IoMT-Enabled Computer-Aided Diagnosis of Pulmonary Embolism from Computed Tomography Scans Using Deep Learning
Abstract
1. Introduction
- The paper presents a deep-learning-based PE detection CAD system.
- The proposed method performs feature extraction using DenseNet201 with customized, fully connected decision-making layers.
- The paper also contributes to overcoming the gradient vanishing problem in CNNs for PE detection.
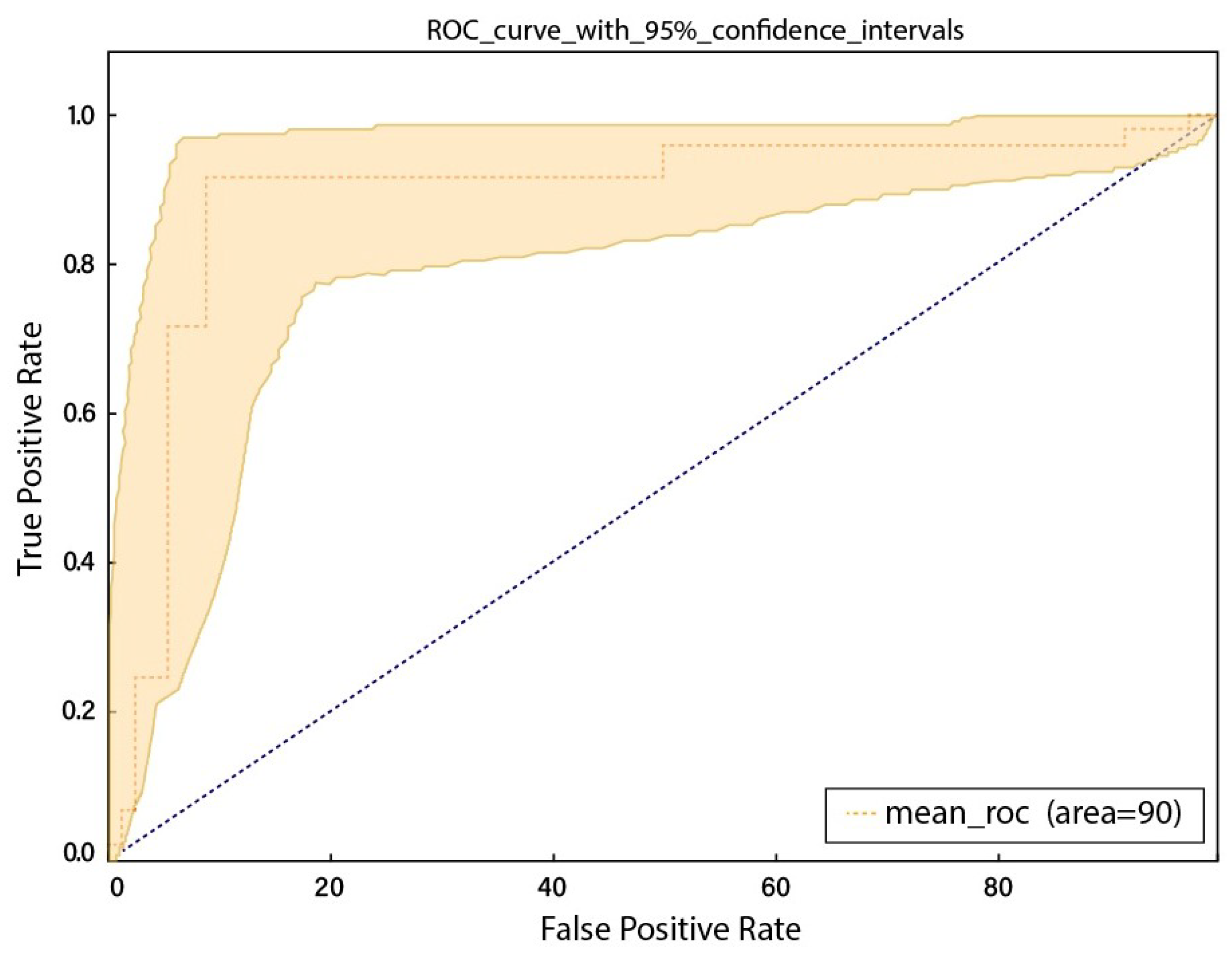
- The customized model was trained on the Radiological Society of North America (RSNA)-Pulmonary Embolism Detection Challenge (2020) Kaggle dataset and showed promising results of 88%, 88%, 89%, and 90% in terms of the accuracy, sensitivity, specificity, and AUC.
- The proposed system reduced the error rate by 3% of CAD in PE detection.
2. Related Work
3. Materials and Methods
3.1. Data Set
3.1.1. Annotation Process
3.1.2. The Final Dataset
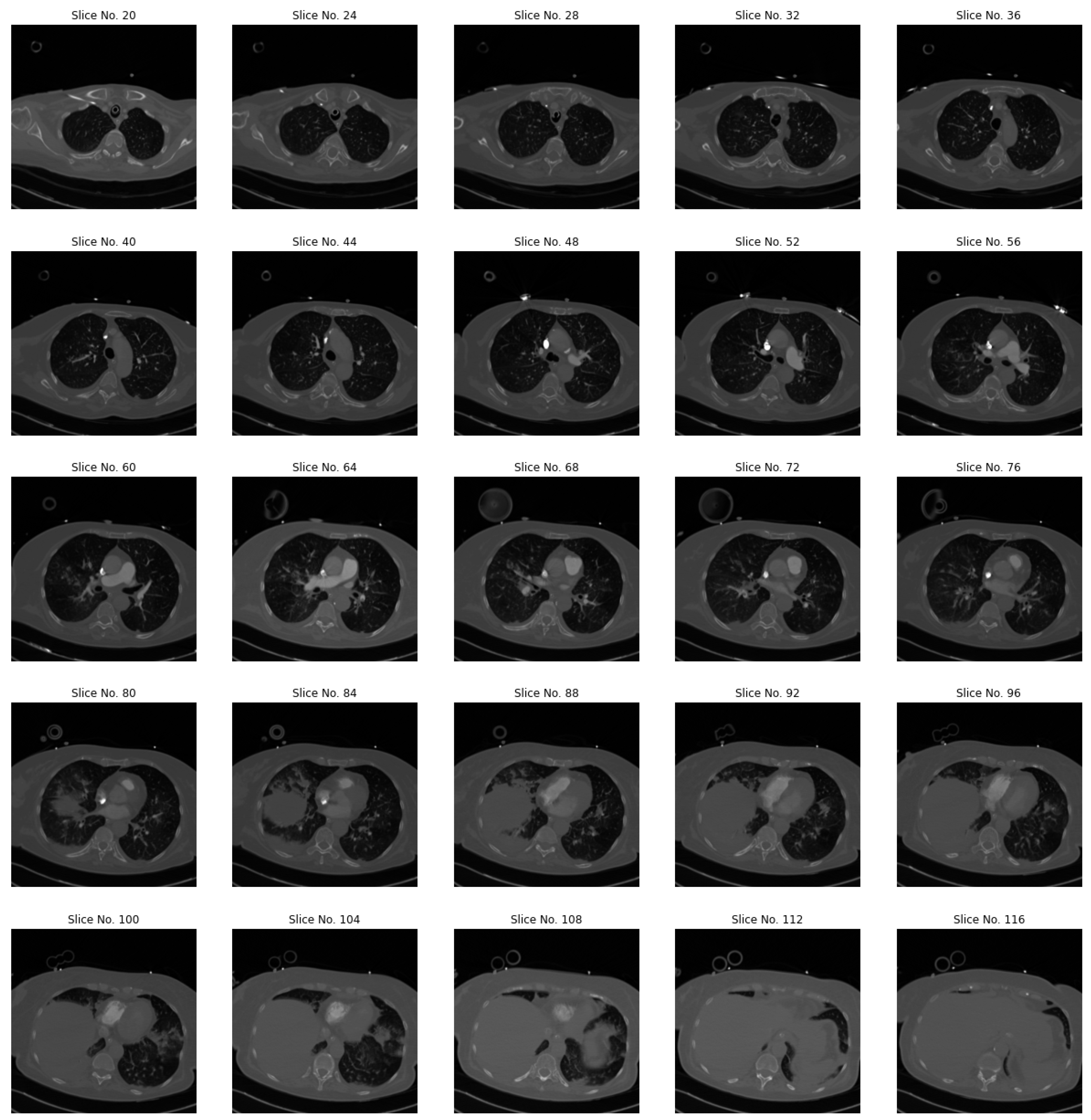
3.2. Slices Selection and Pre-Processing
| Algorithm 1 Adjusting WL and WW for high-contrast setting in CT scans. |
|
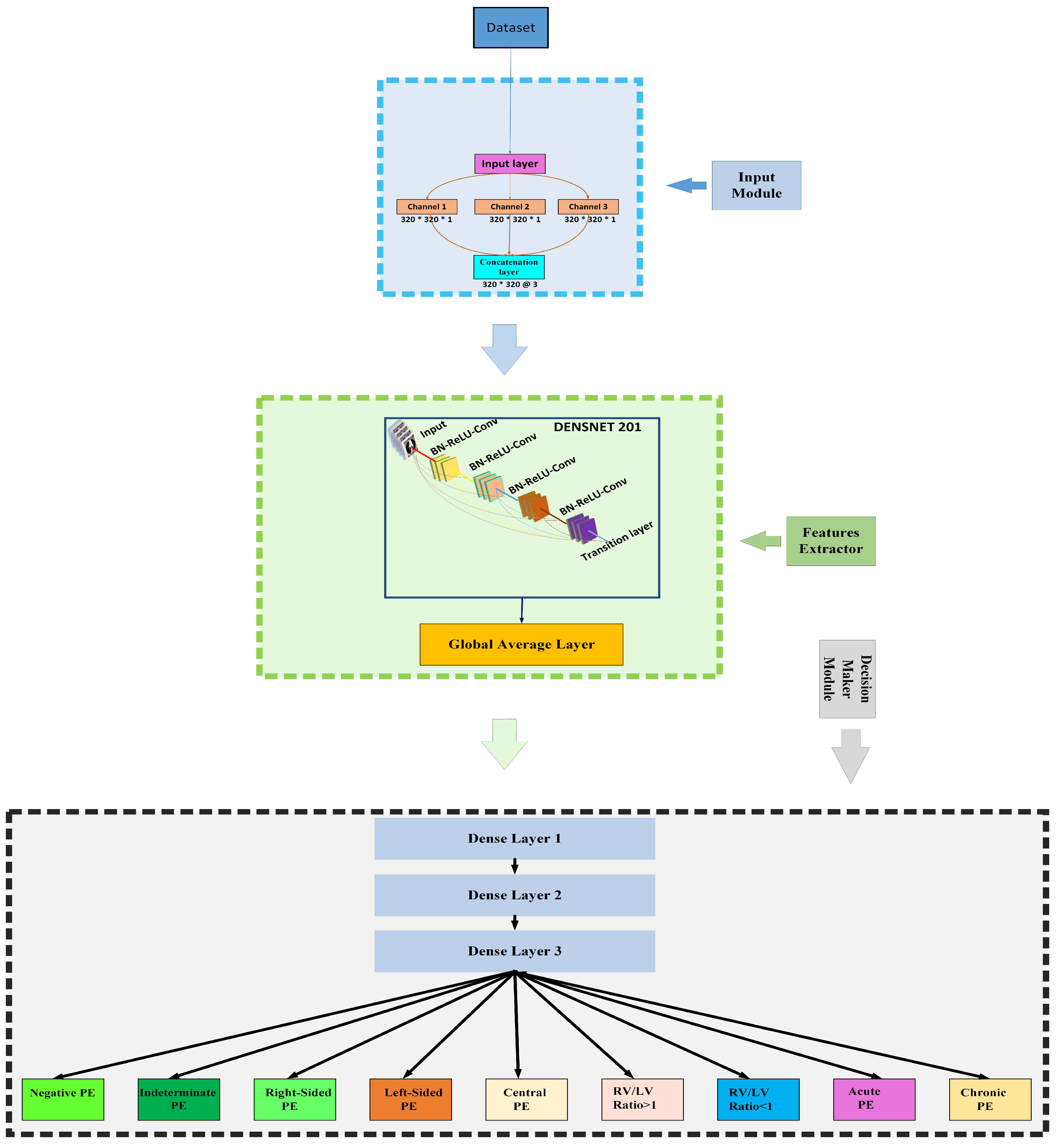
3.3. Proposed Framework
3.3.1. Input Module
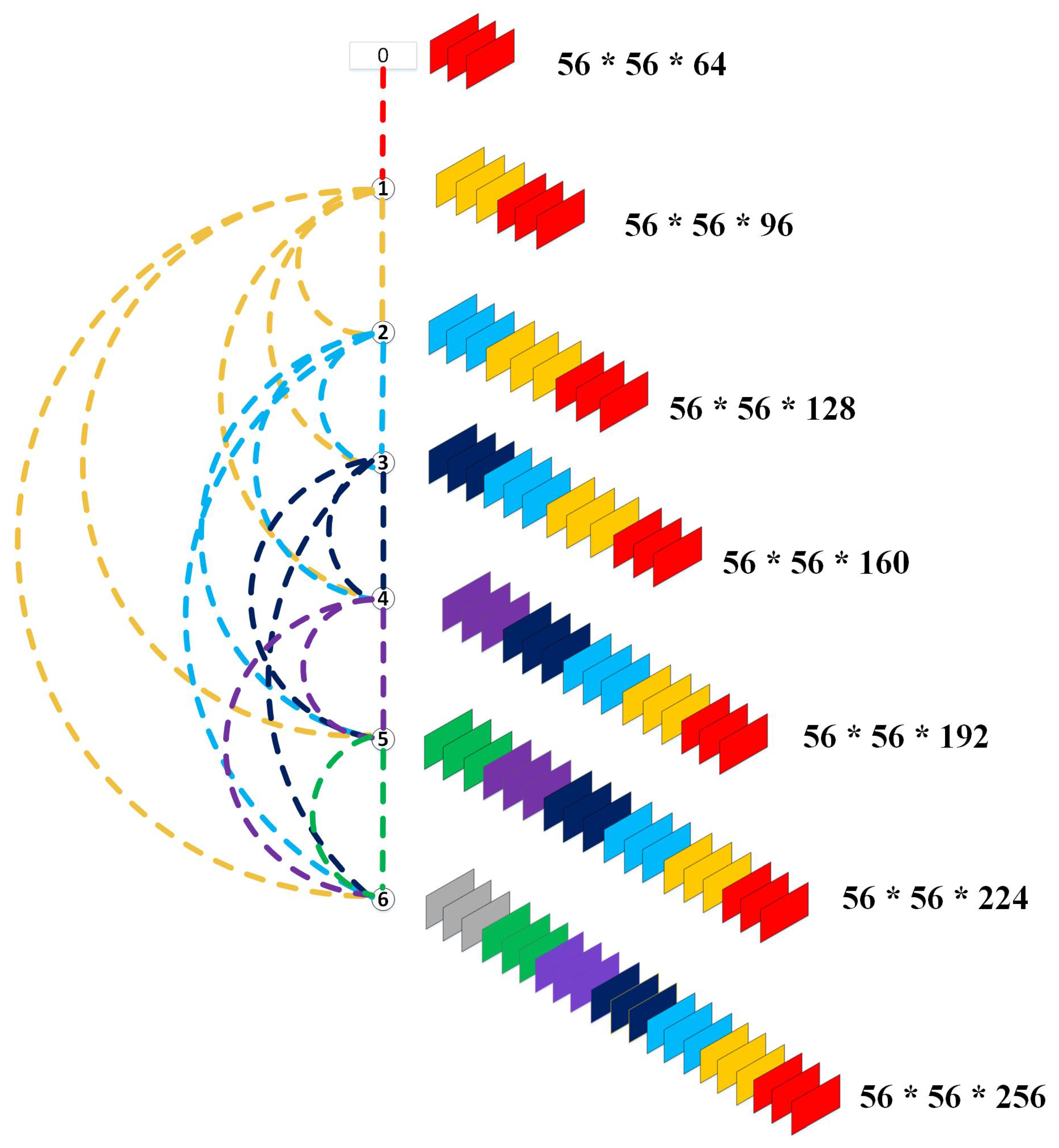
3.3.2. Towards DenseNet
3.3.3. Decision-Making Module
3.4. Experimental Setup
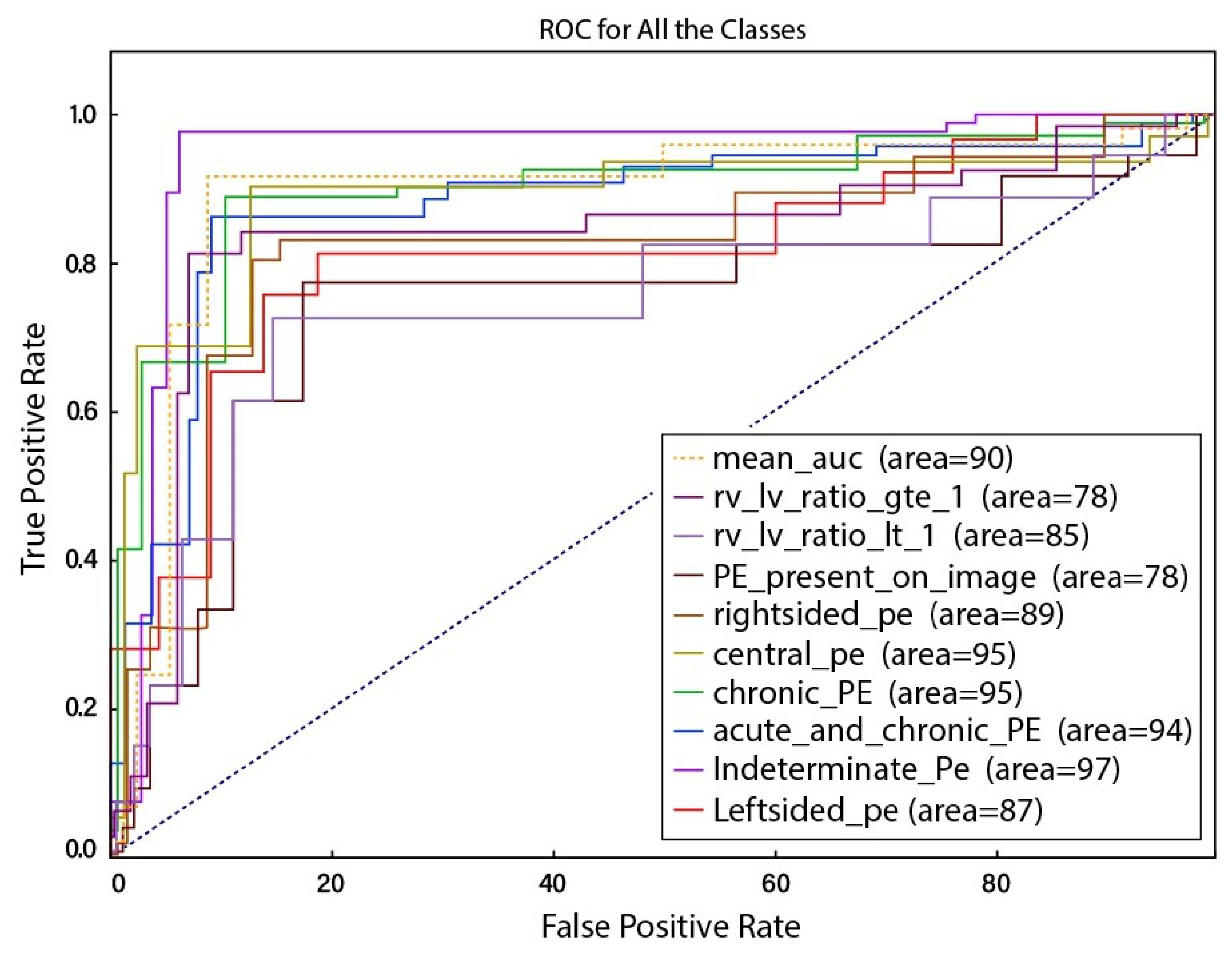
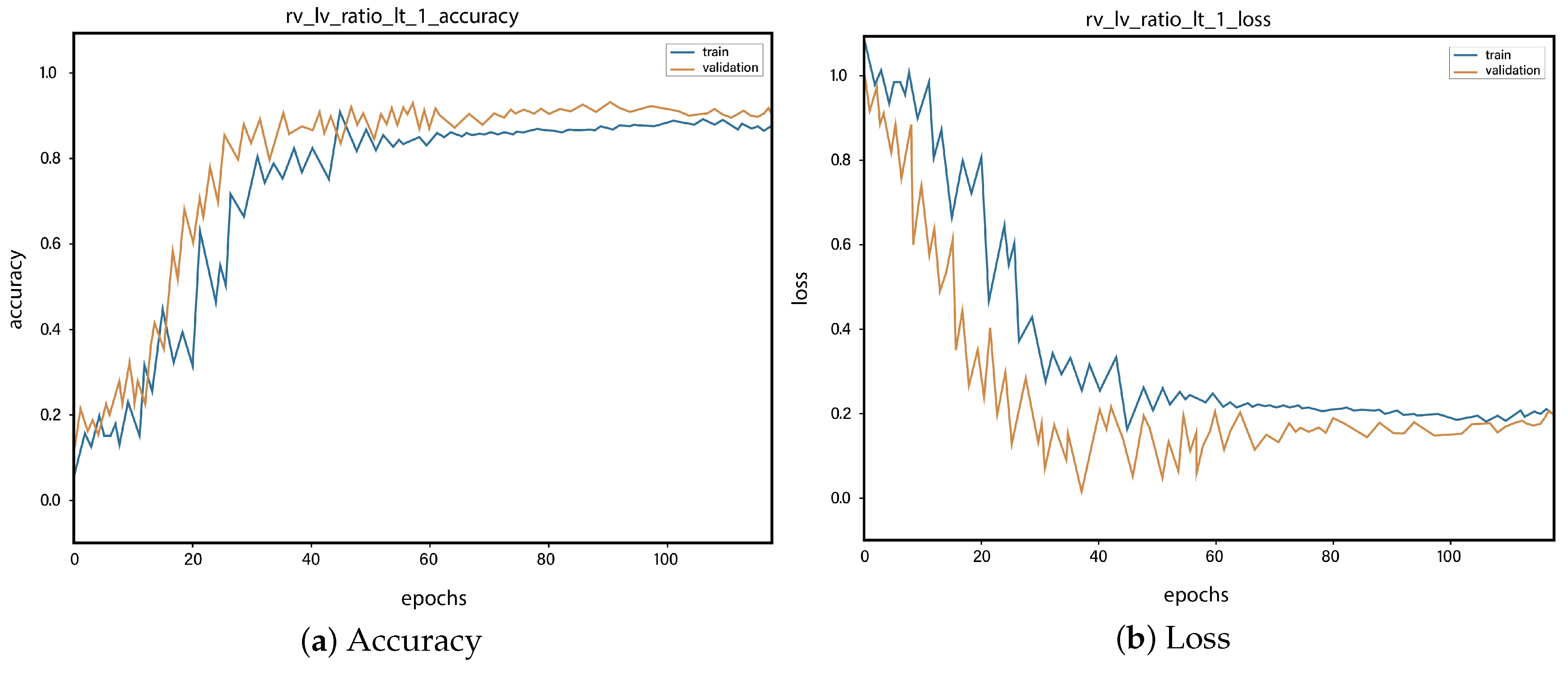
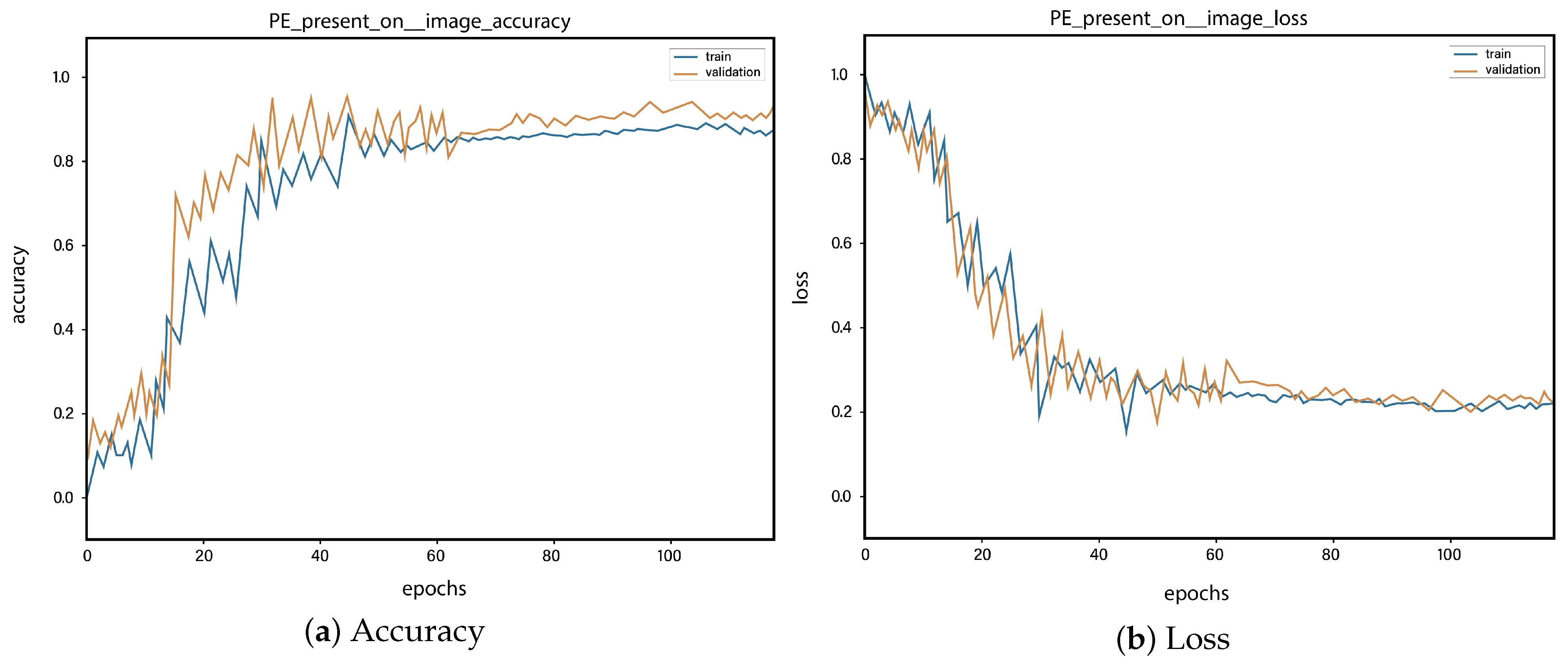
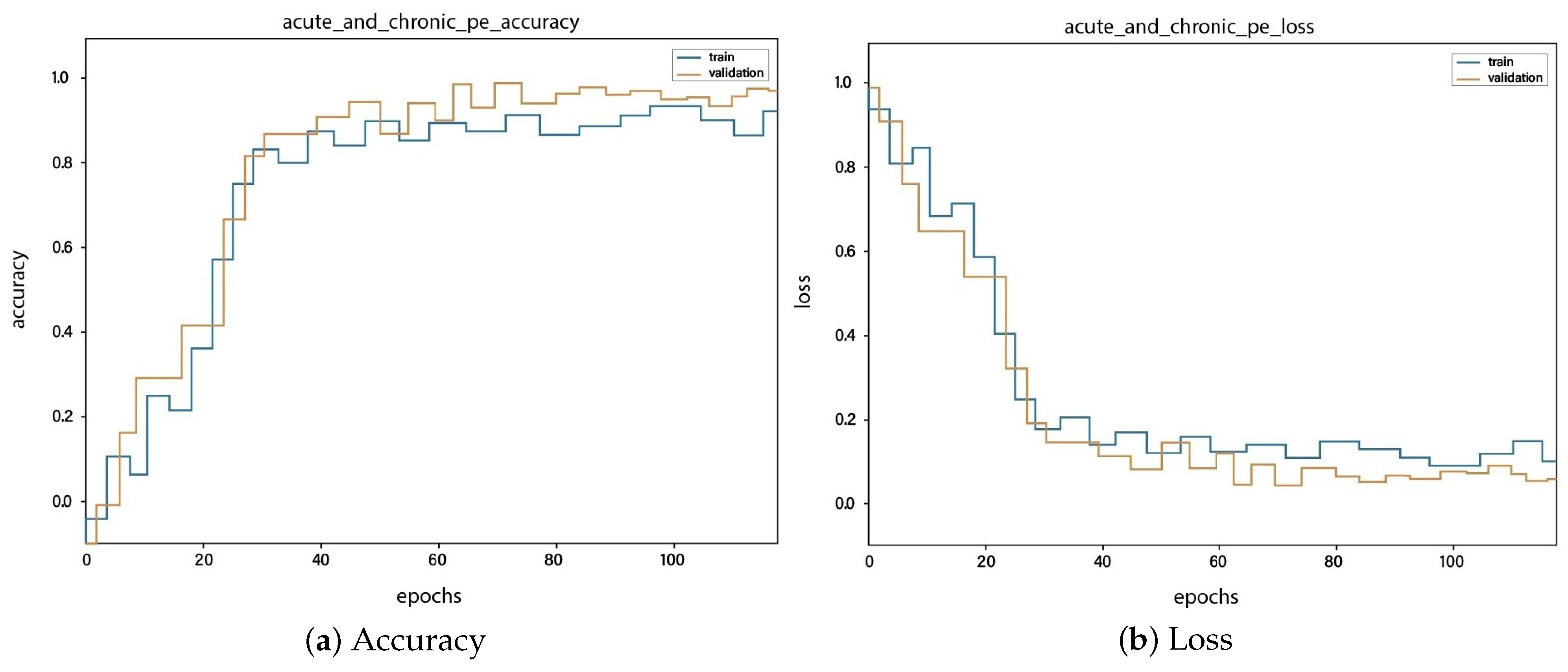
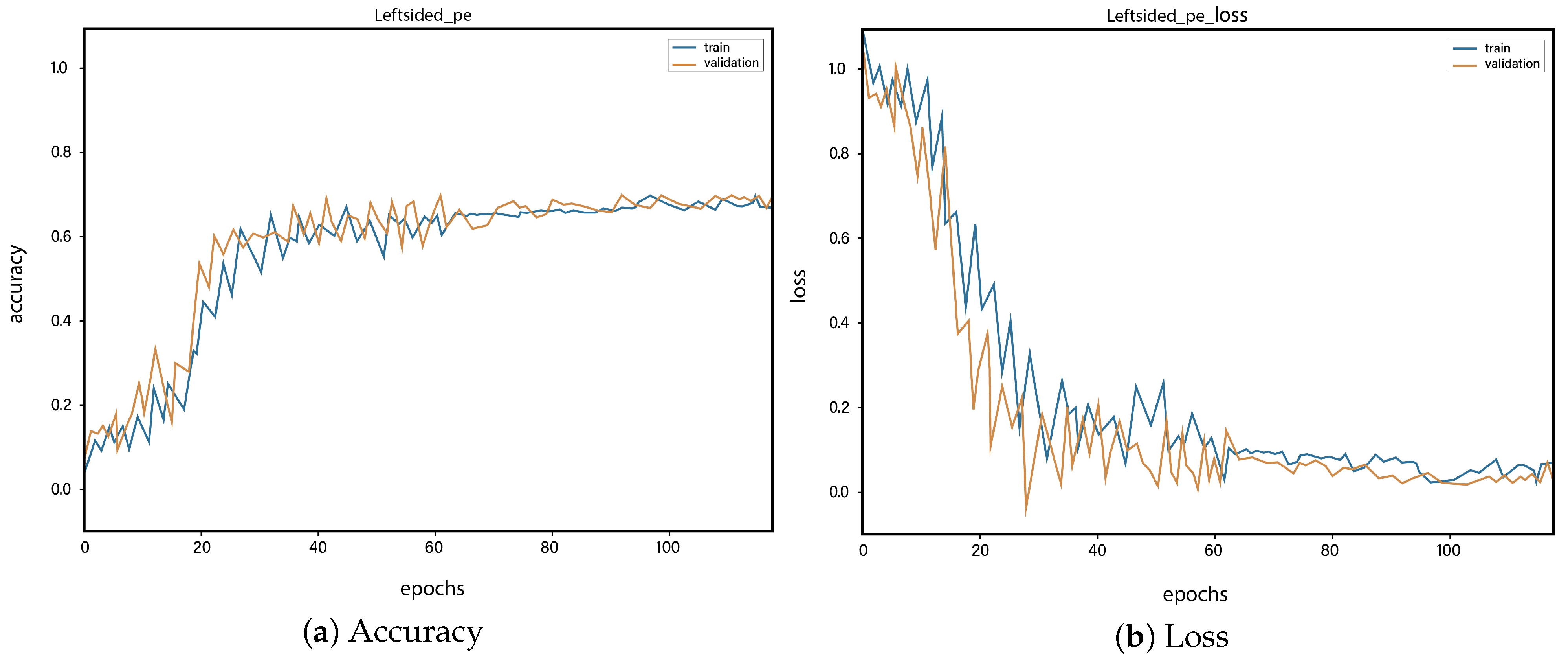
4. Results and Discussions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khan, F.; Tarimer, I.; Taekeun, W. Factor Model for Online Education during the COVID-19 Pandemic Using the IoT. Processes 2022, 10, 1419. [Google Scholar] [CrossRef]
- Al-Kahtani, M.S.; Khan, F.; Taekeun, W. Application of Internet of Things and Sensors in Healthcare. Sensors 2022, 22, 5738. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Zahid, M.; Gürüler, H.; Tarımer, İ.; Whangbo, T. An Efficient and Reliable Multicasting for Smart Cities. CMC-Comput. Mater. Contin. 2022, 72, 663–678. [Google Scholar] [CrossRef]
- Khan, H.; Shah, P.M.; Shah, M.A.; Islam, S.u.; Rodrigues, J.J. Cascading handcrafted features and Convolutional Neural Network for IoT-enabled brain tumor segmentation. Comput. Commun. 2020, 153, 196–207. [Google Scholar] [CrossRef]
- Ahmed, G.; Islam, S.U.; Shahid, M.; Akhunzada, A.; Jabbar, S.; Khan, M.K.; Riaz, M.; Han, K. Rigorous analysis and evaluation of specific absorption rate (SAR) for mobile multimedia healthcare. IEEE Access 2018, 6, 29602–29610. [Google Scholar] [CrossRef]
- Awais, M.; Raza, M.; Singh, N.; Bashir, K.; Manzoor, U.; Islam, S.u.; Rodrigues, J.J.P.C. LSTM based Emotion Detection using Physiological Signals: IoT framework for Healthcare and Distance Learning in COVID-19. IEEE Internet Things J. 2020, 8, 16863–16871. [Google Scholar] [CrossRef]
- Huang, S.C.; Kothari, T.; Banerjee, I.; Chute, C.; Ball, R.L.; Borus, N.; Huang, A.; Patel, B.N.; Rajpurkar, P.; Irvin, J.; et al. PENet—A scalable deep-learning model for automated diagnosis of pulmonary embolism using volumetric CT imaging. NPJ Digit. Med. 2020, 3, 61. [Google Scholar] [CrossRef] [PubMed]
- Islam, N.U.; Gehlot, S.; Zhou, Z.; Gotway, M.B.; Liang, J. Seeking an Optimal Approach for Computer-Aided Pulmonary Embolism Detection. In Proceedings of the International Workshop on Machine Learning in Medical Imaging, Strasbourg, France, 27 September 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 692–702. [Google Scholar]
- Hampson, N.B. Pulmonary embolism: Difficulties in the clinical diagnosis. Semin. Respir. Infect. 1995, 10, 123–130. [Google Scholar]
- Yang, X.; Lin, Y.; Su, J.; Wang, X.; Li, X.; Lin, J.; Cheng, K.T. A two-stage convolutional neural network for pulmonary embolism detection from CTPA images. IEEE Access 2019, 7, 84849–84857. [Google Scholar] [CrossRef]
- Tajbakhsh, N.; Shin, J.Y.; Gotway, M.B.; Liang, J. Computer-aided detection and visualization of pulmonary embolism using a novel, compact, and discriminative image representation. Med. Image Anal. 2019, 58, 101541. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K.; et al. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Shah, P.M.; Zeb, A.; Shafi, U.; Zaidi, S.F.A.; Shah, M.A. Detection of Parkinson disease in brain MRI using convolutional neural network. In Proceedings of the 2018 24th International Conference on Automation and Computing (ICAC), Newcastle upon Tyne, UK, 6–7 September 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 1–6. [Google Scholar]
- Shah, P.M.; Ullah, F.; Shah, D.; Gani, A.; Maple, C.; Wang, Y.; Shahid, A.; Abrar, M.; ul Islam, S. Deep GRU-CNN model for COVID-19 detection from chest X-rays data. IEEE Access 2021, 10, 35094–35105. [Google Scholar] [CrossRef]
- Shah, P.M.; Ullah, H.; Ullah, R.; Shah, D.; Wang, Y.; Islam, S.u.; Gani, A.; Rodrigues, J.J. DC-GAN-based synthetic X-ray images augmentation for increasing the performance of EfficientNet for COVID-19 detection. Expert Syst. 2022, 39, e12823. [Google Scholar] [CrossRef]
- Shah, P.M.; Khan, H.; Shafi, U.; Raza, M.; Le-Minh, H. 2d-cnn based segmentation of ischemic stroke lesions in mri scans. In Proceedings of the International Conference on Computational Collective Intelligence, Da Nang, Vietnam, 30 November–3 December 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 276–286. [Google Scholar]
- Engelke, C.; Schmidt, S.; Bakai, A.; Auer, F.; Marten, K. Computer-assisted detection of pulmonary embolism: Performance evaluation in consensus with experienced and inexperienced chest radiologists. Eur. Radiol. 2008, 18, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Maizlin, Z.V.; Vos, P.M.; Godoy, M.B.; Cooperberg, P.L. Computer-aided detection of pulmonary embolism on CT angiography: Initial experience. J. Thorac. Imaging 2007, 22, 324–329. [Google Scholar] [CrossRef] [PubMed]
- Buhmann, S.; Herzog, P.; Liang, J.; Wolf, M.; Salganicoff, M.; Kirchhoff, C.; Reiser, M.; Becker, C.H. Clinical evaluation of a computer-aided diagnosis (CAD) prototype for the detection of pulmonary embolism. Acad. Radiol. 2007, 14, 651–658. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Tajbakhsh, N.; Gotway, M.B.; Liang, J. Computer-aided pulmonary embolism detection using a novel vessel-aligned multi-planar image representation and convolutional neural networks. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 62–69. [Google Scholar]
- Liu, W.; Liu, M.; Guo, X.; Zhang, P.; Zhang, L.; Zhang, R.; Kang, H.; Zhai, Z.; Tao, X.; Wan, J.; et al. Evaluation of acute pulmonary embolism and clot burden on CTPA with deep learning. Eur. Radiol. 2020, 30, 3567–3575. [Google Scholar] [CrossRef]
- Weikert, T.; Winkel, D.J.; Bremerich, J.; Stieltjes, B.; Parmar, V.; Sauter, A.W.; Sommer, G. Automated detection of pulmonary embolism in CT pulmonary angiograms using an AI-powered algorithm. Eur. Radiol. 2020, 30, 6545–6553. [Google Scholar] [CrossRef]
- Mohan, A.; Gosha, O.C. Pulmonary Embolism Detection. Available online: https://amsks.github.io/CV/MALIS_Final_Report.pdf (accessed on 16 October 2022).
- Rajan, D.; Beymer, D.; Abedin, S.; Dehghan, E. Pi-PE: A pipeline for pulmonary embolism detection using sparsely annotated 3D CT images. In Proceedings of the Machine Learning for Health Workshop, Virtual Event, 11 December 2020; pp. 220–232. [Google Scholar]
- Vainio, T.; Mäkelä, T.; Savolainen, S.; Kangasniemi, M. Performance of a 3D convolutional neural network in the detection of hypoperfusion at CT pulmonary angiography in patients with chronic pulmonary embolism: A feasibility study. Eur. Radiol. Exp. 2021, 5, 45. [Google Scholar] [CrossRef]
- Suman, S.; Singh, G.; Sakla, N.; Gattu, R.; Green, J.; Phatak, T.; Samaras, D.; Prasanna, P. Attention Based CNN-LSTM Network for Pulmonary Embolism Prediction on Chest Computed Tomography Pulmonary Angiograms. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 356–366. [Google Scholar]
- Schmid, R.; Johnson, J.; Ngo, J.; Lamoureux, C.; Baker, B.; Ngo, L. Development and Validation of a Highly Generalizable Deep Learning Pulmonary Embolism Detection Algorithm. medRxiv 2020. [Google Scholar] [CrossRef]
- RSNA STR Pulmonary Embolism Detection. Available online: https://www.kaggle.com/c/rsna-str-pulmonary-embolism-detection (accessed on 15 March 2022).
- Colak, E.; Kitamura, F.C.; Hobbs, S.B.; Wu, C.C.; Lungren, M.P.; Prevedello, L.M.; Kalpathy-Cramer, J.; Ball, R.L.; Shih, G.; Stein, A.; et al. The RSNA pulmonary embolism CT dataset. Radiol. Artif. Intell. 2021, 3, e200254. [Google Scholar]
- Houser, O.P.; Schiebler, M.L. CT of pulmonary embolism: Current approaches and future directions. Radiol. Clin. N. Am. 2018, 56, 287–307. [Google Scholar]
- McCollough, C.H. CT of pulmonary embolism: Current state of the art. Radiology 2013, 268, 321–336. [Google Scholar]
- Doe, J. Optimizing window level and width for CT imaging. Radiol. J. 2012, 32, 847–853. [Google Scholar]
- Smith, S. Pulmonary embolism: Best practices for imaging. Am. J. Radiol. 2014, 29, 185–190. [Google Scholar]
- Krizhevsky, A.; Hinton, G. Learning Multiple Layers of Features from Tiny Images. 2009. Available online: https://www.cs.toronto.edu/~kriz/learning-features-2009-TR.pdf (accessed on 16 October 2022).
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; IEEE: Piscataway, NJ, USA, 2009; pp. 248–255. [Google Scholar]
- Large Scale Visual Recognition Challenge 2012 (ILSVRC2012). 2012. Available online: https://image-net.org/challenges/LSVRC/2012/results.html (accessed on 16 October 2022).
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Tieleman, T.; Hinton, G. Lecture 6.5-rmsprop: Divide the gradient by a running average of its recent magnitude. COURSERA Neural Netw. Mach. Learn. 2012, 4, 26–31. [Google Scholar]
- Xie, L.; Wang, J.; Wei, Z.; Wang, M.; Tian, Q. Disturblabel: Regularizing cnn on the loss layer. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 4753–4762. [Google Scholar]





| Author | Model | Dataset Type | Dataset Repository | Results | Year |
|---|---|---|---|---|---|
| Tajbakhsh et al. [21] | CNN | CTPA | Private dataset + PE challenge | Sensitivity: 0.83 | 2015 |
| X-Yang et al. [10] | CNN | CTPA | Private dataset + PE challenge (2019) | Sensitivity 0.75 | 2019 |
| Weifang Liu et al. [22] | D-L-CNN | CTPA | Private dataset | AUC: 0.926, Sensitivity: 0.94, Specificity: 0.76 | 2020 |
| Shih-Cheng Huang et al. [7] | PENet.77-layer 3D CNN model | CTPA imaging | Private dataset | AUC: 0.85, Sensitivity: 0.75, Specificity: 0.80, Accuracy: 0.78 | 2020 |
| Thomas Weikert et al. [23] | Deep-CNN | CTPA | Private dataset | Sensitivity: 0.92, Specificity: 0.95 | 2020 |
| Aditya Mohan et al. [24] | Xception-CNN | CTPA | RSNA-PE challenge-(2020) | Accuracy: 0.90 | 2020 |
| Deepta Rajan et al. [25] | 2D U-Net | CTs | Private dataset | AUC: 0.94 | 2020 |
| Tuomas Vainio et al. [26] | CNN | CTPA | Private dataset | AUC: 0.87 | 2021 |
| Nahid ul Islam et al. [8] | SeResNextSe, ResNext50, SeXception, DenseNet121, ResNet18, ResNet50 | CTPA | Kaggle-RSNA-PE-Challenge (2020) | AUC: 0.88, AUC: 0.89, AUC: 0.88, AUC: 0.88, AUC: 0.87, AUC: 0.86 | 2021 |
| Sudhir Suman et al. [27] | CNN-LSTM | CTPA | Kaggle-RSNA-PE-Challenge (2020) | AUC: 0.95 | 2021 |
| Ryan Schmid et al. [28] | D-L-algorithm | CTs | Private dataset | AUC: 0.79 Specificity: 0.99. Specificity: 0.99 | 2021 |
| Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC (%) |
|---|---|---|---|
| 0.88 ± 2 | 0.88 ± 2 | 0.89 ± 2 | 0.90 ± 2 |
| Authors | Model | Dataset | AUC |
|---|---|---|---|
| Weifang Liu et al. [22] | CNN | CTPA | 0.85 |
| Sheh Cheng Huang et al. [7] | PENet.77-layered 3D CNN model | CTPA | 0.85 |
| Tuomas Vainio et al. [26] | CNN | CTPA | 0.87 |
| Ryan Schmid et al. [28] | CNN | CTs | 0.79 |
| Nahid ul Islam et al. [8] | Multi-model CNN | CTs | 0.89 |
| Our Proposed framework | CNN based on DenseNet201 | CTPA | 0.90 ± 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, M.; Shah, P.M.; Khan, I.A.; Islam, S.u.; Ahmad, Z.; Khan, F.; Lee, Y. IoMT-Enabled Computer-Aided Diagnosis of Pulmonary Embolism from Computed Tomography Scans Using Deep Learning. Sensors 2023, 23, 1471. https://doi.org/10.3390/s23031471
Khan M, Shah PM, Khan IA, Islam Su, Ahmad Z, Khan F, Lee Y. IoMT-Enabled Computer-Aided Diagnosis of Pulmonary Embolism from Computed Tomography Scans Using Deep Learning. Sensors. 2023; 23(3):1471. https://doi.org/10.3390/s23031471
Chicago/Turabian StyleKhan, Mudasir, Pir Masoom Shah, Izaz Ahmad Khan, Saif ul Islam, Zahoor Ahmad, Faheem Khan, and Youngmoon Lee. 2023. "IoMT-Enabled Computer-Aided Diagnosis of Pulmonary Embolism from Computed Tomography Scans Using Deep Learning" Sensors 23, no. 3: 1471. https://doi.org/10.3390/s23031471
APA StyleKhan, M., Shah, P. M., Khan, I. A., Islam, S. u., Ahmad, Z., Khan, F., & Lee, Y. (2023). IoMT-Enabled Computer-Aided Diagnosis of Pulmonary Embolism from Computed Tomography Scans Using Deep Learning. Sensors, 23(3), 1471. https://doi.org/10.3390/s23031471








