3D Ultrasound Reconstructions of the Carotid Artery and Thyroid Gland Using Artificial-Intelligence-Based Automatic Segmentation—Qualitative and Quantitative Evaluation of the Segmentation Results via Comparison with CT Angiography
Abstract
1. Introduction
2. Materials and Methods
2.1. Ultrasound Data Acquisition
2.2. CT Angiography
2.3. Original 2D Ultrasound Image Segmentation
2.4. CT-Scan Segmentation Methods
2.4.1. Semiautomatic Segmentation Method of Patient CT Scans Using 3D Slicer
- Patient’s DICOM scans were loaded into 3D Slicer.
- The CT angiography volume was selected as the closest 3D representation of the regions of interest: the carotid circulated lumen, the carotid artery wall, carotid artery soft plaque and calcified plaque, and the thyroid gland.
- The rendering mode was adjusted to MR angio to obtain a colored view of the bones and the blood vessels in the volume and identify the regions of interest.
- The regions of interest were selected to capture both the carotid arteries and the entire neck length of the patient, and the volume was cropped to the space inside the regions of interest.
- Segments for the relevant anatomical regions of the volume were created using the segmentation editor: for the carotid circulated lumen, the carotid artery wall, carotid artery soft plaque and calcified plaque, and the thyroid gland.
- Segmentation editor tools such as region growing, the paint tool, the eraser tool, the islands tool, etc. were used to segment various tissue types across various DICOM frames on all 3 axes. At the end, the 3D rendering feature of the region-growing tool was used to visualize and fine-tune the result before it was exported to STL format in order to have it as a comparison reference for the ultrasound segmentation. The segmentation was conducted in such a way so that the segmented result would overlap as closely as possible to the MR-angio visualization of the angiography volume, described above, this visualization being considered the gold-standard 3D representation of tissues of interest.
2.4.2. Methodology for CT Segmentation of Ground Truth
- The circulated lumen was segmented from the CT angiography volume (Figure 2).
- It was possible for some of the hard deposits to be partially or totally included in the circulated-lumen segmentation because the HU range for these, in some cases, depending on deposit density, was similar to the HU range of the contrast substance.
- The intersection volumes between the segmented circulated lumen on the CT angiography volume and the segmented hard deposits on the native CT volume were excluded from the result. This represented the ground truth for the circulated lumen.
- Hard deposits were segmented from the native CT volume. This was the ground truth.
- The thyroid was segmented from either the native or the CT angiography volume.
2.5. Machine-Learning Dataset Preparation
2.6. Ultrasound-Scan Automatic Segmentation Methods
2.7. 3D Ultrasound Reconstructions of the Carotid Arteries
2.8. Qualitative Analysis of the 3D US Reconstructions
2.9. Quantitative Analysis of the 3D US Reconstructions
3. Results
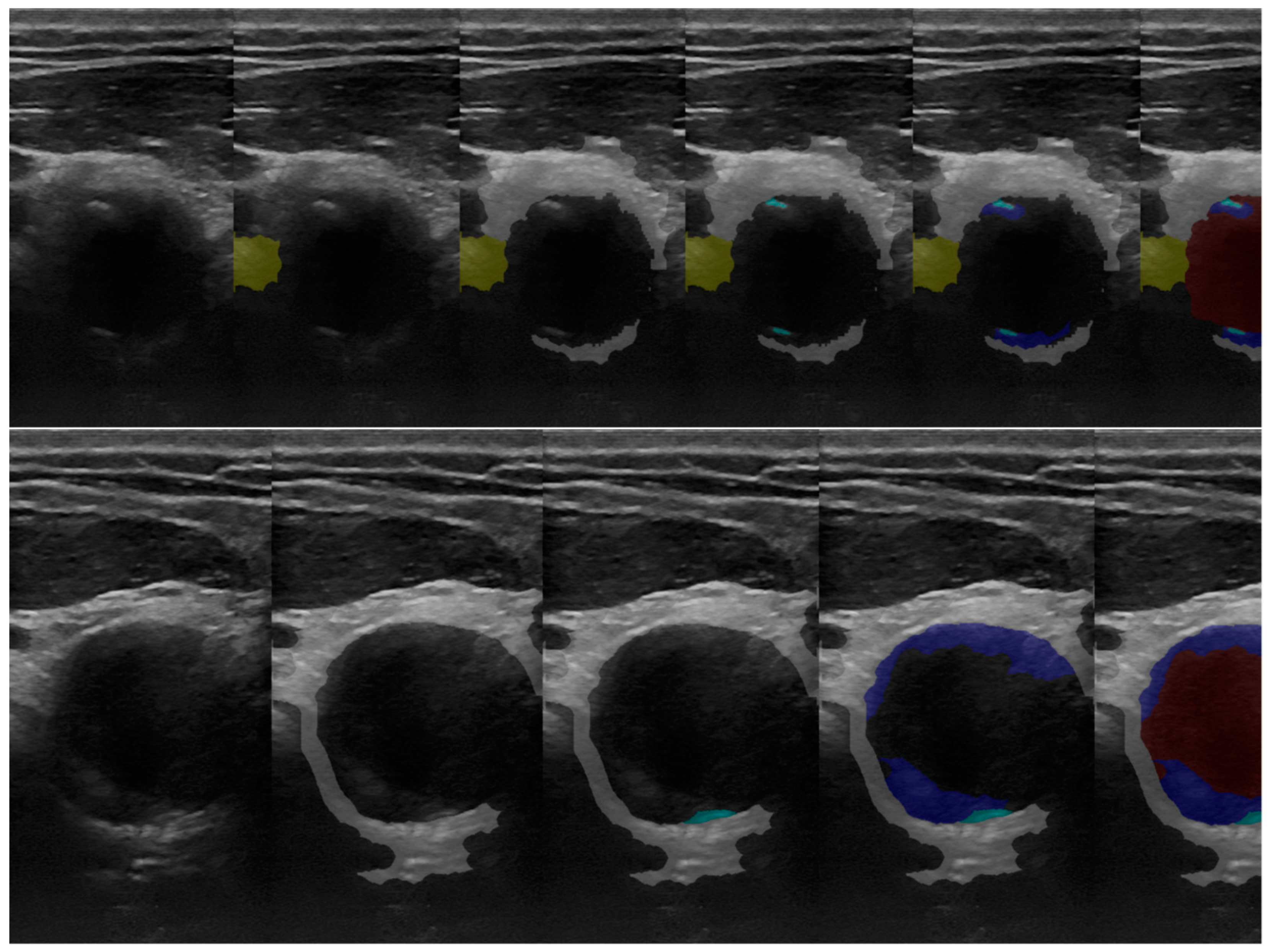
3.1. 2D Automatic Segmentation
3.1.1. Automatic Segmentation Results Compared with the Gold Standard (Operator’s Segmentation)
3.1.2. Automatic Segmentation (MultiRes U-Net) Training Results Metrics
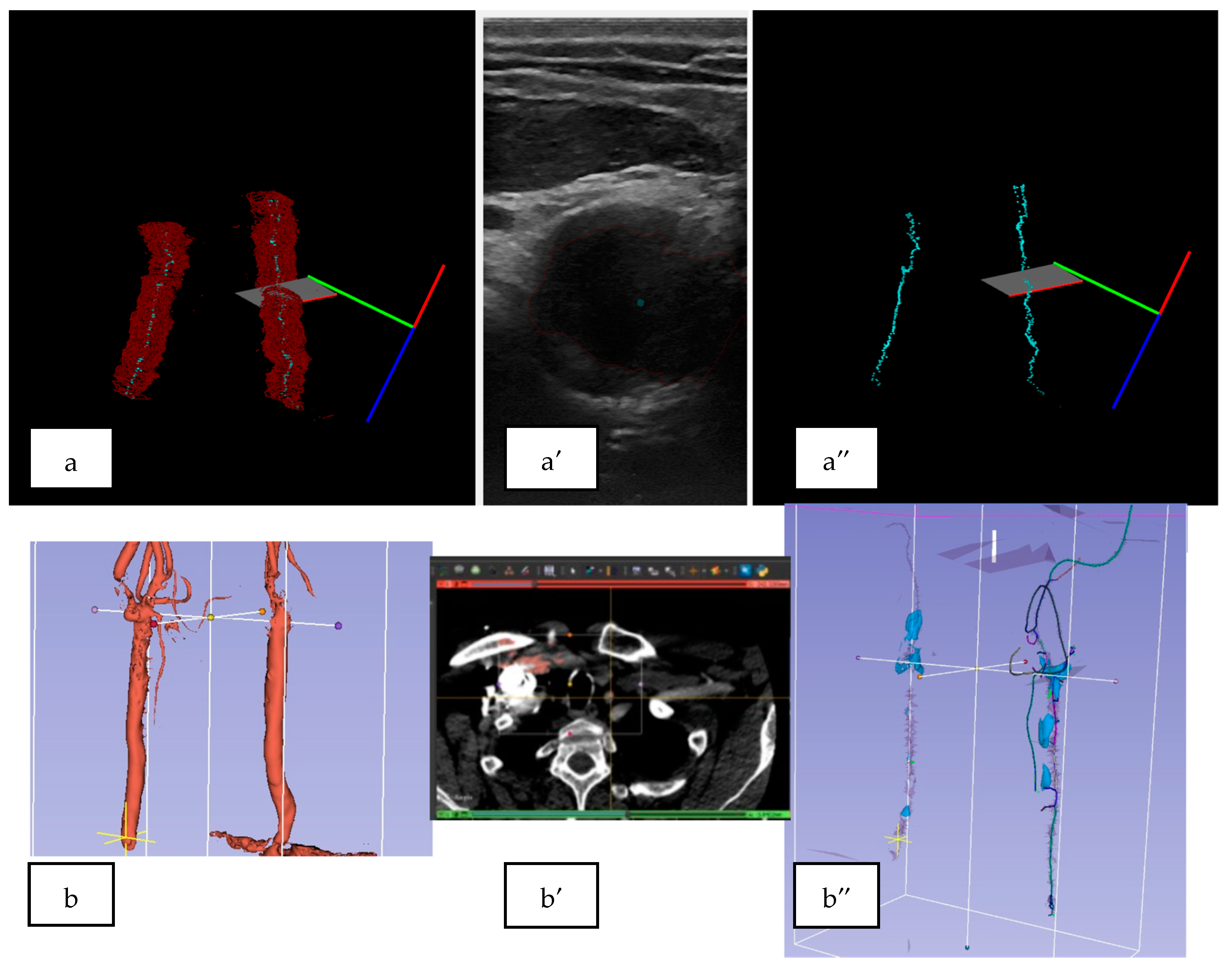
3.2. 3D US Reconstruction Compared with CT Angiography
3.3. Quantitative Analysis for the 3D US Reconstruction Based on Automated Mask Segmentation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, Z.; Yu, W. Update in the treatment of extracranial atherosclerotic disease for stroke prevention. Stroke Vasc. Neurol. 2020, 5, 65–70. [Google Scholar]
- Heyes, A.; Crichton, A.; Rajagopalan, S. Carotid artery disease: Knowing the numbers. Br. J. Hosp. Med. 2022, 83, 1–6. [Google Scholar] [CrossRef]
- Voigt, S.; van Os, H.; van Walderveen, M.; van der Schaaf, I.; Kappelle, L.; Broersen, A.; Velthuis, B.K.; de Jong, P.A.; Kockelkoren, R.; Kruyt, N.D.; et al. Sex differences in intracranial and extracranial atherosclerosis in patients with acute ischemic stroke. Int. J. Stroke 2021, 16, 385–391. [Google Scholar] [CrossRef]
- Saba, L.; Saam, T.; Jäger, H.R.; Yuan, C.; Hatsukami, T.S.; Saloner, D.; Wasserman, B.A.; Bonati, L.H.; Wintermark, M. Imaging biomarkers of vulnerable carotid plaques for stroke risk prediction and their potential clinical implications. Lancet Neurol. 2019, 18, 559–572. [Google Scholar] [CrossRef]
- Saxena, A.; Ng, E.Y.K.; Lim, S.T. Imaging modalities to diagnose carotid artery stenosis: Progress and prospect. Biomed. Eng. OnLine 2019, 18, 66. [Google Scholar] [CrossRef]
- Aursulesei, V. Nature or the natural evolution of plaque: What matters? Maedica 2013, 8, 309–314. [Google Scholar]
- Baradaran, H.; Al-Dasuqi, K.; Knight-Greenfield, A.; Giambrone, A.; Delgado, D.; Ebani, E.; Kamel, H.; Gupta, A. Association between Carotid Plaque Features on CTA and Cerebrovascular Ischemia: A Systematic Review and Meta-Analysis. AJNR Am. J. Neuroradiol. 2017, 38, 2321–2326. [Google Scholar] [CrossRef]
- Munnur, R.K.; Cheng, K.; Laggoune, J.; Talman, A.; Muthalaly, R.; Nerlekar, N.; Baey, Y.W.; Nogic, J.; Lin, A.; Cameron, J.D.; et al. Quantitative plaque characterisation and association with acute coronary syndrome on medium to long term follow up: Insights from computed tomography coronary angiography. Cardiovasc. Diagn. Ther. 2022, 12, 415–425. [Google Scholar] [CrossRef]
- Adla, T.; Adlova, R. Multimodality Imaging of Carotid Stenosis. Int. J. Angiol. 2015, 24, 179–184. [Google Scholar] [CrossRef]
- Samarzija, K.; Milosevic, P.; Jurjevic, Z.; Erdeljac, E. Grading of carotid artery stenosis with computed tomography angiography: Whether to use the narrowest diameter or the cross-sectional area. Insights Imaging 2018, 9, 527–534. [Google Scholar] [CrossRef]
- Saba, L.; Yuan, C.; Hatsukami, T.S.; Balu, N.; Qiao, Y.; DeMarco, J.K.; Saam, T.; Moody, A.R.; Li, D.; Matouk, C.C.; et al. Carotid Artery Wall Imaging: Perspective and Guidelines from the ASNR Vessel Wall Imaging Study Group and Expert Consensus Recommendations of the American Society of Neuroradiology. Am. J. Neuroradiol. 2018, 39, E9–E31. [Google Scholar] [CrossRef]
- Meshram, N.H.; Mitchell, C.C.; Wilbrand, S.; Dempsey, R.J.; Varghese, T. Deep Learning for Carotid Plaque Segmentation using a Dilated U-Net Architecture. Ultrason. Imaging 2020, 42, 221–230. [Google Scholar] [CrossRef]
- Jain, P.K.; Sharma, N.; Giannopoulos, A.A.; Saba, L.; Nicolaides, A.; Suri, J.S. Hybrid deep learning segmentation models for atherosclerotic plaque in internal carotid artery B-mode ultrasound. Comput. Biol. Med. 2021, 136, 104721. [Google Scholar] [CrossRef]
- Zhou, R.; Azarpazhooh, M.R.; Spence, J.D.; Hashemi, S.; Ma, W.; Cheng, X.; Gan, H.; Ding, M.; Fenster, A. Deep Learning-Based Carotid Plaque Segmentation from B-Mode Ultrasound Images. Ultrasound. Med. Biol. 2021, 47, 2723–2733. [Google Scholar] [CrossRef]
- Chifor, R.; Li, M.; Nguyen, K.-C.T.; Arsenescu, T.; Chifor, I.; Badea, A.F.; Badea, M.E.; Hotoleanu, M.; Major, P.W.; Le, L.H. Three-Dimensional Periodontal Investigations Using a Prototype Handheld Ultrasound Scanner with Spatial Positioning Reading Sensor. Med. Ultrason. 2021, 23, 297–304. Available online: https://www.medultrason.ro/medultrason/index.php/medultrason/article/view/2837 (accessed on 20 January 2022).
- Chifor, R.; Marita, T.; Arsenescu, T.; Santoma, A.; Badea, A.F.; Colosi, H.A.; Badea, M.-E.; Chifor, I. Accuracy Report on a Handheld 3D Ultrasound Scanner Prototype Based on a Standard Ultrasound Machine and a Spatial Pose Reading Sensor. Sensors 2022, 22, 3358. [Google Scholar] [CrossRef]
- Pinter, C.; Lasso, A.; Fichtinger, G. Polymorph segmentation representation for medical image computing. Comput. Methods Programs Biomed. 2019, 171, 19–26. [Google Scholar] [CrossRef]
- Ibtehaz, N.; Rahman, M.S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar] [CrossRef]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.-C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef]
- Marita, T.; Brehar, R.; Itu, R.; Trif, P.; Mitrea, D. Deep learning based mases segmentation method in mammograms. J. Autom. Comput. Appl. Math. 2020, 28, 15–21. [Google Scholar]
- Groves, L.A.; VanBerlo, B.; Veinberg, N.; Alboog, A.; Peters, T.M.; Chen, E.C.S. Automatic segmentation of the carotid artery and internal jugular vein from 2D ultrasound images for 3D vascular reconstruction. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 1835–1846. [Google Scholar] [CrossRef]
- Zhuang, S.; Li, F.; Raj, A.N.J.; Ding, W.; Zhou, W.; Zhuang, Z. Automatic segmentation for ultrasound image of carotid intimal-media based on improved superpixel generation algorithm and fractal theory. Comput. Methods Programs Biomed. 2021, 205, 106084. [Google Scholar] [CrossRef]
- Li, Y.; Zou, L.; Xiong, L.; Yu, F.; Jiang, H.; Fan, C.; Cheng, M.; Li, Q. FRDD-Net: Automated Carotid Plaque Ultrasound Images Segmentation Using Feature Remapping and Dense Decoding. Sensors 2022, 22, 887. [Google Scholar] [CrossRef]
- Smits, L.P.; Van Wijk, D.F.; Duivenvoorden, R.; Xu, D.; Yuan, C.; Stroes, E.S.; Nederveen, A.J. Manual versus Automated Carotid Artery Plaque Component Segmentation in High and Lower Quality 3.0 Tesla MRI Scans. PLoS ONE 2016, 11, e0164267. [Google Scholar] [CrossRef]

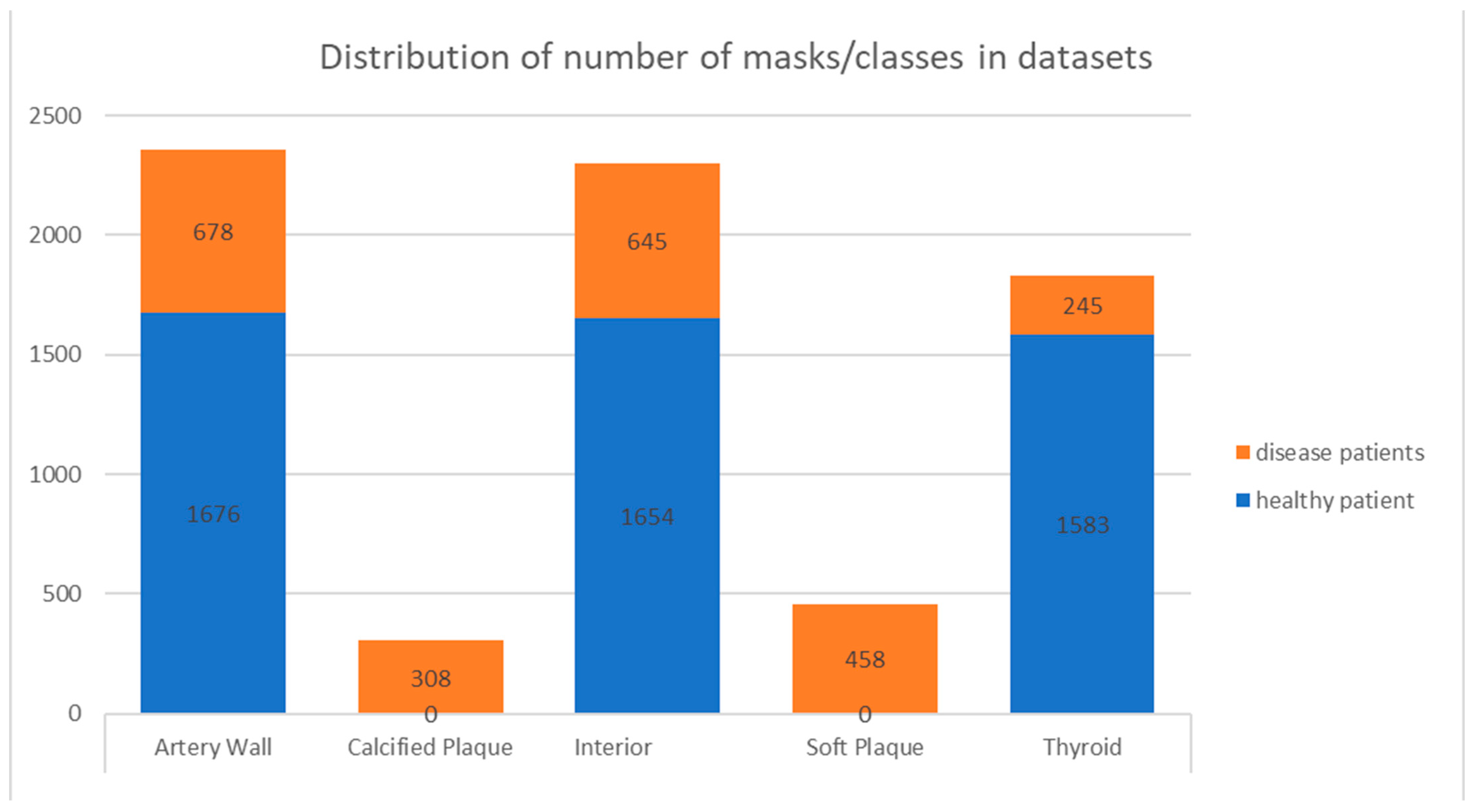


| Dataset | Number of Frames | Patient |
|---|---|---|
| Dataset 1 | 1001 | Healthy (patient 1) |
| Dataset 2 | 971 | Healthy (patient 1) |
| Dataset 3 | 959 | Healthy (patient 1) |
| Dataset 4 | 531 | Carotid disease (patient 2) |
| Dataset 5 | 222 | Carotid disease (patient 3) |
| US Scan | Frame Interval | Number of Frames | Type of Segmentation |
|---|---|---|---|
| Healthy Patient Scan 1–4 | - | 2931 | Manual |
| Carotid Disease Patient1 Scan 1 | 530–1060 | 531 | Manual |
| Carotid Disease Patient1 Scan 2 | 330–1100 | 770 | AI Prediction |
| Carotid Disease Patient1 Scan 3 | 230–1200 | 970 | AI Prediction |
| Carotid Disease Patient1 Scan 4 | 280–1210 | 930 | AI Prediction |
| Carotid Disease Patient2 Scan 1 | 400–622 | 222 | Manual |
| Carotid Disease Patient2 Scan 2 | 330–1080 | 750 | AI Prediction |
| Carotid Disease Patient2 Scan 3 | 280–1000 | 720 | AI Prediction |
| Carotid Disease Patient2 Scan 4 | 310–1195 | 885 | AI Prediction |
| Per Class Prediction’s Performance Based on Refernce Training Data. | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dataset | Class Name | tp | tn | fp | fn | Accuracy | Specificity | Recall | Precision | Misclassification Rate | f1_Score (Dice) | Avg_iou | Std_ dev |
| Carotid Disease Patients | Artery Wall | 637 | 73 | 0 | 43 | 0.94290 | 1.00000 | 0.93676 | 1.00000 | 0.05710 | 0.96735 | 0.76728 | 0.16596 |
| Carotid Disease Patients | Hard Plaque | 290 | 439 | 6 | 18 | 0.96813 | 0.98652 | 0.94156 | 0.97973 | 0.03187 | 0.96026 | 0.64748 | 0.19819 |
| Carotid Disease Patients | Interior | 622 | 105 | 2 | 24 | 0.96547 | 0.98131 | 0.96285 | 0.99679 | 0.03453 | 0.97953 | 0.93094 | 0.05395 |
| Carotid Disease Patients | Soft plaque | 444 | 280 | 15 | 14 | 0.96149 | 0.94915 | 0.96943 | 0.96732 | 0.03851 | 0.96838 | 0.75507 | 0.17420 |
| Carotid Disease Patients | Thyroid | 187 | 494 | 11 | 61 | 0.90438 | 0.97822 | 0.75403 | 0.94444 | 0.09562 | 0.83857 | 0.85365 | 0.18058 |
| Carotid Disease Patients | All Classes | 0.80296 | 0.14231 | ||||||||||
| Aligned 3D US Reconstructions | Mean Distance | Std Deviation | Scale | Theoretical Overlap | RMS |
|---|---|---|---|---|---|
| Patient 1 Scan1 + Scan2, CCOSS-Aligned | 10.9656 | 25.5394 | 0.96326 | 100% | 25.0231 |
| Patient 1 Scan1 + Scan3, CCOSS-Aligned | 24.5671 | 34.3423 | 0.943269 | 100% | 16.064 |
| Patient 1 Scan1 + Scan4, CCOSS-Aligned | 5.03695 | 8.62778 | 1.01003 | 100% | 25.2053 |
| Patient 2 Scan1 + Scan2, CCOSS-Aligned | Corrupt data | Corrupt data | Corrupt data | Corrupt data | Corrupt data |
| Patient 2 Scan1 + Scan3, CCOSS-Aligned | 7.62545 | 15.7469 | 0.628546 | 100% | 50.2331 |
| Patient 2 Scan1 + Scan4, CCOSS-Aligned | 6.07168 | 16.0121 | 0.638966 | 100% | 18.9179 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arsenescu, T.; Chifor, R.; Marita, T.; Santoma, A.; Lebovici, A.; Duma, D.; Vacaras, V.; Badea, A.F. 3D Ultrasound Reconstructions of the Carotid Artery and Thyroid Gland Using Artificial-Intelligence-Based Automatic Segmentation—Qualitative and Quantitative Evaluation of the Segmentation Results via Comparison with CT Angiography. Sensors 2023, 23, 2806. https://doi.org/10.3390/s23052806
Arsenescu T, Chifor R, Marita T, Santoma A, Lebovici A, Duma D, Vacaras V, Badea AF. 3D Ultrasound Reconstructions of the Carotid Artery and Thyroid Gland Using Artificial-Intelligence-Based Automatic Segmentation—Qualitative and Quantitative Evaluation of the Segmentation Results via Comparison with CT Angiography. Sensors. 2023; 23(5):2806. https://doi.org/10.3390/s23052806
Chicago/Turabian StyleArsenescu, Tudor, Radu Chifor, Tiberiu Marita, Andrei Santoma, Andrei Lebovici, Daniel Duma, Vitalie Vacaras, and Alexandru Florin Badea. 2023. "3D Ultrasound Reconstructions of the Carotid Artery and Thyroid Gland Using Artificial-Intelligence-Based Automatic Segmentation—Qualitative and Quantitative Evaluation of the Segmentation Results via Comparison with CT Angiography" Sensors 23, no. 5: 2806. https://doi.org/10.3390/s23052806
APA StyleArsenescu, T., Chifor, R., Marita, T., Santoma, A., Lebovici, A., Duma, D., Vacaras, V., & Badea, A. F. (2023). 3D Ultrasound Reconstructions of the Carotid Artery and Thyroid Gland Using Artificial-Intelligence-Based Automatic Segmentation—Qualitative and Quantitative Evaluation of the Segmentation Results via Comparison with CT Angiography. Sensors, 23(5), 2806. https://doi.org/10.3390/s23052806







