Diversity Learning Based on Multi-Latent Space for Medical Image Visual Question Generation
Abstract
1. Introduction
- We propose a novel medical VQG model based on diversity learning with a multi-latent space to solve the conventional informativeness problem of the VQG task and the multi-latent space is trained by the newly introduced MIC loss.
- We introduce semantic similarity as an evaluation metric of the medical VQG model to address the limitations of previous evaluation metrics.
2. Related Works
2.1. Medical Visual Question Answering
2.2. Medical Visual Question Generation
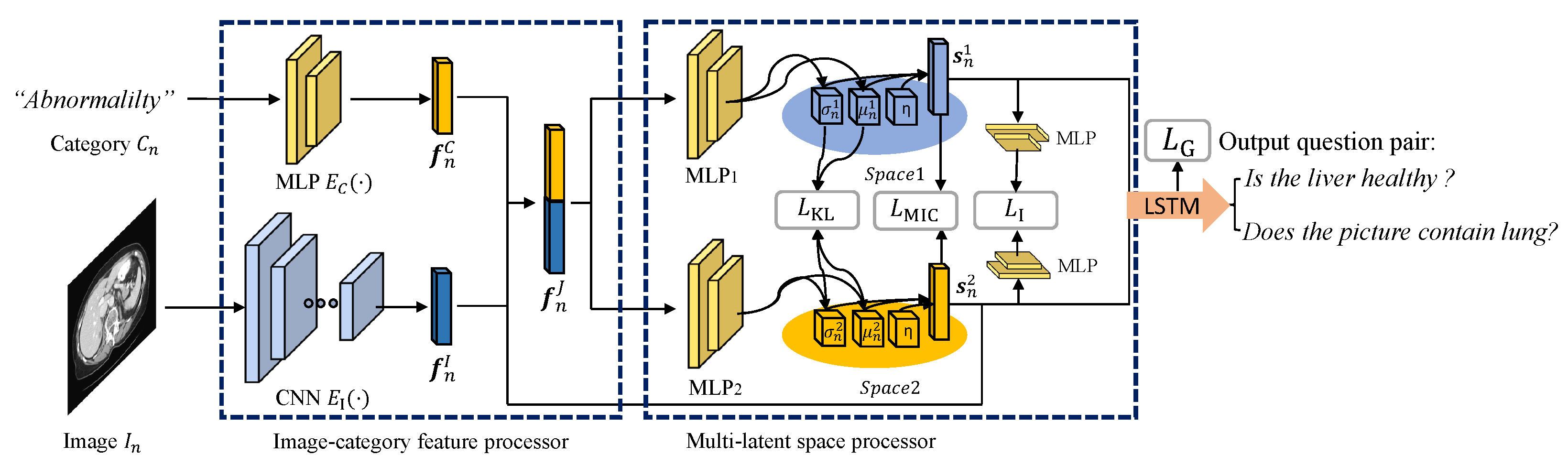
3. Model Framework
3.1. Image-Category Feature Processor
3.2. Multi-Latent Space Processor
3.3. Question Set Generation Based on LSTM
3.4. Total Loss Function
4. Experiments
4.1. Conditions
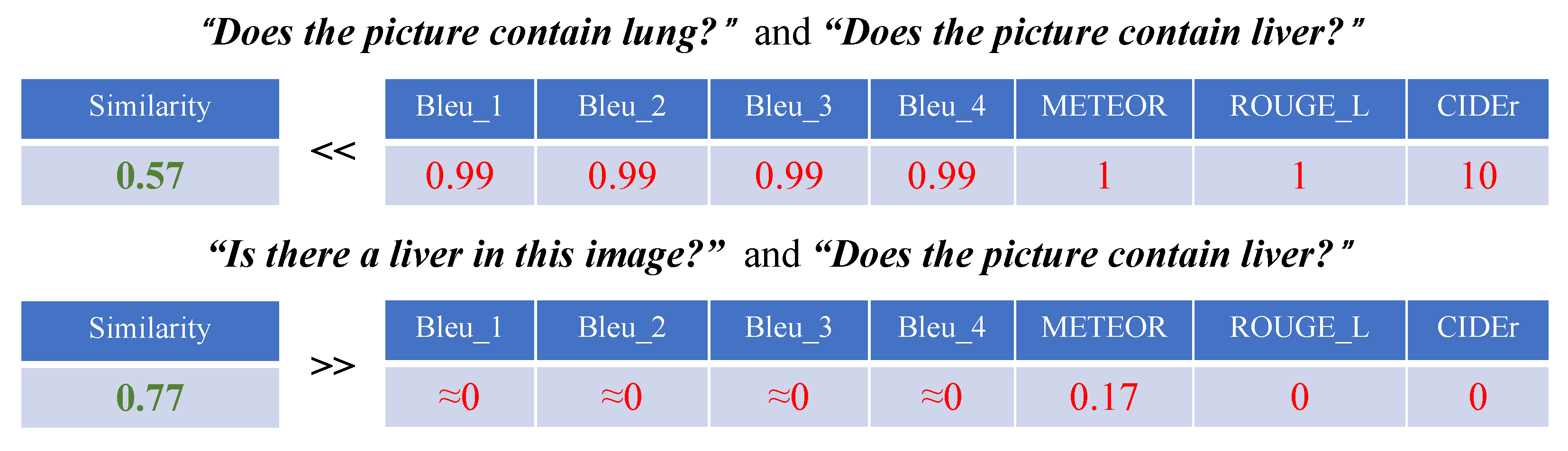
4.2. Evaluation Metrics
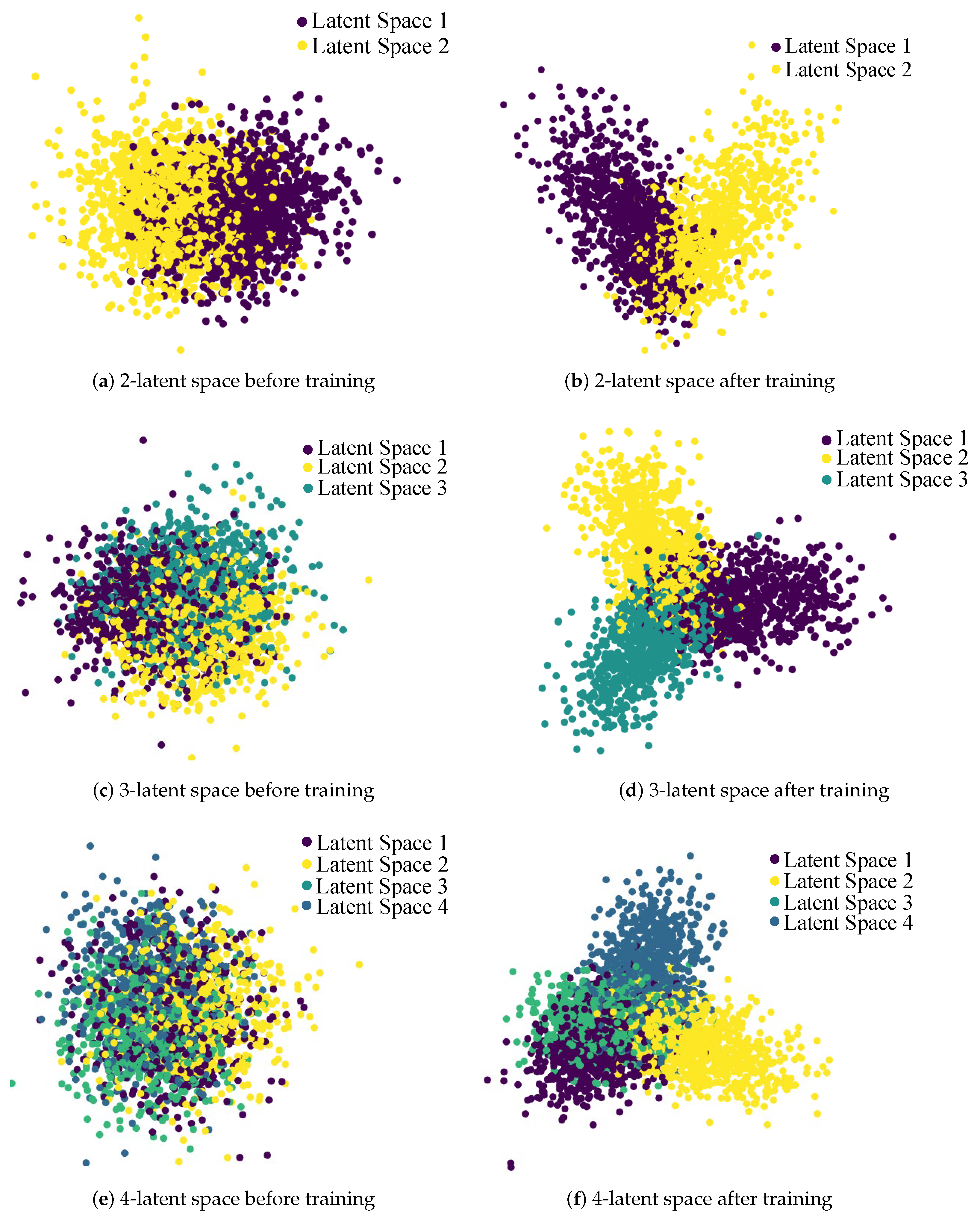
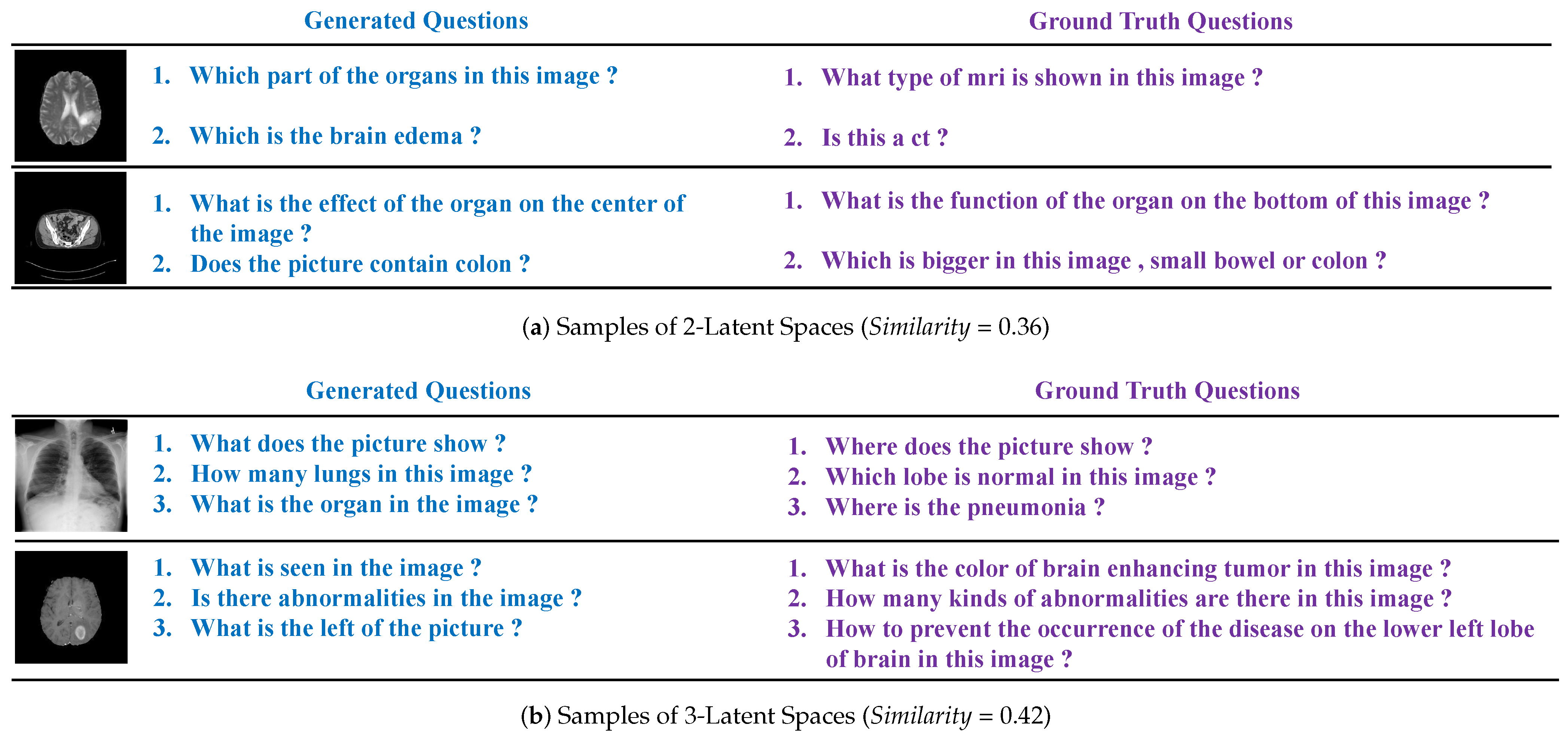
4.3. Evaluation Results
5. Discussion
5.1. Limitations
5.2. Future Work
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Architecture | Depth | Output Size | ||
|---|---|---|---|---|
| S0 | Conv (7 × 7, 64), stride 2 | 1 | 112 × 112 | |
| S1 | Max pooling (3 × 3), stride 2 | 1 | 56 × 56 | |
| Conv (1 × 1, 64), BN, ReLU Conv (3 × 3, 64), BN, ReLU Conv (1 × 1, 256), BN shortcut, ReLU | 3 | |||
| Encoder | S2 | Conv (1 × 1, 128), BN, ReLU Conv (3 × 3, 128), BN, ReLU Conv (1 × 1, 512), BN shortcut, ReLU | 4 | 28 × 28 |
| S3 | Conv (1 × 1, 256), BN, ReLU Conv (3 × 3, 256), BN, ReLU Conv (1 × 1, 1024), BN shortcut, ReLU | 6 | 14 × 14 | |
| S4 | Conv (1 × 1, 512), BN, ReLU Conv (3 × 3, 512), BN, ReLU Conv (1 × 1, 2048), BN shortcut, ReLU | 3 | 7 × 7 |
| Parameters | Value |
|---|---|
| Number of features in hidden state | 256 |
| Number of recurrent layers | 3 |
| Dropout probability applied to the model | 0.3 |
| Dropout probability applied to the inputs of the model | 0.3 |
| Maximum sequence length for outputs | 20 |
| Maximum sequence length for inputs | 20 |
References
- Wu, Q.; Teney, D.; Wang, P.; Shen, C.; Dick, A.; van den Hengel, A. Visual question answering: A survey of methods and datasets. Comput. Vis. Image Underst. 2017, 163, 21–40. [Google Scholar] [CrossRef]
- Antol, S.; Agrawal, A.; Lu, J.; Mitchell, M.; Batra, D.; Zitnick, C.L.; Parikh, D. VQA: Visual question answering. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 2425–2433. [Google Scholar] [CrossRef]
- Mostafazadeh, N.; Misra, I.; Devlin, J.; Mitchell, M.; He, X.; Vanderwende, L. Generating Natural Questions About an Image. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Berlin, Germany, 7–12 August 2016. [Google Scholar] [CrossRef]
- Sarrouti, M.; El Alaoui, S.O. A passage retrieval method based on probabilistic information retrieval model and UMLS concepts in biomedical question answering. J. Biomed. Inform. 2017, 68, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Kim, J.; Kim, G. A Joint Sequence Fusion Model for Video Question Answering and Retrieval. In Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018. [Google Scholar] [CrossRef]
- Yanagi, R.; Togo, R.; Ogawa, T.; Haseyama, M. Database-adaptive Re-ranking for Enhancing Cross-modal Image Retrieval. In Proceedings of the 29th ACM International Conference on Multimedia, Virtual Event, China, 20–24 October 2021; pp. 3816–3825. [Google Scholar] [CrossRef]
- McDonald, R.J.; Schwartz, K.M.; Eckel, L.J.; Diehn, F.E.; Hunt, C.H.; Bartholmai, B.J.; Erickson, B.J.; Kallmes, D.F. The effects of changes in utilization and technological advancements of cross-sectional imaging on radiologist workload. Acad. Radiol. 2015, 22, 1191–1198. [Google Scholar] [CrossRef] [PubMed]
- Takada, S.; Togo, R.; Ogawa, T.; Haseyama, M. Estimation Of Visual Contents Based On Question Answering From Human Brain Activity. In Proceedings of the 2020 IEEE International Conference on Image Processing, Abu Dhabi, United Arab Emirates, 25–28 October 2020; pp. 61–65. [Google Scholar] [CrossRef]
- Cai, L.; Gao, J.; Zhao, D. A review of the application of deep learning in medical image classification and segmentation. Ann. Transl. Med. 2020, 8, 713. [Google Scholar] [CrossRef]
- Masood, S.; Sharif, M.; Masood, A.; Yasmin, M.; Raza, M. A survey on medical image segmentation. Curr. Med. Imaging 2015, 11, 3–14. [Google Scholar] [CrossRef]
- Miranda, E.; Aryuni, M.; Irwansyah, E. A survey of medical image classification techniques. In Proceedings of the 2016 International Conference on Information Management and Technology, Bandung, Indonesia, 16–18 November 2016; pp. 56–61. [Google Scholar] [CrossRef]
- Wei, W.; Yang, D.; Li, L.; Xia, Y. An Intravascular Catheter Bending Recognition Method for Interventional Surgical Robots. Machines 2022, 10, 42. [Google Scholar] [CrossRef]
- Patil, C.; Patwardhan, M. Visual Question Generation: The State of the Art. ACM Comput. Surv. 2020, 53, 22. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations 2015, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Yang, Y.; Li, Y.; Fermüller, C.; Aloimonos, Y. Neural Self Talk: Image Understanding via Continuous Questioning and Answering. arXiv 2015, arXiv:1512.03460. [Google Scholar]
- Zaremba, W.; Sutskever, I.; Vinyals, O. Recurrent neural network regularization. arXiv 2014, arXiv:1409.2329. [Google Scholar]
- Geman, D.; Geman, S.; Hallonquist, N.; Younes, L. Visual turing test for computer vision systems. Proc. Natl. Acad. Sci. USA 2015, 112, 3618–3623. [Google Scholar] [CrossRef] [PubMed]
- Sarrouti, M.; Ben Abacha, A.; Demner-Fushman, D. Visual Question Generation from Radiology Images. In Proceedings of the First Workshop on Advances in Language and Vision Research, Online, 9 July 2020; pp. 12–18. [Google Scholar] [CrossRef]
- Kingma, D.P.; Welling, M. Auto-encoding variational bayes. arXiv 2013, arXiv:1312.6114. [Google Scholar]
- Ben Abacha, A.; Datla, V.V.; Hasan, S.A.; Demner-Fushman, D.; Müller, H. Overview of the VQA-Med task at ImageCLEF 2020. In Proceedings of the CLEF 2020-Conference and Labs of the Evaluation Forum, Thessaloniki, Greece, 22–25 September 2020. [Google Scholar]
- Ionescu, B.; Müller, H.; Péteri, R.; Abacha, A.B.; Sarrouti, M.; Demner-Fushman, D.; Hasan, S.A.; Kozlovski, S.; Liauchuk, V.; Cid, Y.D.; et al. Overview of the ImageCLEF 2021: Multimedia retrieval in medical, nature, internet and social media applications. In Proceedings of the International Conference of the Cross-Language Evaluation Forum for European Languages, Bologna, Italy, 5–8 September 2021; pp. 345–370. [Google Scholar]
- Krishna, R.; Bernstein, M.; Fei-Fei, L. Information maximizing visual question generation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 2008–2018. [Google Scholar] [CrossRef]
- Papineni, K.; Roukos, S.; Ward, T.; Zhu, W.J. BLEU: A method for automatic evaluation of machine translation. In Proceedings of the 40th Annual Meeting of the Association for Computational Linguistics, Philadelphia, PA, USA, 7–12 July 2002; pp. 311–318. [Google Scholar] [CrossRef]
- Banerjee, S.; Lavie, A. METEOR: An automatic metric for MT evaluation with improved correlation with human judgments. In Proceedings of the ACL Workshop on Intrinsic and Extrinsic Evaluation Measures for Machine Translation and/or Summarization, Ann Arbor, MI, USA, 29–30 June 2005; pp. 65–72. [Google Scholar] [CrossRef]
- Ma, Z.; Zheng, W.; Chen, X.; Yin, L. Joint embedding VQA model based on dynamic word vector. PeerJ Comput. Sci. 2021, 7, e353. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; Volume 30. [Google Scholar]
- Andreas, J.; Rohrbach, M.; Darrell, T.; Klein, D. Neural module networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 39–48. [Google Scholar]
- Natarajan, A.; Motani, M.; de Silva, B.; Yap, K.K.; Chua, K.C. Investigating network architectures for body sensor networks. In Proceedings of the 1st ACM SIGMOBILE International Workshop on Systems and Networking Support for Healthcare and Assisted Living Environments, New York, NY, USA, 11 June 2007; pp. 19–24. [Google Scholar] [CrossRef]
- Jain, U.; Zhang, Z.; Schwing, A.G. Creativity: Generating diverse questions using variational autoencoders. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 6485–6494. [Google Scholar] [CrossRef]
- Ren, M.; Kiros, R.; Zemel, R. Exploring models and data for image question answering. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 7–12 December 2015; Volume 28. [Google Scholar]
- Hudson, D.A.; Manning, C.D. GQA: A new dataset for real-world visual reasoning and compositional question answering. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 6700–6709. [Google Scholar] [CrossRef]
- Chen, D.; Manning, C.D. A fast and accurate dependency parser using neural networks. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing, Doha, Qatar, 25–29 October 2014; pp. 740–750. [Google Scholar] [CrossRef]
- Uppal, S.; Madan, A.; Bhagat, S.; Yu, Y.; Shah, R.R. C3VQG: Category consistent cyclic visual question generation. In Proceedings of the 2nd ACM International Conference on Multimedia in Asia, Singapore, 7–9 March 2021; pp. 1–7. [Google Scholar] [CrossRef]
- Al-Sadi, A.; Hana’Al-Theiabat, A.A.M.; Al-Ayyoub, M. The Inception Team at VQA-Med 2020: Pretrained VGG with Data Augmentation for Medical VQA and VQG. In Proceedings of the CLEF (Working Notes), Thessaloniki, Greece, 22–25 September 2020. [Google Scholar]
- Sarrouti, M. NLM at VQA-Med 2020: Visual Question Answering and Generation in the Medical Domain. In Proceedings of the CLEF (Working Notes), Thessaloniki, Greece, 22–25 September 2020. [Google Scholar]
- Eslami, S.; de Melo, G.; Meinel, C. TeamS at VQA-Med 2021: BBN-Orchestra for Long-tailed Medical Visual Question Answering. In Proceedings of the CLEF (Working Notes), Bucharest, Romania, 21–24 September 2021; pp. 1211–1217. [Google Scholar]
- Lau, J.J.; Gayen, S.; Abacha, A.B.; Demner-Fushman, D. A dataset of clinically generated visual questions and answers about radiology images. Sci. Data 2018, 5, 1–10. [Google Scholar] [CrossRef]
- Liu, B.; Zhan, L.M.; Xu, L.; Ma, L.; Yang, Y.; Wu, X.M. SLAKE: A Semantically-Labeled Knowledge-Enhanced Dataset for Medical Visual Question Answering. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging, Nice, France, 13–16 April 2021; pp. 1650–1654. [Google Scholar] [CrossRef]
- Simpson, A.L.; Antonelli, M.; Bakas, S.; Bilello, M.; Farahani, K.; Van Ginneken, B.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B.; et al. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv 2019, arXiv:1902.09063. [Google Scholar]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-Ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2097–2106. [Google Scholar] [CrossRef]
- Kavur, A.E.; Gezer, N.S.; Barış, M.; Aslan, S.; Conze, P.H.; Groza, V.; Pham, D.D.; Chatterjee, S.; Ernst, P.; Özkan, S.; et al. CHAOS challenge-combined (CT-MR) healthy abdominal organ segmentation. Med. Image Anal. 2021, 69, 101950. [Google Scholar] [CrossRef]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006, 31, 1116–1128. [Google Scholar] [CrossRef]
- Cer, D.; Yang, Y.; Kong, S.y.; Hua, N.; Limtiaco, N.; John, R.S.; Constant, N.; Guajardo-Cespedes, M.; Yuan, S.; Tar, C.; et al. Universal sentence encoder for English. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing: System Demonstrations, Brussels, Belgium, 31 October–4 November 2018; pp. 169–174. [Google Scholar] [CrossRef]
- Lin, C.Y.; Hovy, E. Automatic evaluation of summaries using n-gram co-occurrence statistics. In Proceedings of the 2003 Human Language Technology Conference of the North American Chapter of the Association for Computational Linguistics, Edmonton, AB, Canada, 27 May–1 June 2003; pp. 150–157. [Google Scholar] [CrossRef]
- Chen, X.; Fang, H.; Lin, T.Y.; Vedantam, R.; Gupta, S.; Dollár, P.; Zitnick, C.L. Microsoft COCO captions: Data collection and evaluation server. arXiv 2015, arXiv:1504.00325. [Google Scholar]
- Vedantam, R.; Lawrence Zitnick, C.; Parikh, D. CIDEr: Consensus-based image description evaluation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 4566–4575. [Google Scholar] [CrossRef]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Radford, A.; Kim, J.W.; Hallacy, C.; Ramesh, A.; Goh, G.; Agarwal, S.; Sutskever, I. Learning transferable visual models from natural language supervision. In Proceedings of the International Conference on Machine Learning, Online, 18–24 July 2021; pp. 8748–8763. [Google Scholar]
- Valanarasu, J.M.J.; Oza, P.; Hacihaliloglu, I.; Patel, V.M. Medical transformer: Gated axial-attention for medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Virtual Event, 27 September–1 October 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 36–46. [Google Scholar]
- Gu, Y.; Tinn, R.; Cheng, H.; Lucas, M.; Usuyama, N.; Liu, X.; Naumann, T.; Gao, J.; Poon, H. Domain-specific language model pretraining for biomedical natural language processing. ACM Trans. Comput. Healthc. 2021, 3, 1–23. [Google Scholar] [CrossRef]
- Marino, K.; Rastegari, M.; Farhadi, A.; Mottaghi, R. Ok-vqa: A visual question answering benchmark requiring external knowledge. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 3195–3204. [Google Scholar]
- Zheng, W.; Yin, L.; Chen, X.; Ma, Z.; Liu, S.; Yang, B. Knowledge base graph embedding module design for Visual question answering model. Pattern Recognit. 2021, 120, 108153. [Google Scholar] [CrossRef]
- Zheng, W.; Liu, X.; Ni, X.; Yin, L.; Yang, B. Improving Visual Reasoning Through Semantic Representation. IEEE Access 2021, 9, 91476–91486. [Google Scholar] [CrossRef]
- Becker, G.S.; Lovas, R. Uniformity Correction of CMOS Image Sensor Modules for Machine Vision Cameras. Sensors 2022, 22, 9733. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, C.; Zhang, W.; Fang, C.; Xia, Y.; Liu, Y.; Dong, H. Object-Based Reliable Visual Navigation for Mobile Robot. Sensors 2022, 22, 2387. [Google Scholar] [CrossRef]



| Category | Question Number |
|---|---|
| Abnormality | 312 |
| Modality | 290 |
| Organ | 290 |
| Plane | 484 |
| Position | 742 |
| Others | 1772 |
| Language Modeling | Diversity | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| BLEU-1 | BLEU-2 | BLEU-3 | BLEU-4 | METEOR | ROUGE-L | CIDEr | Inventiveness (↑) | Similarity (↓) | |
| CNN+RNN | 38.3 | 22.2 | 14.6 | 10.4 | 15.4 | 37.8 | 55.4 | 0.10 | 0.61 |
| VQGR [21] | 40.3 | 23.8 | 15.7 | 11.3 | 15.9 | 39.7 | 64.1 | 31.1 | 0.65 |
| C3VQG [36] | 24.2 | 9.4 | 3.8 | 2.2 | 8.0 | 30.7 | 14.6 | 20.1 | 0.41 |
| Krishna [25] | 42.1 | 25.9 | 18.3 | 13.7 | 17.7 | 41.2 | 83.1 | 17.0 | 0.48 |
| Our model-2 | 46.2 | 29.9 | 20.0 | 13.8 | 18.1 | 45.6 | 61.8 | 32.5 | 0.37 |
| Our model-3 | 48.7 | 33.0 | 23.2 | 16.0 | 19.6 | 50.0 | 78.6 | 33.0 | 0.43 |
| Our model-4 | 46.5 | 31.6 | 23.2 | 16.8 | 18.3 | 46.7 | 73.0 | 33.9 | 0.35 |
| BLEU-1 | BLEU-2 | BLEU-3 | BLEU-4 | METEOR | ROUGE-L | CIDEr | |
|---|---|---|---|---|---|---|---|
| Our model-2 w M | 46.2 | 29.9 | 20.0 | 13.8 | 18.1 | 45.6 | 61.8 |
| Our model-2 w/o M | 42.6 | 26.4 | 16.3 | 11.2 | 16.5 | 43.1 | 44.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, H.; Togo, R.; Ogawa, T.; Haseyama, M. Diversity Learning Based on Multi-Latent Space for Medical Image Visual Question Generation. Sensors 2023, 23, 1057. https://doi.org/10.3390/s23031057
Zhu H, Togo R, Ogawa T, Haseyama M. Diversity Learning Based on Multi-Latent Space for Medical Image Visual Question Generation. Sensors. 2023; 23(3):1057. https://doi.org/10.3390/s23031057
Chicago/Turabian StyleZhu, He, Ren Togo, Takahiro Ogawa, and Miki Haseyama. 2023. "Diversity Learning Based on Multi-Latent Space for Medical Image Visual Question Generation" Sensors 23, no. 3: 1057. https://doi.org/10.3390/s23031057
APA StyleZhu, H., Togo, R., Ogawa, T., & Haseyama, M. (2023). Diversity Learning Based on Multi-Latent Space for Medical Image Visual Question Generation. Sensors, 23(3), 1057. https://doi.org/10.3390/s23031057






