kernel 1) and sigmoid operation on the final output, we obtain the final segmentation result graph.