Convergence of Artificial Intelligence and Neuroscience towards the Diagnosis of Neurological Disorders—A Scoping Review
Abstract
1. Introduction
2. Objective
- How significant is the relationship between AI and neuroscience?
- How do other existing surveys focus on this topic?
- How does neuroscience inspire the design of AI?
- How does AI help in the advancement of neuroscience?
- What are the applications of AI in neuroimaging methods and tools?
- How does AI help in the diagnosis of neurological disorders?
- What are the challenges associated with the implementation of AI-based applications for neurological diseases?
- What are the directions for future research?
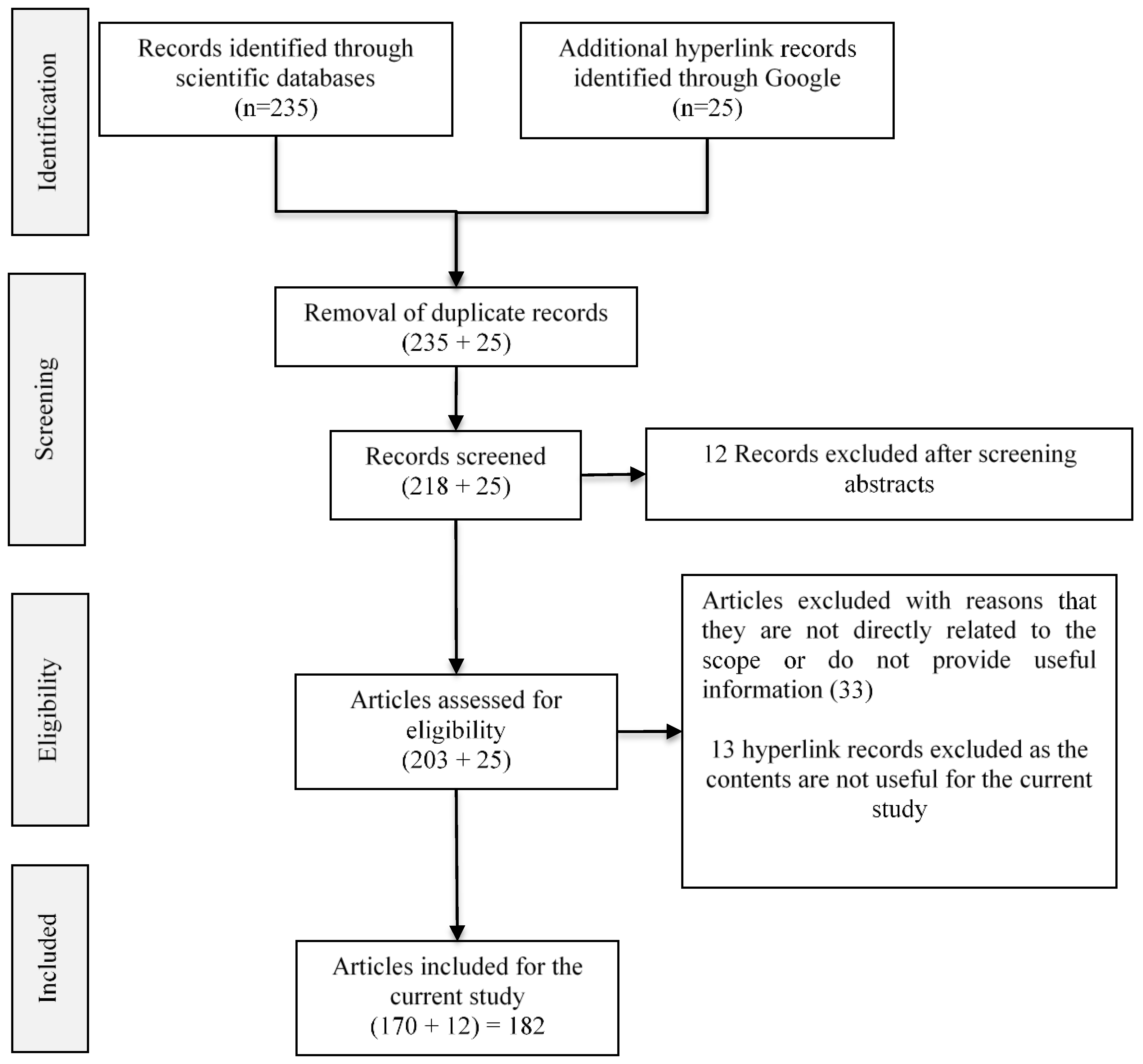
3. Review Method
4. Neuroscience for AI
4.1. ANN
4.2. Multilayer Perceptron (MLP)
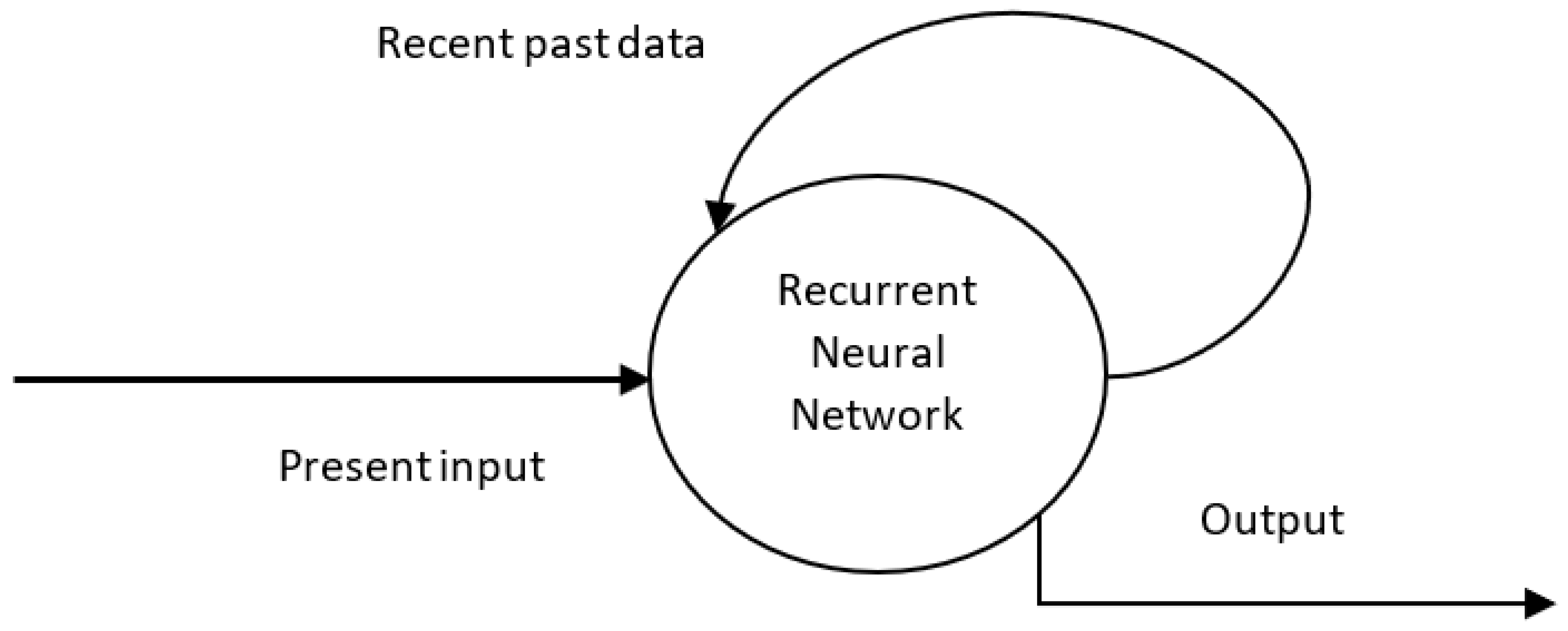
4.3. Recurrent Neural Network (RNN)
4.4. Convolutional Neural Network
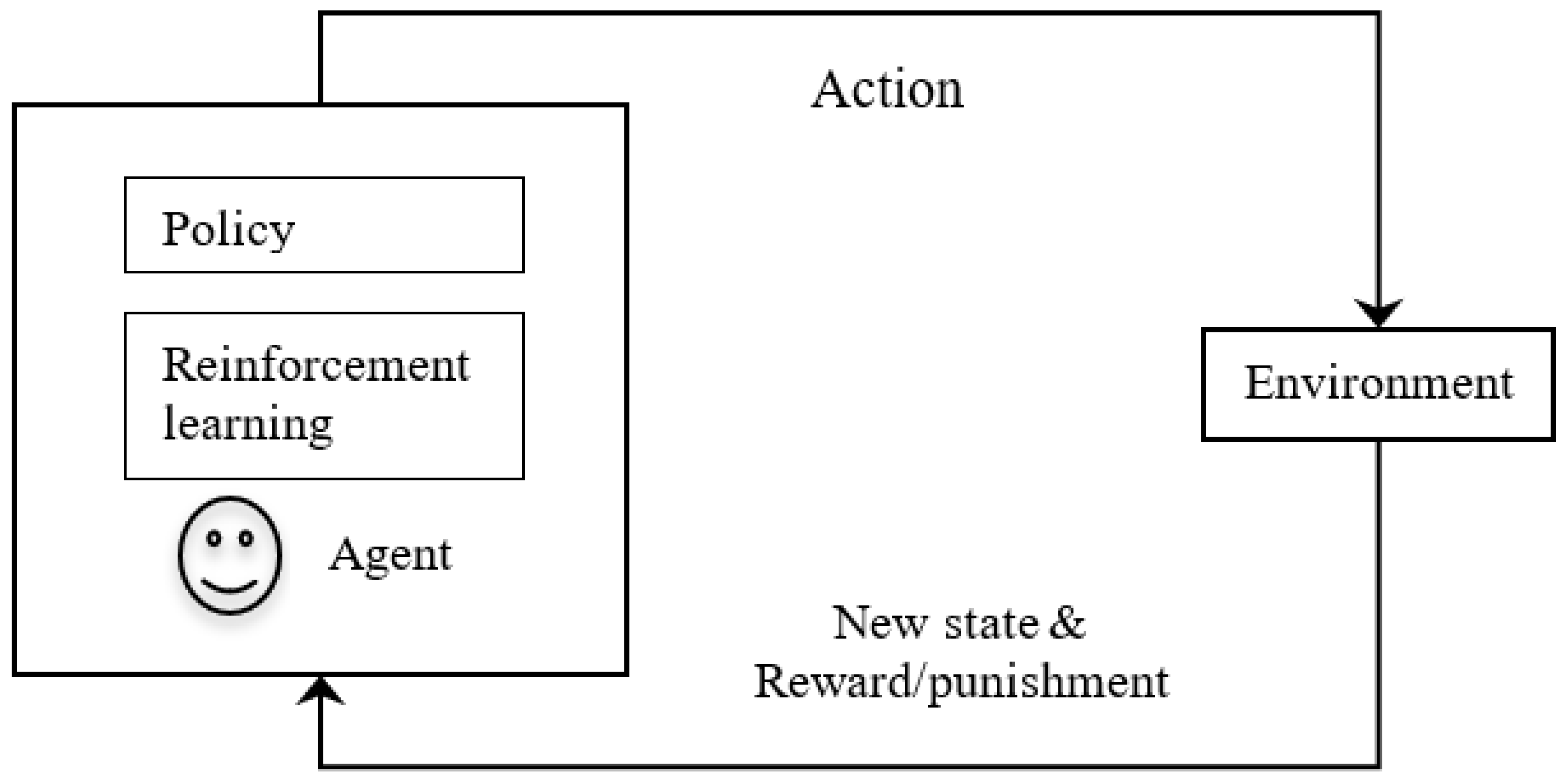
4.5. Reinforcement Learning (RL)
4.6. Deep Reinforcement Learning
4.7. Spiking Neural Network (SNN)
5. AI for the Development of Neuroscience
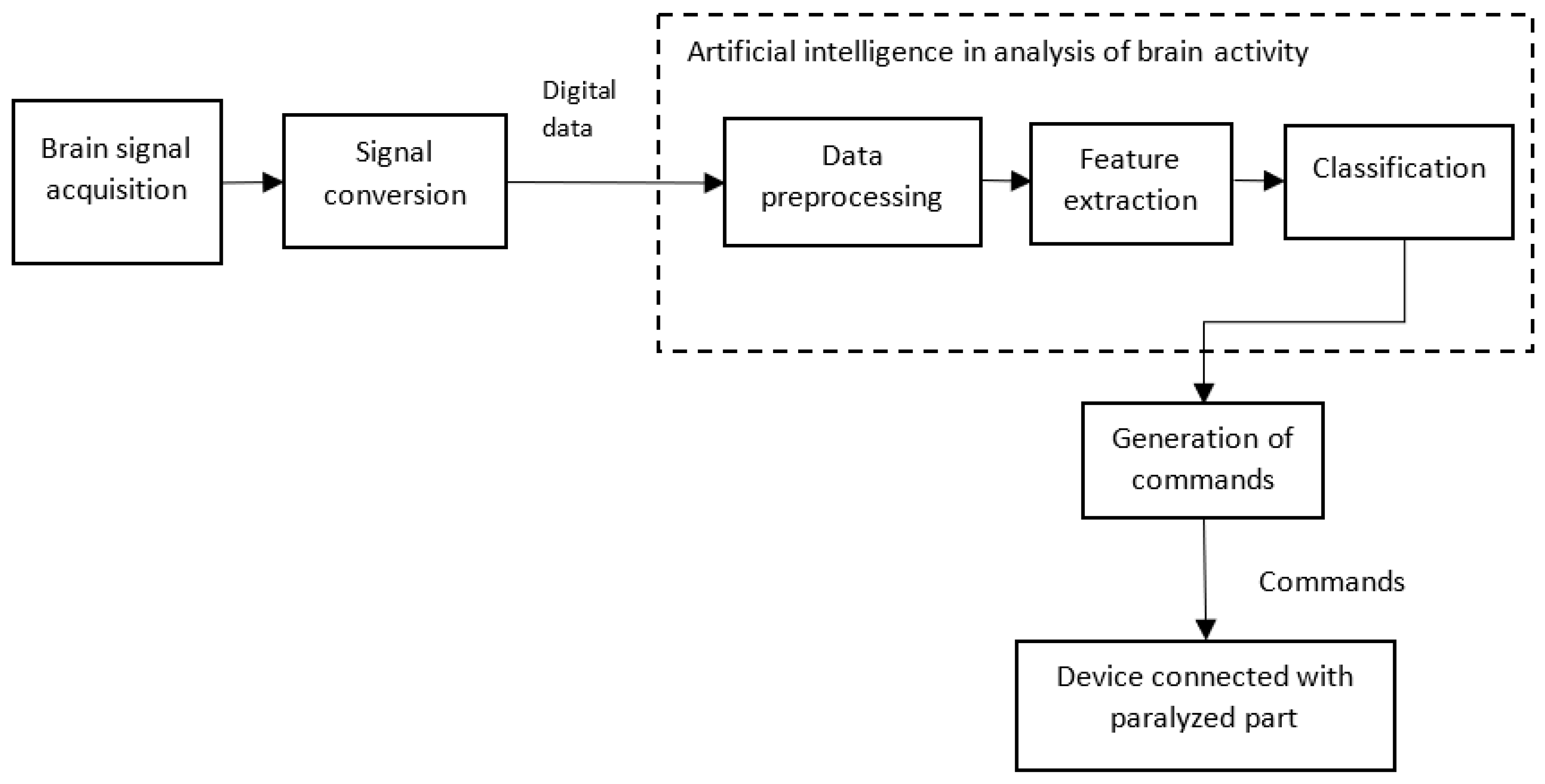
5.1. AI helps in Brain Computer/Machine Interface (BCI)
5.2. AI helps in Stimulation Studies and in the Analysis of Neurons at the Genetic Level
5.3. AI helps in the Study of the Connectome
5.4. AI helps in Neuroimaging Analysis
- Improving signal-to-noise ratio—MRI images often suffer from a low signal-to-noise ratio, and AI-based methods are used to eliminate noise [80]. Low-resolution images can be converted into high-resolution images using deep convolutional networks, as discussed in [81]. Further, in [82], the authors discussed how the quality of MRI and CT images could be improved by using different techniques, namely “noise and artifact reduction”, “super resolution”, and “image acquisition and reconstruction”. How the two major limitations of PET imaging, namely, high noise and low-spatial resolution, are effectively handled by AI methods is discussed in [83];
- Image registration—AI methods are used in image registration or image alignment, where multiple images are aligned for spatial correspondence [86]. Further, in the case of DMRI, during image registration, along with spatial correspondence, the spatial agreement of fiber orientation among different subjects is also involved, and deep learning methods for image registration have improved accuracy and reduced computation time [87]. Improved image registration using deep learning methods for fast and accurate registration among DMRI datasets is presented in [88];
- Dose optimization—as discussed in [89], AI is being used in every stage of CT imaging to obtain high-quality images and help reduce noise and optimize radiation dosage [90]. Moreover, AI-based methods have found application in predicting radiation dosages, as described in [91]. AI enables the interpretation of low-dose MRI scans, which can be adopted for individuals who have kidney diseases or contrast allergies [14];
- Synthetic generation of CT scans—deep convolutional neural networks are useful for converting MRI images into equivalent CT images (called synthetic CT) for dose calculation and patient positioning [92]. Further, AI has been increasingly applied to problems in medical imaging, such as generating CT scans for attenuation correction, segmentation, diagnosis (of diseases), and making outcome predictions [93];
- Translation of EEG data—AI-based dynamic brain imaging methods that can translate EEG data in neural network circuit activity without human activity have been discussed [94];
- Quality assessment of MRI—a fast, automated deep neural network-based method is discussed in [95] for assessing the quality of MRIs and determining whether an image is clinically usable or if a repeated MRI is required;
- As described in [96], explainable AI provides reasons for the decisions in neuroimaging data.
5.5. AI in the Study of Brain Aging
6. Applications of AI for Neurological Disorders
- Tumors;
- Seizure disorders;
- Disorders of development;
- Degenerative disorders;
- Headaches and facial pain;
- Cerebrovascular accidents;
- Neurological infections.
6.1. AI in Tumors
6.2. AI in Seizures
6.3. AI in Intellectual and Developmental Disabilities
6.4. AI in Neurodegenerative Disorders
6.5. AI in Headaches
6.6. AI in Cerebrovascular Accidents
6.7. AI in Neurological Infections
- For most infections, no specific treatment is available, and the reversal of immune suppression is the only available, viable treatment;
- Infections can be caused by unusual pathogens, and laboratories are not equipped to detect such pathogens;
- Imaging techniques represent the most common diagnostic method, and the major challenge here is that most infectious diseases are likely to produce only nonspecific patterns;
- Many of the infected individuals may not have any symptoms, and such infections can even remain undiagnosed;
- Infections may be seasonal, and such infections require specialized laboratories for diagnostics;
- A wide range of pathogens are able to trigger immune disorders, and identifying the exact pathogen for an immune disorder is itself tedious;
- Prevention strategies also remain unknown in many cases;
- More importantly, this infectious disease can trigger other neurodegenerative and other neurological disorders.
7. Challenges and Future Directions
7.1. Challenges in the Creation of Interlinked Datasets Due to the Working Culture of Teams in Isolation
7.2. Challenges Associated with Depth of Understanding in Neuro-inspired AI
7.3. Challenges Associated with the Interpretation and Assessment of AI-Based Solutions
7.4. Challenges with Standards and Regulations
7.5. Methodological and Ethical Challenges
- AI-based solutions are associated with inherent methodological and epistemic issues due to possible malfunctioning and uncertainty of such solutions;
- The over-optimization and over-fitting of AI-based solutions are likely to introduce biases in the results;
- There is another ethical dilemma; it is unclear whether AI models should be used to assist physicians when making decisions or should be used for automatic decision-making;
- Though AI-based solutions help in improving the quality of life of patients with motor and cognitive disabilities, it is also inherent that the AI-based solutions have autonomy and impact the cognitive liberty of the individuals;
- AI models reveal the analyzed results transparently, irrespective of the risk or sensitivity associated with the results;
- The training data on which the algorithms are trained will introduce a neurodiscrimination issue for the individuals concerned in the data, and this is basically due to the range of coverage of the data at hand.
7.6. Challenges with Neuroimaging Techniques
7.7. Challenges with Data Availability and Privacy
7.8. Challenges with Interpretation
- Failure to consult prior studies or reports;
- Limitations of an imaging technique (inappropriate or incomplete protocols);
- Inaccurate or incomplete history;
- Location of lesions outside the region of interest on an image;
- Failure to search systematically beyond the first abnormality discovered (“satisfaction of search”);
- Failure to recognize a normal variant.
- Making interlinked datasets from diverse and large collaborative teams from neuroscience, computing, and biology;
- Preparing quality data up to the standards of clinical practices;
- Establishing standards and regulations for data sharing;
- Validating AI models with prospective data;
- Establishing performance metrics for AI models (up to the clinical effectiveness);
- Developing methods and techniques for integrating data from heterogeneous neuroimaging methods;
- Developing software to facilitate data fusion from multimodal neuroimaging;
- Establishing huge data repositories for the effective training of AI algorithms (as training the algorithms with huge training data enables the model to understand the problem at hand efficiently and enhances the accuracy of the models);
- Bringing in interoperability standards from the different organizations involved in the neurological disease sector.
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Malik, N.; Solanki, A. Simulation of Human Brain: Artificial Intelligence-Based Learning. In Impact of AI Technologies on Teaching, Learning, and Research in Higher Education; Verma, S., Tomar, P., Eds.; IGI Global: Hershey, PA, USA, 2021; pp. 150–160. [Google Scholar] [CrossRef]
- Zeigler, B.; Muzy, A.; Yilmaz, L. Artificial Intelligence in Modeling and Simulation. In Encyclopedia of Complexity and Systems Science; Meyers, R., Ed.; Springer: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Muhammad, L.J.; Algehyne, E.A.; Usman, S.S.; Ahmad, A.; Chakraborty, C.; Mohammed, I.A. Supervised machine learning models for prediction of COVID-19 infection using epidemiology dataset. SN Comput. Sci. 2021, 2, 11. [Google Scholar] [CrossRef]
- Surianarayanan, C.; Chelliah, P.R. Leveraging Artificial Intelligence (AI) Capabilities for COVID-19 Containment. New Gener. Comput. 2021, 39, 717–741. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Wang, B.; Xu, H.; Luo, C.; Wei, L.; Zhao, W.; Hou, X.; Ma, W.; Zhengqing, X.; Zheng, Z.; et al. AI-assisted CT imaging analysis for COVID-19 screening: Building and deploying a medical AI system in four weeks. medRxiv 2020. [Google Scholar] [CrossRef]
- Rana, A.; Rawat, A.S.; Bijalwan, A.; Bahuguna, H. Application of multi-layer (perceptron) artificial neural network in the diagnosis system: A systematic review. In Proceedings of the 2018 International Conference on Research in Intelligent and Computing in Engineering (RICE), San Salvador, El Salvador, 22–24 August 2018; pp. 1–6. [Google Scholar]
- Berggren, K.K.; Xia, Q.; Likharev, K.K.; Strukov, D.B.; Jiang, H.; Mikolajick, T.; Querlioz, D.; Salinga, M.; Erickson, J.R.; Pi, S.; et al. Roadmap on emerging hardware and technology for machine learning. Nanotechnology 2020, 32, 012002. [Google Scholar] [CrossRef]
- Fan, J.; Fang, L.; Wu, J.; Guo, Y.; Dai, Q. From brain science to artificial intelligence, Engineering. Engineering 2020, 6, 248–252. [Google Scholar] [CrossRef]
- Samancı, B.M.; Yıldızhan, E.; Tüzün, E. Neuropsychiatry in the Century of Neuroscience. Noro. Psikiyatr. Ars. 2022, 59 (Suppl. S1), S1–S2. [Google Scholar] [CrossRef] [PubMed]
- Kaur, K. A study of Neuroscience. In Neurodevelopmental Disorders and Treatment; Pulsus Group: London, UK, 2021; Available online: https://www.pulsus.com/abstract/a-study-of-neuroscience-8559.html (accessed on 24 January 2023).
- Morita, T.; Asada, M.; Naito, E. Contribution of Neuroimaging Studies to Understanding Development of Human Cognitive Brain Functions. Front. Hum. Neurosci. 2016, 10, 464. [Google Scholar] [CrossRef]
- Nahirney, P.C.; Tremblay, M.-E. Brain Ultrastructure: Putting the Pieces Together. Front. Cell Dev. Biol. 2021, 9, 629503. [Google Scholar] [CrossRef]
- Jorgenson, L.A.; Newsome, W.T.; Anderson, D.J.; Bargmann, C.I.; Brown, E.N.; Deisseroth, K.; Donoghue, J.P.; Hudson, K.L.; Ling, G.S.; MacLeish, P.R.; et al. The BRAIN Initiative: Developing technology to catalyse neuroscience discovery. Philos. Trans. Soc. Lond. B Biol. Sci. 2015, 370, 20140164. [Google Scholar] [CrossRef]
- Monsour, R.; Dutta, M.; Mohamed, A.Z.; Borkowski, A.; Viswanadhan, N.A. Neuroimaging in the Era of Artificial Intelligence: Current Applications. Fed. Pract. 2022, 39 (Suppl. S1), S14–S20. [Google Scholar] [CrossRef]
- Van Horn, J.D.; Woodward, J.B.; Simonds, G.; Vance, B.; Grethe, J.S.; Montague, M.; Aslam, J.; Rus, D.; Rockmore, D.; Gazzaniga, M.S. The fMRI Data Center: Software Tools for Neuroimaging Data Management, Inspection, and Sharing. In Neuroscience Databases; Kötter, R., Ed.; Springer: Boston, MA, USA, 2003. [Google Scholar] [CrossRef]
- van der Velde, F. Where Artificial Intelligence and Neuroscience Meet: The Search for Grounded Architectures of Cognition. Adv. Artif. Intell. 2010, 2010, 918062. [Google Scholar] [CrossRef]
- Hassabis, D.; Kumaran, D.; Summerfield, C.; Botvinick, M. Neuroscience-Inspired Artificial Intelligence. Neuron 2017, 95, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Macpherson, T.; Churchland, A.; Sejnowski, T.; DiCarlo, J.; Kamitani, Y.; Takahashi, H.; Hikida, T. Natural and Artificial Intelligence: A brief introduction to the interplay between AI and neuroscience research. Neural Netw. 2021, 144, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Ullman, S. Using neuroscience to develop artificial intelligence. Science 2019, 363, 692–693. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Chitlangia, S.; Baths, V. Reinforcement learning and its connections with neuroscience and psychology. Neural Netw. 2022, 145, 271–287. [Google Scholar] [CrossRef] [PubMed]
- Li, Z. Intelligence: From Invention to Discovery. Neuron 2020, 105, 413–415. [Google Scholar] [CrossRef]
- Potter, S.M. What Can AI Get from Neuroscience? In 50 Years of Artificial Intelligence; Lecture Notes in Computer Science; Lungarella, M., Iida, F., Bongard, J., Pfeifer, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 4850. [Google Scholar] [CrossRef]
- Fellous, J.-M.; Sapiro, G.; Rossi, A.; Mayberg, H.; Ferrante, M. Explainable Artificial Intelligence for Neuroscience: Behavioral Neurostimulation. Front. Neurosci. 2019, 13, 1346. [Google Scholar] [CrossRef]
- Chauhan, N.K.; Singh, K. A Review on Conventional Machine Learning vs Deep Learning. In Proceedings of the 2018 International Conference on Computing, Power and Communication Technologies (GUCON), Greater Noida, India, 28–29 September 2018; pp. 347–352. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; The MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Janiesch, C.; Zschech, P.; Heinrich, K. Machine learning and deep learning. Electron. Mark. 2021, 31, 685–695. [Google Scholar] [CrossRef]
- Available online: https://www.zendesk.com/in/blog/machine-learning-and-deep-learning/ (accessed on 1 March 2023).
- Available online: https://www.deepmind.com/blog/ai-and-neuroscience-a-virtuous-circle (accessed on 17 February 2023).
- Available online: https://www.linkedin.com/pulse/shared-vision-machine-learning-neuroscience-harshit-goyal/ (accessed on 17 February 2023).
- Nwadiugwu, M.C. Neural Networks, Artificial Intelligence and the Computational Brain. Available online: https://arxiv.org/ftp/arxiv/papers/2101/2101.08635.pdf (accessed on 24 January 2023).
- Hebb, D.O. The Organization of Behavior; Wiley: New York, NY, USA, 1949. [Google Scholar]
- Casarella, J.M.; Frank Rosenblatt; Alan, M. Turing, Connectionism, and Artificial Intelligence. In Proceedings of the Student-Faculty Research Day, CSIS, Pace University, New York, NY, USA, 6 May 2011; Available online: http://csis.pace.edu/~ctappert/srd2011/d4.pdf (accessed on 1 March 2023).
- Rosenblatt, F. The perceptron: A probabilistic model for information storage and organization in the brain. Psychol. Rev. 1958, 65, 386–408. [Google Scholar] [CrossRef] [PubMed]
- Owens, M.T.; Tanner, K.D. Teaching as Brain Changing: Exploring Connections between Neuroscience and Innovative Teaching. CBE Life Sci. Educ. 2017, 16, fe2. [Google Scholar] [CrossRef]
- Werbos, P.J. Beyond Regression: New Tools for Prediction and Analysis in the Behavioral Sciences. Applied Mathematics. Ph.D. Thesis, Harvard University, Cambridge, MA, USA, 1974. [Google Scholar]
- Whittington, J.C.R.; Bogacz, R. An Approximation of the Error Backpropagation Algorithm in a Predictive Coding Network with Local Hebbian Synaptic Plasticity. Neural Comput. 2017, 29, 1229–1262. [Google Scholar] [CrossRef] [PubMed]
- Bengio, Y.; Mesnard, T.; Fischer, A.; Zhang, S.; Wu, Y. STDP-Compatible approximation of backpropagation in an energy-based model. Neural Comput. 2017, 29, 555–577. [Google Scholar] [CrossRef]
- Xie, Y.; Liu, Y.H.; Constantinidis, C.; Zhou, X. Neural Mechanisms of Working Memory Accuracy Revealed by Recurrent Neural Networks. Front. Syst. Neurosci. 2022, 16, 760864. [Google Scholar] [CrossRef]
- Eichenbaum, H. Prefrontal–hippocampal interactions in episodic memory. Nat. Rev. Neurosci. 2017, 18, 547–558. [Google Scholar] [CrossRef]
- Conway, A.R.; Kane, M.J.; Engle, R.W. Working memory capacity and its relation to general intelligence. Trends Cogn. Sci. 2003, 7, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Goulas, A.; Damicelli, F.; Hilgetag, C. Bio-instantiated recurrent neural networks. Neural Netw. 2021, 142, 608–618. [Google Scholar] [CrossRef] [PubMed]
- Sawant, Y.; Kundu, J.N.; Radhakrishnan, V.B.; Sridharan, D. A Midbrain Inspired Recurrent Neural Network Model for Robust Change Detection. J. Neurosci. 2022, 42, 8262–8283. [Google Scholar] [CrossRef]
- Close Krizhevsky, A.; Sutskever, I.; Hinton, G. ImageNet classification with deep convolutional neural networks. In Proceedings of the Advances in Neural Information Processing Systems 25, Lake Tahoe, NV, USA, 3–6 December 2012; pp. 1097–1105, ISBN 9781627480031. [Google Scholar]
- Thomas, S.; Lior, W.; Stanley, B.; Maximilian, R.; Tomasoa, P. Robust object recognition with cortex-like mechanisms. IEEE Trans. Pattern Anal. Mach. Intell. 2007, 29, 411–426. [Google Scholar]
- Maia, T.V. Reinforcement learning, conditioning, and the brain: Successes and challenges. Cogn. Affect. Behav. Neurosci. 2009, 9, 343–364. [Google Scholar] [CrossRef]
- Available online: https://www.akc.org/expert-advice/training/operant-conditioning-the-science-behind-positive-reinforcement-dog-training/ (accessed on 12 February 2023).
- Black, S.; Mareschal, D. A complementary learning systems approach to temporal difference learning. Neural Netw. 2020, 122, 218–230. [Google Scholar] [CrossRef]
- Seo, D.L.H.; Jung, M.W. Neural Basis of Reinforcement Learning and Decision Making. Annu. Rev. Neurosci. 2012, 35, 287–308. [Google Scholar]
- Dabney, W.; Kurth-Nelson, Z.; Uchida, N.; Starkweather, C.K.; Hassabis, D.; Munos, R.; Botvinick, M. A distributional code for value in dopamine-based reinforcement learning. Nature 2020, 577, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Tesauro, G. Temporal difference learning and td-gammon. Commun. ACM 1995, 38, 58–68. [Google Scholar] [CrossRef]
- Kumaran, D.; Hassabis, D.; McClelland, J.L. What learning systems do intelligent agents need? Complementary learning systems theory updated. Trends Cogn. Sci. 2016, 20, 512–534. [Google Scholar] [CrossRef]
- Parisi, G.I.; Kemker, R.; Part, J.L.; Kanan, C.; Wermter, S. Continual lifelong learning with neural networks: A review. Neural Netw. 2019, 113, 54–71. [Google Scholar] [CrossRef] [PubMed]
- Blakeman, S.; Mareschal, D. Generating Explanations from Deep Reinforcement Learning Using Episodic Memory. arXiv 2022, arXiv:2205.08926. [Google Scholar]
- Hu, H.; Ye, J.; Zhu, G.; Ren, Z.; Zhang, C. Generalizable Episodic Memory for Deep Reinforcement Learning. In Proceedings of the 38th International Conference on Machine Learning, Virtual Event, 18–24 July 2021. PMLR 139. [Google Scholar]
- Kim, T.; Hu, S.; Kim, J.; Kwak, J.Y.; Park, J.; Lee, S.; Kim, I.; Park, J.-K.; Jeong, Y. Spiking Neural Network (SNN) With Memristor Synapses Having Non-linear Weight Update. Front. Comput. Neurosci. 2021, 15, 646125. [Google Scholar] [CrossRef]
- Pfeiffer, M.; Pfeil, T. Deep Learning With Spiking Neurons: Opportunities and Challenges. Front. Neurosci. 2018, 12, 774. [Google Scholar] [CrossRef]
- Zhan, G.; Song, Z.; Fang, T.; Zhang, Y.; Le, S.; Zhang, X.; Wang, S.; Lin, Y.; Jia, J.; Zhang, L.; et al. Applications of Spiking Neural Network in Brain Computer Interface. In Proceedings of the 2021 9th International Winter Conference on Brain-Computer Interface (BCI), Gangwon, Republic of Korea, 22–24 February 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Available online: https://www.healtheuropa.com/the-role-of-artificial-intelligence-in-neuroscience/116572/ (accessed on 17 February 2023).
- Frye, J.; Ananthanarayanan, R.; Modha, D.S. Towards Real-Time, Mouse-Scale Cortical Simulations. IBM Research Report RJ10404. 2007. Available online: https://dominoweb.draco.res.ibm.com/reports/rj10404.pdf (accessed on 24 January 2023).
- Shih, J.J.; Krusienski, D.J.; Wolpaw, J.R. Brain-computer interfaces in medicine. Mayo Clin. Proc. 2012, 87, 268–279. [Google Scholar] [CrossRef]
- Available online: https://www.linkedin.com/pulse/artificial-intelligence-can-make-brain-computer-more-chhabra/?trk=public_profile_article_view (accessed on 14 February 2023).
- Zhang, X.; Ma, Z.; Zheng, H.; Li, T.; Chen, K.; Wang, X.; Liu, C.; Xu, L.; Wu, X.; Lin, D.; et al. The combination of brain-computer interfaces and artificial intelligence: Applications and challenges. Ann. Transl. Med. 2020, 8, 11. [Google Scholar] [CrossRef]
- Dias, R.; Torkamani, A. Artificial intelligence in clinical and genomic diagnostics. Genome Med. 2019, 11, 70. [Google Scholar] [CrossRef] [PubMed]
- Markram, H. The blue brain project. Nat. Rev. Neurosci. 2006, 7, 153–160. [Google Scholar] [CrossRef]
- Almeida, J.E.; Teixeira, C.; Morais, J.; Oliveira, E.; Couto, L. Applications of Artificial Intelligence in Neuroscience Research: An Overview. Available online: http://www.kriativ-tech.com/wp-content/uploads/2022/06/JoaoAlmeida_IA_Neurociencias-EN-2.pdf (accessed on 24 January 2023).
- Toga, A.W.; Clark, K.A.; Thompson, P.M.; Shattuck, D.W.; Van Horn, J.D. Mapping the human connectome. Neurosurgery 2012, 71, 1–5. [Google Scholar] [CrossRef]
- Brown, C.; Hamarneh, G. Machine Learning on Human Connectome Data from MRI. arXiv 2016, arXiv:1611.08699v1. [Google Scholar]
- Zhu, G.; Jiang, B.; Tong, L.; Xie, Y.; Zaharchuk, G.; Wintermark, M. Applications of Deep Learning to Neuro-Imaging Techniques. Front. Neurol. 2019, 10, 869. [Google Scholar] [CrossRef]
- Helmstaedter, M. The Mutual Inspirations of Machine Learning and Neuroscience. Neuroview 2015, 86, 25–28. [Google Scholar] [CrossRef]
- Boland, G.W.L.; Guimaraes, A.S.; Mueller, P.R. The radiologist’s conundrum: Benefits and costs of increasing CT capacity and utilization. Eur. Radiol. 2009, 19, 9–12. [Google Scholar] [CrossRef] [PubMed]
- McDonald, R.J.; Schwartz, K.M.; Eckel, L.J.; Diehn, F.E.; Hunt, C.H.; Bartholmai, B.J.; Erickson, B.J.; Kallmes, D.F. The effects of changes in utilization and technological advancements of cross-sectional imaging on radiologist workload. Acad. Radiol. 2015, 22, 1191–1198. [Google Scholar] [CrossRef]
- Fitzgerald, R. Error in radiology. Clin. Radiol. 2001, 56, 938–946. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Ghafoorian, M.; Karssemeijer, N.; Heskes, T.; van Uden, I.W.M.; Sanchez, C.I.; Litjens, G.; de Leeuw, F.E.; van Ginneken, B.; Marchiori, E.; Platel, B. Location sensitive deep convolutional neural networks for segmentation of white matter hyperintensities. Sci. Rep. 2017, 7, 5110. [Google Scholar] [CrossRef]
- Chen, H.; Zheng, Y.; Park, J.H.; Heng, P.A.; Zhou, S.K. Iterative Multi-domain Regularized Deep Learning for Anatomical Structure Detection and Segmentation from Ultrasound Images. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016; MICCAI 2016; Lecture Notes in Computer Science; Ourselin, S., Joskowicz, L., Sabuncu, M., Unal, G., Wells, W., Eds.; Springer: Cham, Germany, 2016; Volume 9901. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef] [PubMed]
- McBee, M.P.; Awan, O.A.; Colucci, A.T.; Ghobadi, C.W.; Kadom, N.; Kansagra, A.P.; Auffermann, W.F. Deep learning in radiology. Acad. Radiol. 2018, 25, 1472–1480. [Google Scholar] [CrossRef] [PubMed]
- Liang, N. MRI Image Reconstruction Based on Artificial Intelligence. J. Phys. Conf. Ser. 2021, 1852, 022077. [Google Scholar] [CrossRef]
- Zhao, C.; Shao, M.; Carass, A.; Li, H.; Dewey, B.E.; Ellingsen, L.M.; Woo, J.; Guttman, M.A.; Blitz, A.M.; Stone, M.; et al. Applications of a deep learning method for anti-aliasing and super-resolution in MRI. Magn. Reason. Imaging 2019, 64, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Dou, W.; Vosters, L.; Xu, X.; Sun, Y.; Tan, T. Denoising of 3D magnetic resonance images with multi-channel residual learning of convolutional neural network. Jpn. J. Radiol. 2018, 36, 566–574. [Google Scholar] [CrossRef]
- Dong, C.; Loy, C.C.; He, K.; Tang, X. Image super-resolution using deep convolutional networks. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 38, 295–307. [Google Scholar] [CrossRef]
- Higaki, T.; Nakamura, Y.; Tatsugami, F.; Nakaura, T.; Awai, K. Improvement of image quality at CT and MRI using deep learning. Jpn. J. Radiol. 2019, 37, 73–80. [Google Scholar] [CrossRef]
- Liu, J.; Malekzadeh, M.; Mirian, N.; Song, T.A.; Liu, C.; Dutta, J. Artificial Intelligence-Based Image Enhancement in PET Imaging: Noise Reduction and Resolution Enhancement. PET Clin. 2021, 16, 553–576. [Google Scholar] [CrossRef]
- Hansen, M.S.; Kellman, P. Image reconstruction: An overview for clinicians. J. Magn. Reson. Imaging 2015, 41, 573–585. [Google Scholar] [CrossRef]
- Lin, D.J.; Johnson, P.M.; Knoll, F.; Lui, Y.W. Artificial Intelligence for MR Image Reconstruction: An Overview for Clinicians. J. Magn. Reson. Imaging 2021, 53, 1015–1028. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Cao, X.; Wang, Q.; Yapa, P.T.; Shen, D. Adversarial learning for mono-or multi-modal registration. Med. Image Anal. 2019, 58, 101545. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Lei, Y.; Wang, T.; Curran, W.J.; Liu, T.; Yang, X. Deep learning in medical image registration: A review. Phys. Med. Biol. 2020, 65, 20TR01. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wells, W.M.; O’Donnell, L.J. Deep Diffusion MRI Registration (DDMReg): A Deep Learning Method for Diffusion MRI Registration. IEEE Trans. Med. Imaging 2022, 41, 1454–1467. [Google Scholar] [CrossRef]
- McCollough, C.H.; Leng, S. Use of artificial intelligence in computed tomography dose optimization. Ann. ICRP 2020, 49 (Suppl. S1), 113–125. [Google Scholar] [CrossRef]
- Ng, C.K.C. Artificial Intelligence for Radiation Dose Optimization in Pediatric Radiology: A Systematic Review. Children 2022, 9, 1044. [Google Scholar] [CrossRef]
- Fan, J.; Wang, J.; Chen, Z.; Hu, C.; Zhang, Z.; Hu, W. Automatic treatment planning based on three-dimensional dose distribution predicted from deep learning technique. Med. Phys. 2019, 46, 370–381. [Google Scholar] [CrossRef]
- Palmér, E.; Nordström, F.; Karlsson, A.; Petruson, K.; Ljungberg, M.; Sohlin, M. Head and neck cancer patient positioning using synthetic CT data in MRI-only radiation therapy. J. Appl. Clin. Med. Phys. 2022, 23, e13525. [Google Scholar] [CrossRef]
- Boyle, A.J.; Gaudet, V.C.; Black, S.E.; Vasdev, N.; Rosa-Neto, P.; Zukotynski, K.A. Artificial intelligence for molecular neuroimaging. Ann. Transl. Med. 2021, 9, 822. [Google Scholar] [CrossRef]
- Available online: https://engineering.cmu.edu/news-events/news/2022/07/29-brain-imaging.html (accessed on 18 February 2023).
- Sreekumari, A.; Shanbhag, D.; Yeo, D.; Foo, T.; Pilitsis, J.; Polzin, J.; Patil, U.; Coblentz, A.; Kapadia, A.; Khinda, J.; et al. A deep learning-based approach to reduce rescan and recall rates in clinical MRI examinations. AJNR Am. J. Neuroradiol. 2019, 40, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Chuang, K.V.; DeCarli, C.; Jin, L.-W.; Beckett, L.; Keiser, M.J.; Dugger, B.N. Interpretable classification of Alzheimer’s disease pathologies with a convolutional neural network pipeline. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.K.; Singh, K.K. A Review of Publicly Available Automatic Brain Segmentation Methodologies, Machine Learning Models, Recent Advancements, and Their Comparison. Ann. Neurosci. 2021, 28, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Behroozi, M.; Daliri, M.R. Software Tools for the Analysis of Functional Magnetic Resonance Imaging. Basic Clin. Neurosci. 2012, 3, 71–83. [Google Scholar]
- Available online: https://www.bitbrain.com/blog/ai-eeg-data-processing (accessed on 18 February 2023).
- Goebel, R. BrainVoyager—Past, present, future. Neuroimage 2012, 62, 748–756. [Google Scholar] [CrossRef]
- Fischl, B. FreeSurfer. NeuroImage 2012, 62, 774–781. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, C.S.; Ramprashad, A.; Thompson, C.; Botti, J.-A.; Coman, I.L.; Kates, W.R. A comparison of FreeSurfer-generated data with and without manual intervention. Front. Neurosci. 2015, 9, 379. [Google Scholar] [CrossRef]
- Tae, W.S.; Kim, S.S.; Lee, K.U.; Nam, E.C.; Kim, K.W. Validation of hippocampal volumes measured using a manual method and two automated methods (FreeSurfer and IBASPM) in chronic major depressive disorder. Neuroradiology 2008, 50, 569–581. [Google Scholar] [CrossRef]
- Henschel, L.; Conjeti, S.; Estrada, S.; Diers, K.; Fischl, B.; Reuter, M. FastSurfer—A fast and accurate deep learning based neuroimaging pipeline. NeuroImage 2020, 219, 117012. [Google Scholar] [CrossRef]
- Ghazia, M.M.; Nielsen, M. FAST-AID Brain: Fast and Accurate Segmentation Tool using Artificial Intelligence Developed for Brain. arXiv 2022, arXiv:2208.14360v1. [Google Scholar]
- Levakov, G.; Rosenthal, G.; Shelef, I.; Raviv, T.R.; Avidan, G. From a deep learning model back to the brain-Identifying regional predictors and their relation to aging. Hum. Brain Mapp. 2020, 41, 3235–3252. [Google Scholar] [CrossRef]
- Available online: https://news.usc.edu/204691/ai-brain-aging-risk-of-cognitive-decline-alzheimers/ (accessed on 19 February 2023).
- Khan, R.S.; Ahmed, M.R.; Khalid, B.; Mahmood, A.; Hassan, R. Biomarker Detection of Neurological Disorders through Spectroscopy Analysis. Int. Dent. Med. J. Adv. Res. 2018, 4, 1–9. [Google Scholar] [CrossRef]
- Akinyelu, A.A.; Zaccagna, F.; Grist, J.T.; Castelli, M.; Rundo, L. Brain Tumor Diagnosis Using Machine Learning, Convolutional Neural Networks, Capsule Neural Networks and Vision Transformers, Applied to MRI: A Survey. J. Imaging 2022, 8, 205. [Google Scholar] [CrossRef] [PubMed]
- Jena, B.; Nayak, G.K.; Saxena, S. An empirical study of different machine learning techniques for brain tumor classification and subsequent segmentation using hybrid texture feature. Mach. Vis. Appl. 2022, 33, 6. [Google Scholar] [CrossRef]
- Sajjad, M.; Khan, S.; Muhammad, K.; Wu, W.; Ullah, A.; Baik, S.W. Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J. Comput. Sci. 2019, 30, 174–182. [Google Scholar] [CrossRef]
- Abd El Kader, I.; Xu, G.; Shuai, Z.; Saminu, S.; Javaid, I.; Salim Ahmad, I. Differential deep convolutional neural network model for brain tumor classification. Brain Sci. 2021, 11, 352. [Google Scholar] [CrossRef]
- Mzoughi, H.; Njeh, I.; Wali, A.; Slima, M.B.; BenHamida, A.; Mhiri, C.; Mahfoudhe, K.B. Deep multi-scale 3D convolutional neural network (CNN) for MRI gliomas brain tumor classification. J. Digit. Imaging 2020, 33, 903–915. [Google Scholar] [CrossRef]
- Patrick, M.K.; Adekoya, A.F.; Mighty, A.A.; Edward, B.Y. Capsule networks—A survey. J. King Saud Univ. Inf. Sci. 2022, 34, 1295–1310. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S. An image is worth 16 × 16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Shoeb, A.H. Application of Machine Learning to Epileptic Seizure Onset Detection and Treatment; Massachusetts Institute of Technology: Cambridge, MA, USA, 2009; Available online: https://dspace.mit.edu/handle/1721.1/54669 (accessed on 24 January 2023).
- Orhan, U.; Hekim, M.; Ozer, M. EEG signals classification using the K-means clustering and a multilayer perceptron neural network model. Exp. Syst. Appl. 2011, 38, 13475–13481. [Google Scholar] [CrossRef]
- HaiderBanka, A.K. Local pattern transformation based feature extraction techniques for classification of epileptic EEG signals. Biomed. Signal Process. Control 2017, 34, 81–92. [Google Scholar]
- Hossain, M.S.; Amin, S.U.; Alsulaiman, M.; Muhammad, G. Applying deep learning for epilepsy seizure detection and brain mapping visualization. ACM Transact. Multimed. Comput. Commun. Appl. 2019, 15, 1–17. [Google Scholar] [CrossRef]
- Hu, W.; Cao, J.; Lai, X.; Liu, J. Mean amplitude spectrum based epileptic state classification for seizure prediction using convolutional neural networks. J. Ambient Intell. Human. Comput. 2019. [Google Scholar] [CrossRef]
- Abdelhameed, A.; Bayoumi, M. A Deep Learning Approach for Automatic Seizure Detection in Children with Epilepsy. Front. Comput. Neurosci. 2021, 15, 650050. [Google Scholar] [CrossRef] [PubMed]
- Heinsfeld, A.S.; Franco, A.R.; Craddock, R.C.; Buchweitz, A.; Meneguzzi, F. Identification of autism spectrum d0isorder using deep learning and the ABIDE dataset. Neuroimage Clin. 2018, 17, 16–23. [Google Scholar] [CrossRef]
- Chen, H.; Song, Y.; Li, X. Use of deep learning to detect personalized spatial-frequency abnormalities in EEGs of children with ADHD. J. Neural Eng. 2019, 16, 066046. [Google Scholar] [CrossRef]
- Movaghar, A.; Page, D.; Brilliant, M.; Mailick, M. Advancing artificial intelligence-assisted pre-screening for fragile X syndrome. BMC Med. Inform. Decis. Mak. 2022, 22, 152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wang, Z.; Qiu, H.; Zhou, W.; Wang, M.; Cheng, G. Machine learning applied to serum and cerebrospinal fluid metabolomes revealed altered arginine metabolism in neonatal sepsis with meningoencephalitis. Comput. Struct. Biotechnol. J. 2021, 19, 3284–3292. [Google Scholar] [CrossRef]
- Michel, P.P.; Hirsch, E.C.; Hunot, S. Understanding dopaminergic cell death pathways in Parkinson disease. Neuron 2016, 90, 675–691. [Google Scholar] [CrossRef]
- Klöppel, S.; Stonnington, C.M.; Chu, C.; Draganski, B.; Scahill, R.I.; Rohrer, J.D.; Fox, N.C.; Jack, C.R.; Ashburner, J.; Frackowiak, R.S.J. Automatic classification of MR scans in Alzheimer’s disease. Brain 2008, 131, 681–689. [Google Scholar] [CrossRef]
- Korolev, S.; Safiullin, A.; Belyaev, M.; Dodonova, Y. Residual and plain convolutional neural networks for 3D brain MRI classification. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, VIC, Australia, 18–21 April 2017; pp. 835–838. [Google Scholar] [CrossRef]
- Moradi, E.; Pepe, A.; Gaser, C.; Huttunen, H.; Tohka, J. Machine learning framework for early MRI based Alzheimer’s conversion prediction in MCI subjects. Neuroimage 2015, 104, 398–412. [Google Scholar] [CrossRef]
- Magnin, B.; Mesrob, L.; Kinkingnéhun, S.; Pélégrini-Issac, M.; Colliot, O.; Sarazin, M.; Dubois, B.; Lehéricy, S.; Benali, H. Support vector machine- based classification of Alzheimer’s disease from whole- brain anatomical MRI. Neuroradiology 2009, 51, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Alty, J.; Cosgrove, J.; Thorpe, D.; Kempster, P. How to use pen and paper tasks to aid tremor diagnosis in the clinic. Pract. Neurol. 2017, 17, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Kotsavasiloglou, C.; Kostikis, N.; Hristu- Varsakelis, D.; Arnaoutoglou, M. Machine learning- based classification of simple drawing movements in Parkinson’s disease. Biomed. Signal. Process. Control 2017, 31, 174–180. [Google Scholar] [CrossRef]
- Orimaye, S.O.; Wong, J.S.-M.; Golden, K.J. Learning Predictive Linguistic Features for Alzheimer’s Disease and related Dementias using Verbal Utterances. In Workshop on Computational Linguistics and Clinical Psychology: From Linguistic Signal to Clinical Reality; Association for Computational Linguistics: Baltimore, MD, USA, 2014; pp. 78–87. [Google Scholar]
- Bron, E.E.; Smits, M.; Niessen, W.J.; Klein, S. Feature Selection Based on the SVM Weight Vector for Classification of Dementia. IEEE J. Biomed. Health Inform. 2015, 19, 1617–1626. [Google Scholar] [CrossRef]
- Zhao, A.; Qi, L.; Dong, J.; Yu, H. Dual channel LSTM based multi- feature extraction in gait for diagnosis of neurodegenerative diseases. Knowl. Syst. 2018, 145, 91–97. [Google Scholar] [CrossRef]
- Ferroni, P.; Zanzotto, F.M.; Scarpato, N.; Spila, A.; Fofi, L.; Egeo, G.; Rullo, A.; Palmirotta, R.; Barbanti, P.; Guadagni, F. Machine learning approach to predict medication overuse in migraine patients. Comput. Struct. Biotechnol. J. 2020, 18, 1487–1496. [Google Scholar] [CrossRef]
- Kwon, J.; Lee, H.; Cho, S.; Chung, C.-S.; Lee, M.J.; Park, H. Machine learning-based automated classification of headache disorders using patient-reported questionnaires. Sci. Rep. 2020, 10, 14062. [Google Scholar] [CrossRef]
- Menon, B.; Pillai, A.S.; Mathew, P.S.; Bartkowiak, A.M. Artificial intelligence–assisted headache classification: A review. In Augmenting Neurological Disorder Prediction and Rehabilitation Using Artificial Intelligence; Academic Press: Cambridge, MA, USA, 2022; pp. 145–162. [Google Scholar] [CrossRef]
- Vandenbussche, N.; Van Hee, C.; Hoste, V.; Paemeleire, K. Using natural language processing to automatically classify written self-reported narratives by patients with migraine or cluster headache. J. Headache Pain 2022, 23, 129. [Google Scholar] [CrossRef]
- Cheng, N.T.; Kim, A.S. Intravenous Thrombolysis for Acute Ischemic Stroke within 3 hours versus between 3 and 4.5 Hours of Symptom Onset. Neurohospitalist 2015, 5, 101–109. [Google Scholar] [CrossRef]
- Mosalov, O.P.; Rebrova, O.; Red’ko, V. Neuroevolutionary method of stroke diagnosis. Opt. Mem. Neural Netw. 2007, 16, 99–103. [Google Scholar] [CrossRef]
- Olabode, O.; Olabode, B.T. Cerebrovascular Accident Attack Classification Using Multilayer Feed Forward Artificial Neural Network with Back Propagation Error. J. Comput. Sci. 2012, 8, 18–25. [Google Scholar] [CrossRef][Green Version]
- Lee, E.J.; Kim, Y.H.; Kim, N.; Kang, D.W. Deep into the Brain: Artificial Intelligence in Stroke Imaging. J. Stroke 2017, 19, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Giacalone, M.; Rasti, P.; Debs, N.; Frindel, C.; Cho, T.-H.; Grenier, E.; Rousseau, D. Local spatio-temporal encoding of raw perfusion MRI for the prediction of final lesion in stroke. Med. Image Anal. 2018, 50, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Dhar, R.; Heitsch, L.; Ford, A.; Fernandez-Cadenas, I.; Carrera, C.; Montaner, J.; Lin, W.; Shen, D.; An, H.; et al. Automated quantification of cerebral edema following hemispheric infarction: Application of a machine-learning algorithm to evaluate CSF shifts on serial head CTs. NeuroImage Clin. 2016, 12, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Alwell, K.; Moomaw, C.J.; Woo, D.; Adeoye, O.; Flaherty, M.L.; Ferioli, S.; Mackey, J.; Rosa, F.D.L.R.L.; Martini, S.; et al. Towards phenotyping stroke: Leveraging data from a large-scale epidemiological study to detect stroke diagnosis. PLoS ONE 2018, 13, e0192586. [Google Scholar] [CrossRef]
- Hayden, D.T.; Hannon, N.; Callaly, E.; Ní Chróinín, D.; Horgan, G.; Kyne, L.; Duggan, J.; Dolan, E.; O’Rourke, K.; Williams, D.; et al. Rates and determinants of 5-year outcomes after atrial fibrillation-related stroke: A population study. Stroke 2015, 46, 3488–3493. [Google Scholar] [CrossRef]
- Li, S.; Nguyen, I.P.; Urbanczyk, K. Common infectious diseases of the central nervous system-clinical features and imaging characteristics. Quant. Imaging Med. Surg. 2020, 10, 2227–2259. [Google Scholar] [CrossRef]
- Reese, H.E.; Ronveaux, O.; Mwenda, J.M.; Bita, A.; Cohen, A.L.; Novak, R.T.; Fox, L.M.; Soeters, H.M. Invasive Meningococcal Disease in Africa’s Meningitis Belt: More Than Just Meningitis? J. Infect. Dis. 2019, 220, S263–S265. [Google Scholar] [CrossRef]
- Posnakoglou, L.; Siahanidou, T.; Syriopoulou, V.; Michos, A. Impact of cerebrospinal fluid syndromic testing in the management of children with suspected central nervous system infection. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 2379–2386. [Google Scholar] [CrossRef]
- Mentis, A.A.; Garcia, I.; Jiménez, J.; Paparoupa, M.; Xirogianni, A.; Papandreou, A.; Tzanakaki, G. Artificial Intelligence in Differential Diagnostics of Meningitis: A Nationwide Study. Diagnostics 2021, 11, 602. [Google Scholar] [CrossRef]
- Šeho, L.; Šutković, H.; Tabak, V.; Tahirović, S.; Smajović, A.; Bečić, E.; Deumić, A.; SpahićBećirović, L.; GurbetaPokvić, L.; Badnjević, A. Using Artificial Intelligence in Diagnostics of Meningitis. IFAC-Pap. OnLine 2022, 55, 56–61. [Google Scholar] [CrossRef]
- Jash, S.; Sharma, S. Pathogenic Infections during Pregnancy and the Consequences for Fetal Brain Development. Pathogens 2022, 11, 193. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Dong, X.; Zeng, C.; Liu, J.; Liu, H.; Hu, X.; Feng, J.; Du, S.; Wang, J.; Han, Y.; et al. Clinical Variables, Deep Learning and Radiomics Features Help Predict the Prognosis of Adult Anti-N-methyl-D-aspartate Receptor Encephalitis Early: A Two-Center Study in Southwest China. Front. Immunol. 2022, 13, 913703. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Du, L.; Li, Q.; Li, F.; Wang, B.; Zhao, Y.; Meng, Q.; Li, W.; Pan, J.; Xia, J.; et al. Deep learning-based relapse prediction of neuromyelitis optica spectrum disorder with anti-aquaporin-4 antibody. Front. Neurol. 2022, 13, 947974. [Google Scholar] [CrossRef]
- Muzumdar, D.; Jhawar, S.; Goel, A. Brain abscess: An overview. Int. J. Surg. 2011, 9, 136–144. [Google Scholar] [CrossRef]
- Bo, L.; Zhang, Z.; Jiang, Z.; Yang, C.; Huang, P.; Chen, T.; Wang, Y.; Yu, G.; Tan, X.; Cheng, Q.; et al. Differentiation of Brain Abscess From Cystic Glioma Using Conventional MRI Based on Deep Transfer Learning Features and Hand-Crafted Radiomics Features. Front. Med. 2021, 8, 748144. [Google Scholar] [CrossRef] [PubMed]
- Venkatasubba Rao, C.P.; Suarez, J.I.; Martin, R.H.; Bauza, C.; Georgiadis, A.; Calvillo, E.; Hemphill, J.C., 3rd; Sung, G.; Oddo, M.; Taccone, F.S.; et al. Global survey of outcomes of neurocritical care patients: Analysis of the PRINCE study part 2. Neurocrit. Care 2020, 32, 88–103. [Google Scholar] [CrossRef] [PubMed]
- Van de Beek, D.; Drake, J.M.; Tunkel, A.R. Nosocomial bacterial meningitis. N. Engl. J. Med. 2010, 362, 146–154. [Google Scholar] [CrossRef]
- Savin, I.; Ershova, K.; Kurdyumova, N.; Ershova, O.; Khomenko, O.; Danilov, G.; Shifrin, M.; Zelman, V. Healthcare-associated ventriculitis and meningitis in a neuro-ICU: Incidence and risk factors selected by machine learning approach. J. Crit. Care. 2018, 45, 95–104. [Google Scholar] [CrossRef]
- Chaudhry, F.; Hunt, R.J.; Hariharan, P.; Anand, S.K.; Sanjay, S.; Kjoller, E.E.; Bartlett, C.M.; Johnson, K.W.; Levy, P.D.; Noushmehr, H.; et al. Machine Learning Applications in the Neuro ICU: A Solution to Big Data Mayhem? Front. Neurol. 2020, 11, 554633. [Google Scholar] [CrossRef]
- Tabrizi, P.R.; Obeid, R.; Mansoor, A.; Ensel, S.; Cerrolaza, J.J.; Penn, A.; Linguraru, M.G. Cranial ultrasound-based prediction of post hemorrhagic hydrocephalus outcome in premature neonates with intraventricular hemorrhage. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2017, 2017, 169–172. [Google Scholar] [CrossRef]
- Aneja, S.; Chang, E.; Omuro, A. Applications of artificial intelligence in neuro-oncology. Curr. Opin. Neurol. 2019, 32, 850–856. [Google Scholar] [CrossRef] [PubMed]
- Rudie, J.D.; Rauschecker, A.M.; Bryan, R.N.; Davatzikos, C.; Mohan, S. Emerging Applications of Artificial Intelligence in Neuro-Oncology. Radiology 2019, 290, 607–618. [Google Scholar] [CrossRef]
- Markram, H. Seven challenges for neuroscience. Funct. Neurol. 2013, 28, 145–151. [Google Scholar] [CrossRef]
- Chance, F.S.; Aimone, J.B.; Musuvathy, S.S.; Smith, M.R.; Vineyard, C.M.; Wang, F. Crossing the Cleft: Communication Challenges Between Neuroscience and Artificial Intelligence. Front. Comput. Neurosci. 2020, 14, 39. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.J.; Karthikesalingam, A.; Suleyman, M.; Corrado, G.; King, D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019, 17, 195. [Google Scholar] [CrossRef]
- Graham, J. Artificial Intelligence, Machine Learning, and the FDA. 2016. Available online: https://www.forbes.com/sites/theapothecary/2016/08/19/artificial-intelligence-machine-learning-and-the-fda/#4aca26121aa1 (accessed on 24 January 2023).
- Jiang, F.; Jiang, Y.; Zhi, H.; Dong, Y.; Li, H.; Ma, S.; Wang, Y.; Dong, Q.; Shen, H.; Wang, Y. Artificial intelligence in healthcare: Past, present and future. Stroke Vasc. Neurol. 2017, 2, 230–243. [Google Scholar] [CrossRef]
- Ayyali, B.; Knott, D.; Kuiken, S.V. The Big-Data Revolution in US Health Care: Accelerating Value and Innovation. 2013. Available online: http://www.mckinsey.com/industries/healthcare-systems-and-services/our-insights/the-big-data-revolution-in-us-health-care (accessed on 24 January 2023).
- Ienca, M.; Ignatiadis, K. Artificial Intelligence in Clinical Neuroscience: Methodological and Ethical Challenges. Neuroscience 2020, 11, 77–87. [Google Scholar] [CrossRef]
- Tong, L. Evaluation of Different Brain Imaging Technologies. In Advances in Social Science, Education and Humanities Research, Proceedings of the 2021 International Conference on Public Art and Human Development (ICPAHD 2021), Kunming, China, 24–26 December 2021; Atlantis Press: Paris, France, 2021; Volume 638, pp. 692–696. [Google Scholar]
- Warbrick, T. Simultaneous EEG-fMRI: What Have We Learned and What Does the Future Hold? Sensors 2022, 22, 2262. [Google Scholar] [CrossRef]
- Hawsawi, H.B.; Carmichael, D.W.; Lemieux, L. Safety of simultaneous scalp or intracranial EEG during MRI: A review. Front. Phys. 2017, 5, 42. [Google Scholar] [CrossRef]
- Neuner, I.; Rajkumar, R.; Brambilla, C.R.; Ramkiran, S.; Ruch, A.; Orth, L.; Farrher, E.; Mauler, J.; Wyss, C.; Kops, E.R.; et al. Simultaneous PET-MR-EEG: Technology, Challenges and Application in Clinical Neuroscience. IEEE Trans. Radiat. Plasma Med. Sci. 2019, 3, 377–385. [Google Scholar] [CrossRef]
- de Senneville, B.D.; Zachiu, C.; Ries, M.; Moonen, C. EVolution: An edge-based variational method for non-rigid multi-modal image registration. Phys. Med. Biol. 2016, 61, 7377–7396. [Google Scholar] [CrossRef] [PubMed]
- Barkovich, M.J.; Li, Y.; Desikan, R.S.; Barkovich, A.J.; Xu, D. Challenges in pediatric neuroimaging. Neuroimage 2019, 185, 793–801. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.himss.org/resources/interoperability-healthcare (accessed on 24 January 2023).
- White, T.; Blok, E.; Calhoun, V.D. Data sharing and privacy issues in neuroimaging research: Opportunities, obstacles, challenges, and monsters under the bed. Hum. Brain Mapp. 2022, 43, 278–291. [Google Scholar] [CrossRef]
- Samper-González, J.; Burgos, N.; Bottani, S.; Fontanella, S.; Lu, P.; Marcoux, A.; Routier, A.; Guillon, J.; Bacci, M.; Wen, J.; et al. Reproducible evaluation of classification methods in Alzheimer’s disease: Framework and application to MRI and PET data. Neuroimage 2018, 183, 504–521. [Google Scholar] [CrossRef]
- Johnson, B.A. Avoiding diagnostic pitfalls in neuroimaging. Appl. Radiol. 2016, 45, 24–29. [Google Scholar] [CrossRef]
- Preston, W.G. Neuroimaging practice issues for the neurologist. Semin. Neurol. 2008, 28, 590–597. [Google Scholar] [CrossRef]
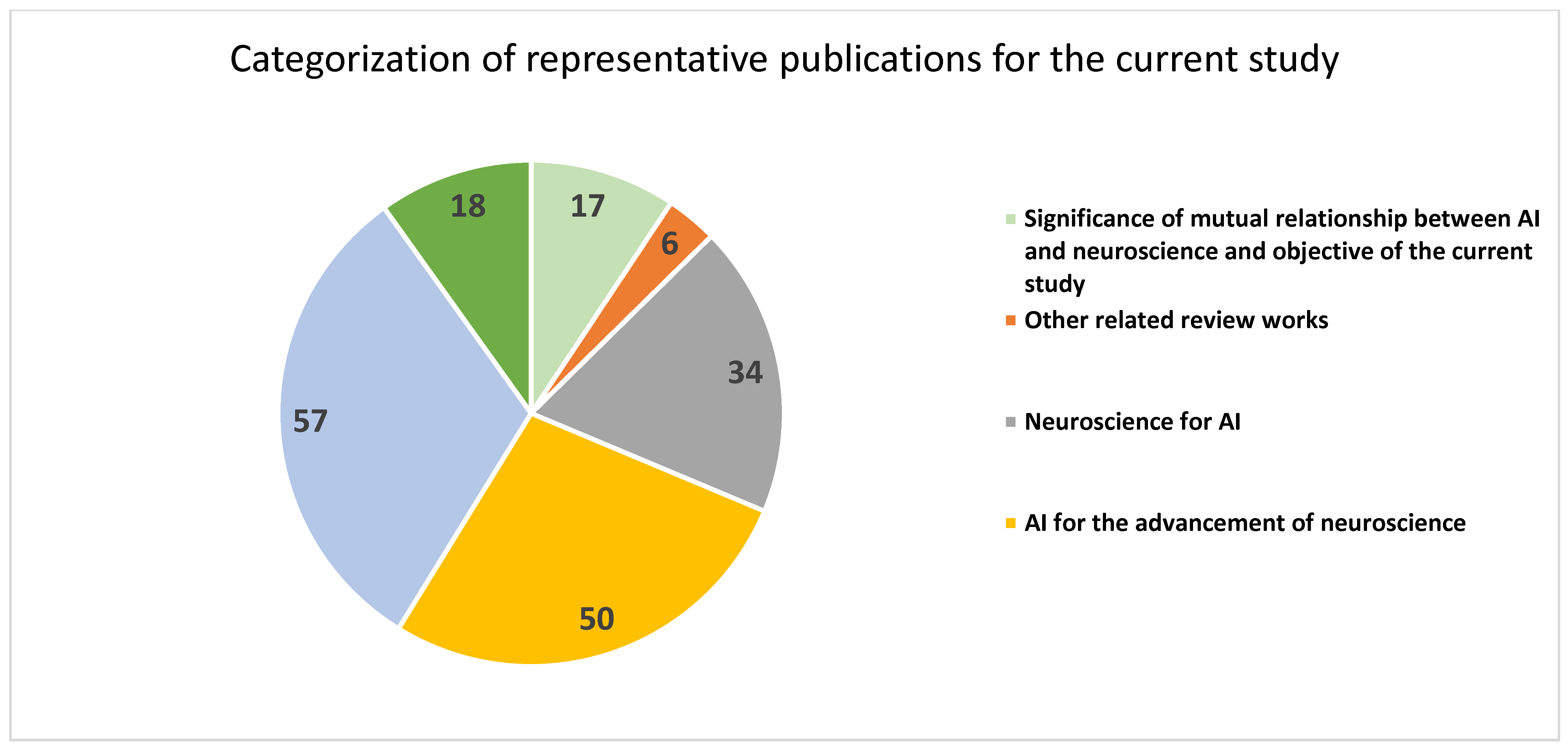



| References | Specific Focus | Number of Publications |
|---|---|---|
| [1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,23] | Significance of mutual relationship between AI and neuroscience and objective of the current study | 17 |
| [17,18,19,20,21,22] | Other related review works | 6 |
| [24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57] | Neuroscience for AI | 34 |
| [58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107] | AI for the advancement of neuroscience | 50 |
| [108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164] | AI for neurological disorders | 57 |
| [165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182] | Challenges and future directions of research | 18 |
| Total | 182 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Surianarayanan, C.; Lawrence, J.J.; Chelliah, P.R.; Prakash, E.; Hewage, C. Convergence of Artificial Intelligence and Neuroscience towards the Diagnosis of Neurological Disorders—A Scoping Review. Sensors 2023, 23, 3062. https://doi.org/10.3390/s23063062
Surianarayanan C, Lawrence JJ, Chelliah PR, Prakash E, Hewage C. Convergence of Artificial Intelligence and Neuroscience towards the Diagnosis of Neurological Disorders—A Scoping Review. Sensors. 2023; 23(6):3062. https://doi.org/10.3390/s23063062
Chicago/Turabian StyleSurianarayanan, Chellammal, John Jeyasekaran Lawrence, Pethuru Raj Chelliah, Edmond Prakash, and Chaminda Hewage. 2023. "Convergence of Artificial Intelligence and Neuroscience towards the Diagnosis of Neurological Disorders—A Scoping Review" Sensors 23, no. 6: 3062. https://doi.org/10.3390/s23063062
APA StyleSurianarayanan, C., Lawrence, J. J., Chelliah, P. R., Prakash, E., & Hewage, C. (2023). Convergence of Artificial Intelligence and Neuroscience towards the Diagnosis of Neurological Disorders—A Scoping Review. Sensors, 23(6), 3062. https://doi.org/10.3390/s23063062






