Rapid Detection of Total Viable Count in Intact Beef Dishes Based on NIR Hyperspectral Hybrid Model
Abstract
:1. Introduction
- Comparison of CS and FS prediction model performance based on NIR-HIS and SVM algorithm.
- Selection of optimal standard sample number.
- Taking advantage of the DS algorithm to perform MT between CS and FS prediction models.
- Performing hybrid modeling of the BS model and the posttransfer model to finish the prediction of the total number of bacteria in stewed beef samples.
2. Materials and Methods
2.1. Experimental Materials and Reagents
2.1.1. Preparation of Experimental Materials
2.1.2. The Main Reagent of the Experiment
2.2. Experimental Method
2.2.1. Determination of Total Bacterial Colony
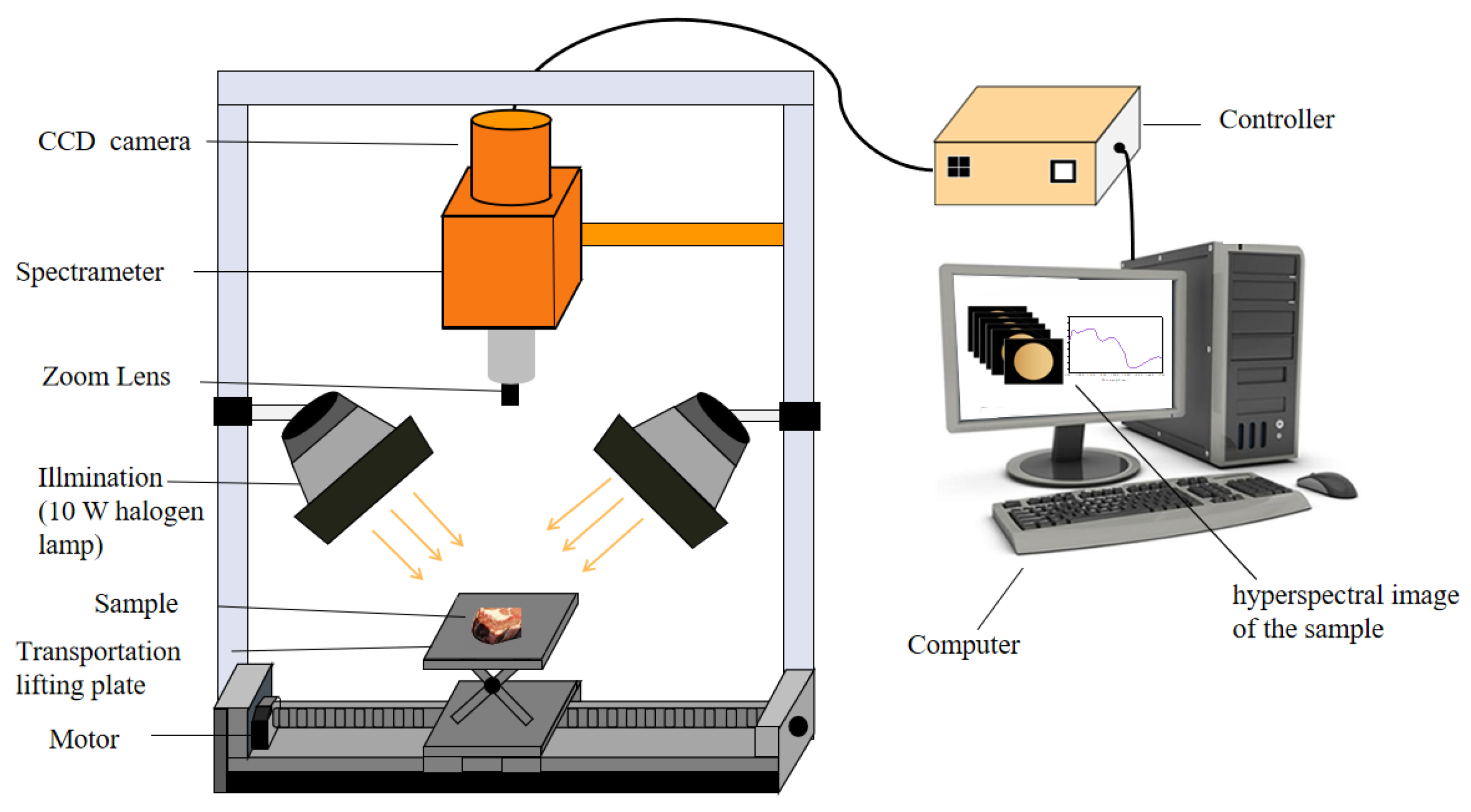
2.2.2. Near-Infrared Hyperspectral Image System
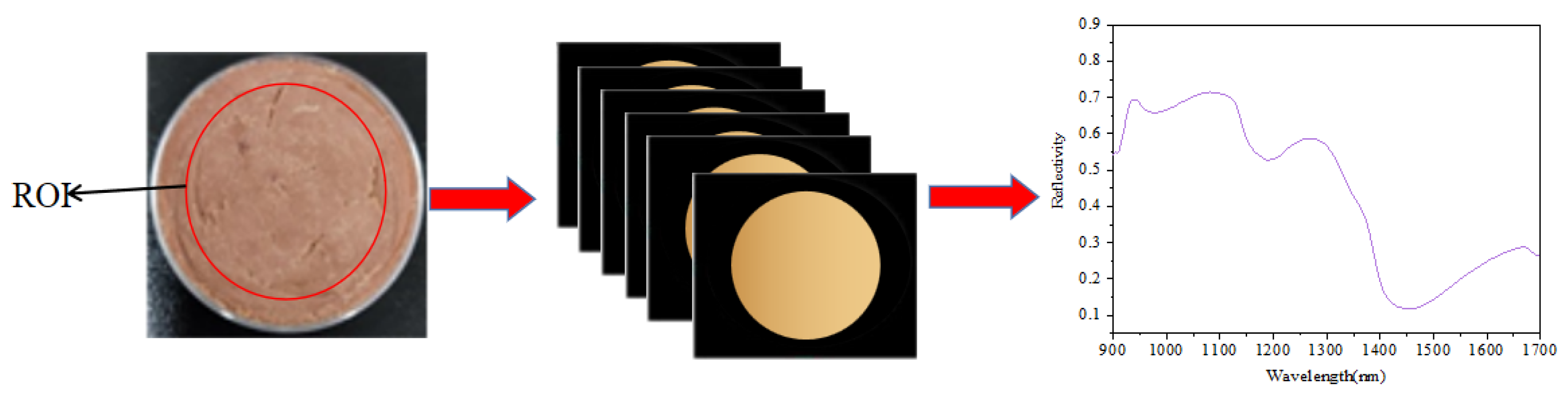
2.2.3. Hyperspectral Image Acquisition and Spectra Extraction
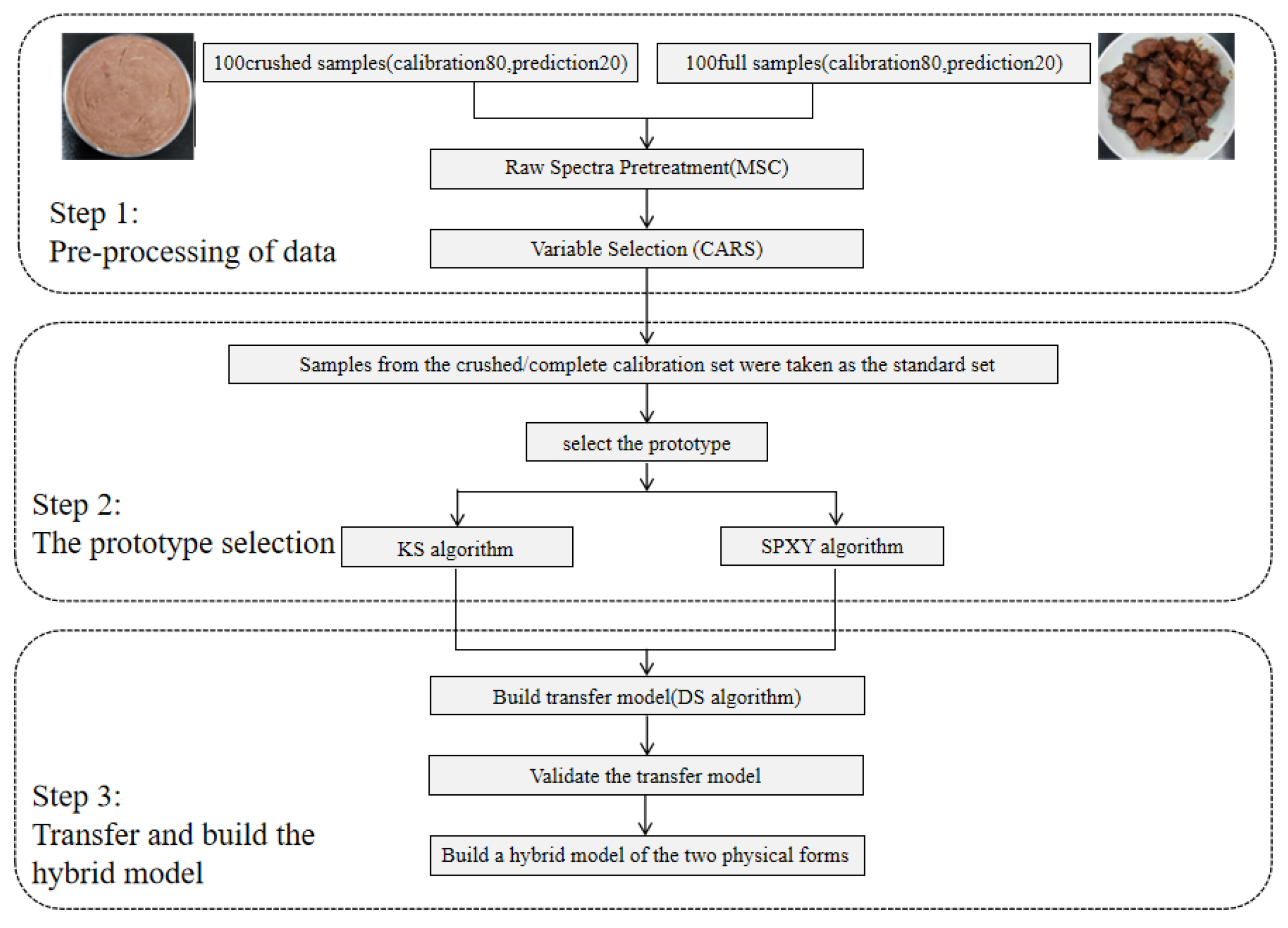
2.3. Date Processing
2.3.1. Preprocessing Method of Original Spectral Data and Model Establishment
2.3.2. Model Performance Evaluation
2.3.3. Model Transfer
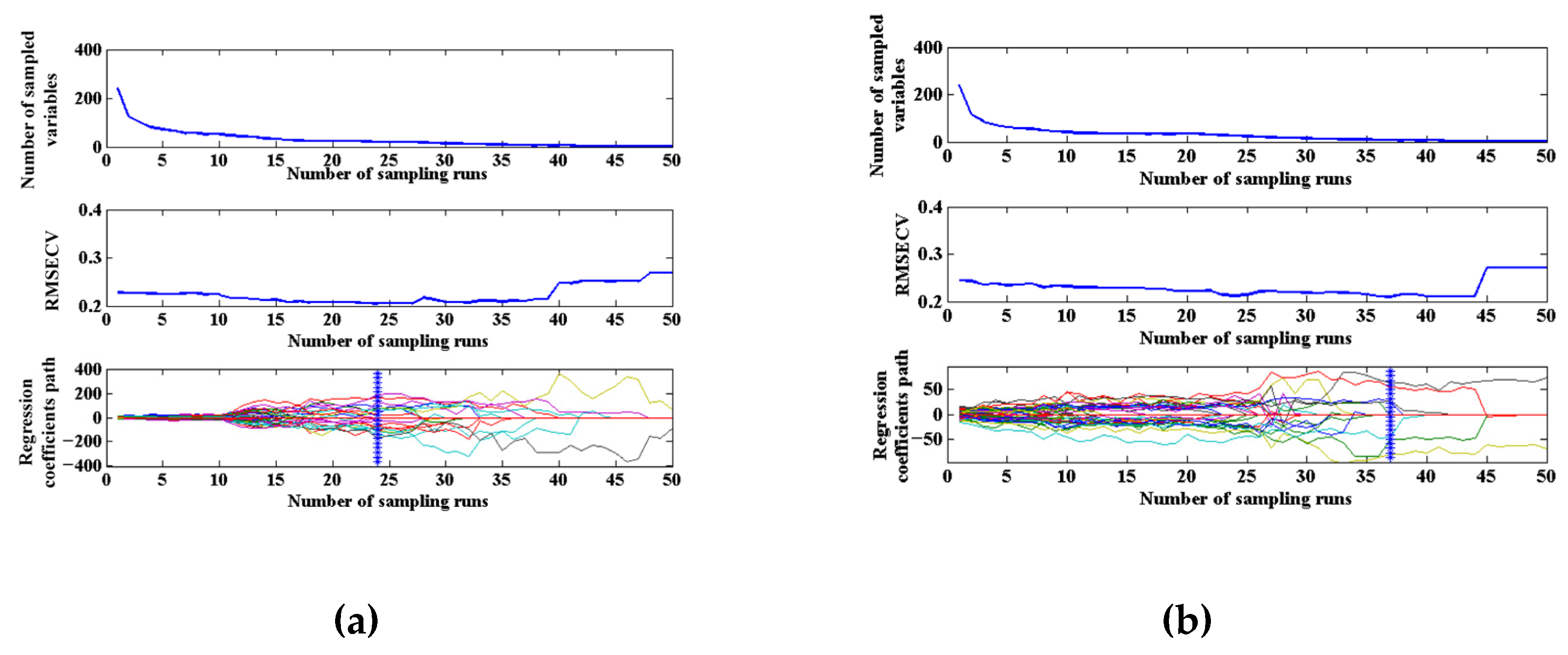
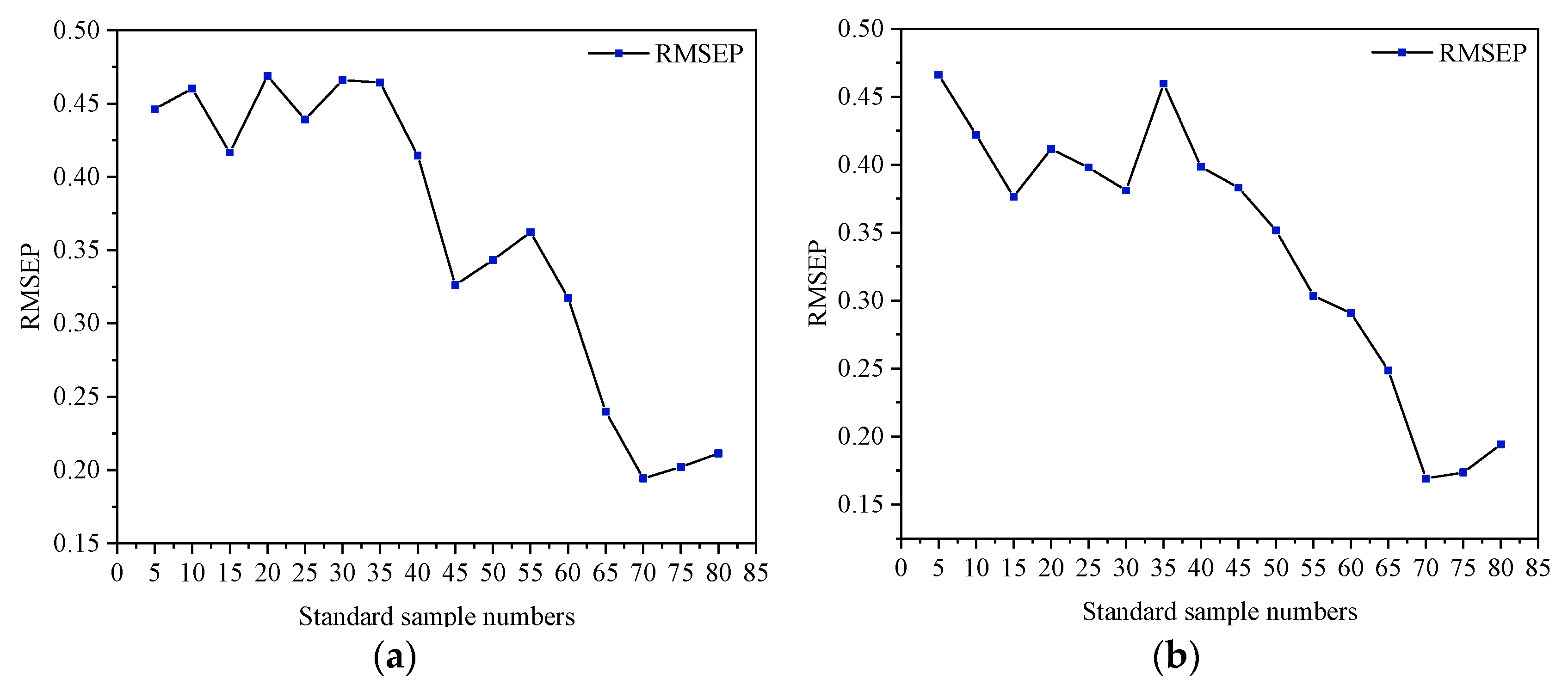
2.3.4. Selection of the Transfer Samples
2.3.5. The Establishment of a Hybrid Model
3. Results
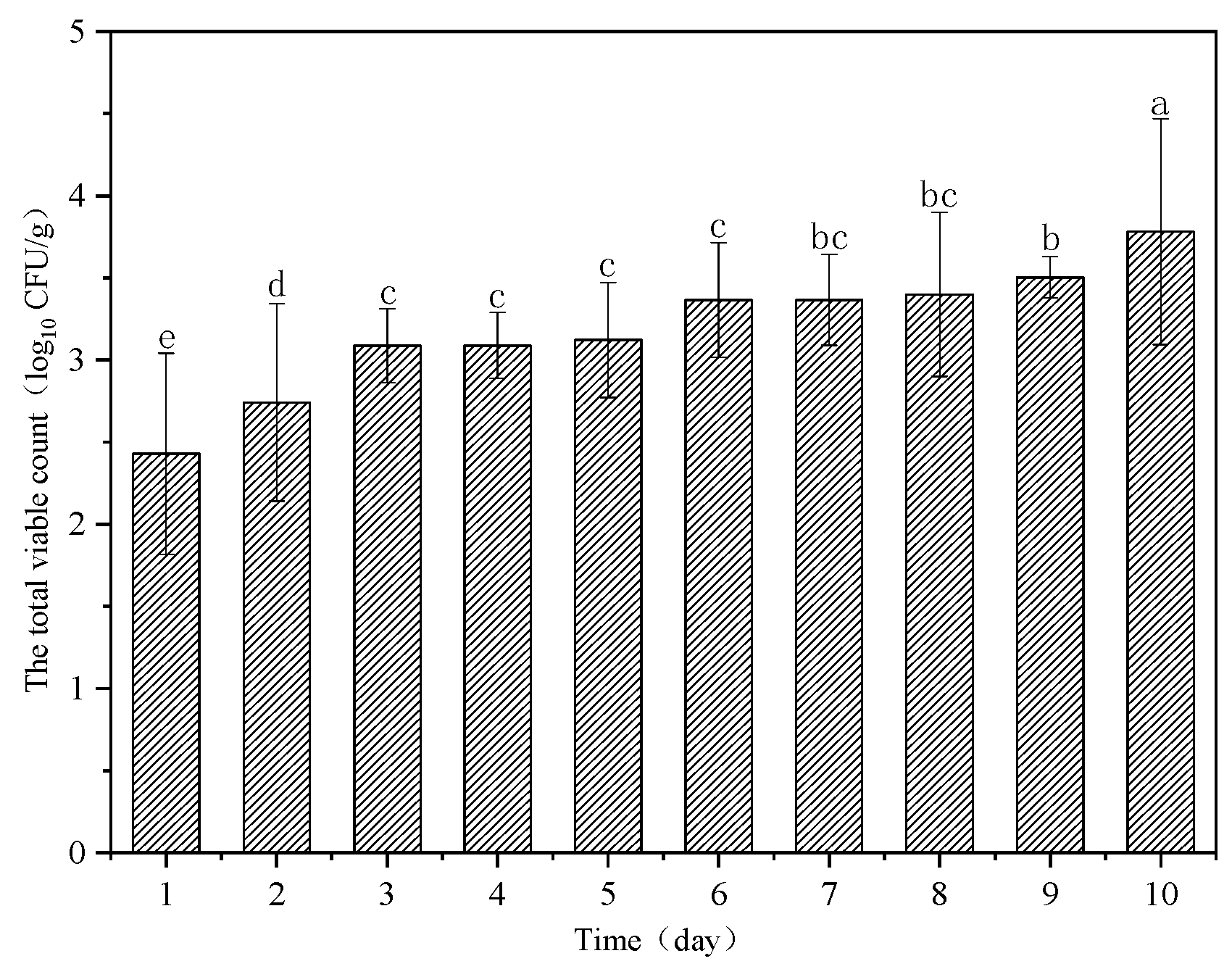
3.1. Results of Microbiological Analyses
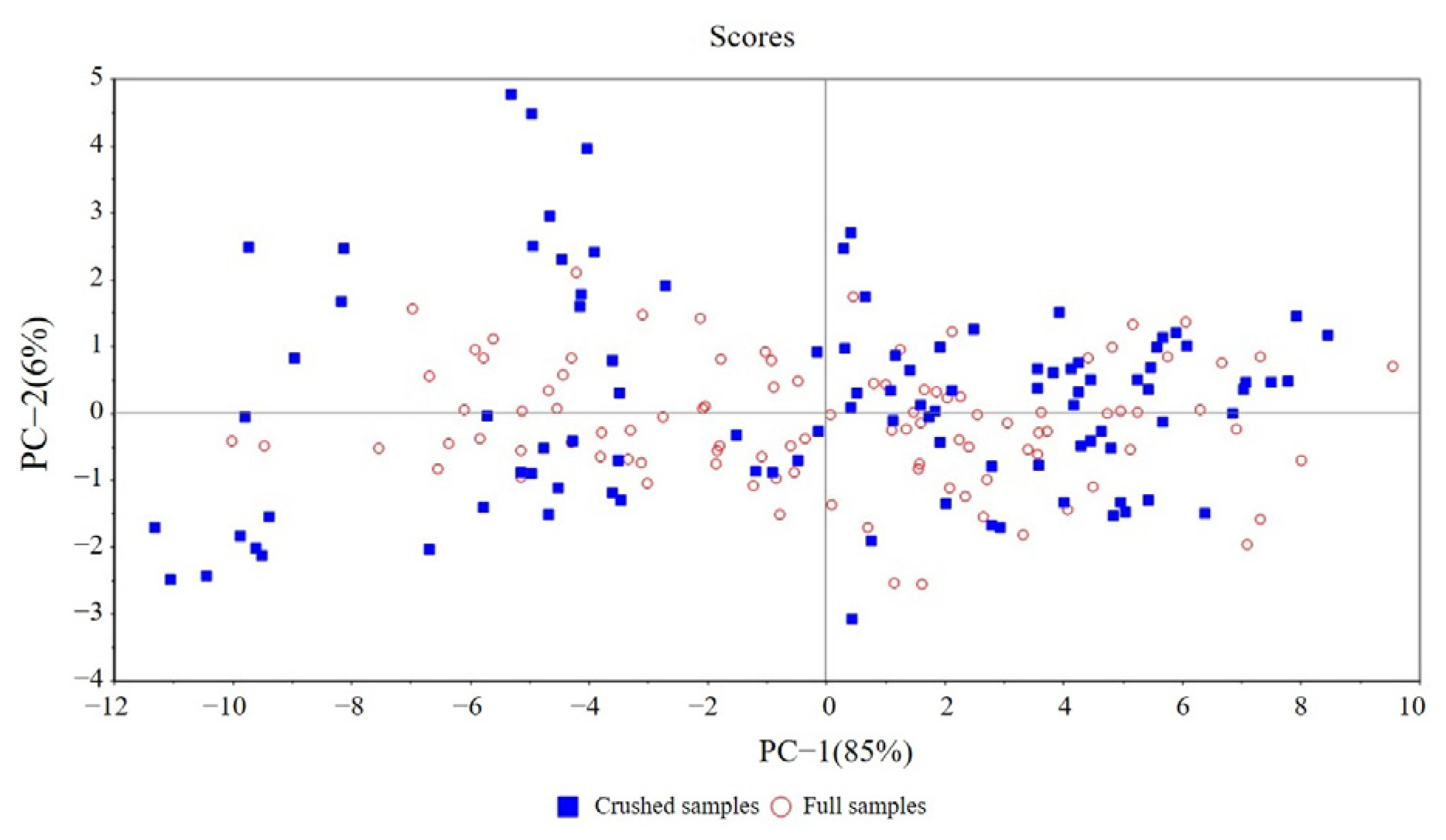
3.2. Difference Analysis of Spectral and Modeling Results
3.3. Model Transfer Analysis
3.3.1. The New Set of Variables and the Predicted Results for Two Physical Forms
3.3.2. Selection of Optimal Standard Sample Number and Establishment of Transfer Model
3.3.3. The Establishment of the Hybrid Model
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Huang, L.; Zhao, J.; Chen, Q.; Zhang, Y. Rapid detection of total viable count (TVC) in pork meat by hyperspectral imaging. Food Res. Int. 2013, 54, 821–828. [Google Scholar] [CrossRef]
- Wen, X.; Zhang, D.; Li, X.; Ding, T.; Liang, C.; Zheng, X.; Yang, W.; Hou, C. Dynamic changes of bacteria and screening of potential spoilage markers of lamb in aerobic and vacuum packaging. Food Microbiol. 2022, 104, 103996. [Google Scholar] [CrossRef] [PubMed]
- Fegan, N.; Jenson, I. The role of meat in foodborne disease: Is there a coming revolution in risk assessment and management? Meat Sci. 2018, 144, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Barbin, D.F.; Elmasry, G.; Sun, D.; Allen, P.; Morsy, N. Non-destructive assessment of microbial contamination in porcine meat using NIR hyperspectral imaging. Innov. Food Sci. Emerg. 2013, 17, 180–191. [Google Scholar] [CrossRef]
- Cao, K.; Wei, W.S.; Xing, S.J.; Ai, X.; Zhao, Z.L.; Zhang, C.J. Determination of the total viable count of Chinese meat dishes by near-infrared spectroscopy: A predictive model. J. Food Process. Preserv. 2021, 45, e16081. [Google Scholar] [CrossRef]
- Ellis, D.I.; Broadhurst, D.; Goodacre, R. Rapid and quantitative detection of the microbial spoilage of beef by fourier transform infrared spectroscopy and machine learning. Anal. Chim. Acta. 2004, 514, 193–201. [Google Scholar] [CrossRef]
- He, H.J.; Sun, D.W.; Wu, D. Rapid and real-time prediction of lactic acid bacteria (LAB) in farmed salmon flesh using near-infrared (NIR) hyperspectral imaging combined with chemometric analysis. Food Res. Int. 2014, 62, 476–483. [Google Scholar] [CrossRef]
- Kodogiannis, V.S.; Pachidis, T.; Kontogianni, E. An intelligent based decision support system for the detection of meat spoilage. Eng. Appl. Artif. Intel. 2014, 34, 23–36. [Google Scholar] [CrossRef]
- Bin, J.; Ai, F.F.; Fan, W.; Zhou, J.H.; Yun, Y.H.; Liang, Y.Z. A modified random forest approach to improve multi-class classification performance of tobacco leaf grades coupled with NIR spectroscopy. RSC Adv. 2016, 6, 30353–30361. [Google Scholar] [CrossRef]
- Ni, L.J.; Zhang, L.G.; Xie, J.; Luo, J.Q. Pattern recognition of Chinese flue-cured tobaccos by an improved and simplified K-nearest neighbors classification algorithm on near infrared spectra. Anal. Chim. Acta 2009, 633, 43–50. [Google Scholar] [CrossRef]
- Zhang, S.; Tan, Z.; Liu, J.; Xu, Z.; Du, Z. Determination of the food dye indigotine in cream by near-infrared spectroscopy technology combined with random forest model. Spectrochim. Acta A 2020, 227, 117551. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, Y.; Zhong, G.; Yu, X. Simultaneous quantitative analysis of protein, carbohydrate and fat in nutritionally complete formulas of medical foods by near-infrared spectroscopy. Infrared Phys. Technol. 2018, 93, 124–129. [Google Scholar] [CrossRef]
- Missori, M.; Pulci, O.; Teodonio, L.; Violante, C.; Kupchak, I.; Bagniuk, J.; Bojewska, J.; Conte, A.M. Optical response of strongly absorbing inhomogeneous materials: Application to paper degradation. Phys. Rev. B 2014, 89, 520–531. [Google Scholar] [CrossRef]
- Blanco, M.; Cueva-Mestanza, R.; Peguero, A. NIR analysis of pharmaceutical samples without reference data: Improving the calibration. Talanta 2011, 85, 2218–2225. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.G.; Shi, C.H. Prediction of grain weight, brown rice weight and amylose content in single rice grains using near-infrared reflectance spectroscopy. Field Crop. Res. 2004, 87, 13–21. [Google Scholar] [CrossRef]
- Mishra, P.; Nikzad-Langerodi, R.; Marini, F.; Roger, J.M.; Biancolillo, A.; Rutledge, D.N.; Lohumi, S. Are standard samples measurements still needed to transfer multivariate calibration models between near-infrared spectrometers? The answer is not always. TrAC Trends Anal. Chem. 2021, 143, 116331. [Google Scholar] [CrossRef]
- Mishra, P.; Passos, D. Deep calibration transfer: Transferring deep learning models between infrared spectroscopy instruments. Infrared Phys. Technol. 2021, 117, 103863. [Google Scholar] [CrossRef]
- Ge, Y.; Morgan, C.L.S.; Grunwald, S.; Brown, D.J.; Sarkhot, D.V. Comparison of soil reflectance spectra and calibration models ob-tained using multiple spectrometers. Geoderma 2011, 161, 202–211. [Google Scholar] [CrossRef]
- Liu, Y.; Cai, W.S.; Shao, X.G. Standardization of near infrared spectra measured on multi-instrument. Anal. Chim. Acta 2014, 836, 18–23. [Google Scholar] [CrossRef]
- Wang, Y.; Veltkamp, D.J.; Kowalski, B.R. Multivariate instrument standardization. Anal. Chem. 1991, 63, 2750–2756. [Google Scholar] [CrossRef]
- Bouveresse, E.; Massart, D.L. Standardisation of near-infrared spectrometric instruments: A review. Vib. Spectrosc. 1996, 11, 3–15. [Google Scholar] [CrossRef]
- Sjöblom, J.; Svensson, O.; Josefson, M.; Kullberg, H.; Wold, S. An evaluation of orthogonal signal correction applied to calibration transfer of near infrared spectra. Chemometr. Intell. Lab. 1998, 44, 229–244. [Google Scholar] [CrossRef]
- Pereira, L.S.A.; Carneiro, M.F.; Botelho, B.G.; Sena, M.M. Calibration transfer from powder mixtures to intact tablets: A new use in pharmaceutical analysis for a known tool. Talanta 2016, 147, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Y.; Peng, Y.; Yan, S.; Zhao, X.; Han, D. Non-destructive and rapid detection of the internal chemical composition of granules samples by spectral transfer. Chemometr. Intell. Lab. 2021, 208, 104174. [Google Scholar] [CrossRef]
- Munnaf, M.A.; Mouazen, A.M. Spectra transfer based learning for predicting and classifying soil texture with short-ranged Vis-NIRS sensor. Soil Tillage Res. 2023, 225, 105545. [Google Scholar] [CrossRef]
- Feng, Y.Z.; Sun, D.W. Determination of total viable count (TVC) in chicken breast fillets by near-infrared hyperspectral imaging and spectroscopic transforms. Talanta 2013, 105, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Isaksson, T.; Naes, T. The Effect of Multiplicative Scatter Correction (MSC) and Linearity Improvement in NIR Spectroscopy. Appl. Spectrosc. 1988, 42, 1273–1284. [Google Scholar] [CrossRef]
- Li, L.S.; Jang, X.G.; Li, B.; Liu, Y.D. Wavelength selection method for near-infrared spectroscopy based on standard-samples calibration transfer of mango and apple. Comput. Electron. Agric. 2021, 190, 106448. [Google Scholar] [CrossRef]
- Shi, Y.; Li, J.; Chu, X. Progress and Applications of Multivariate Calibration Model Transfer Methods. Chin. J. Anal. Chem. 2019, 47, 479–487. [Google Scholar] [CrossRef]
- Ni, L.; Chen, H.; Hong, S.; Zhang, L.; Luan, S. Near infrared spectral calibration model transfer without standards by screening spectral points with scale invariant feature transform from master samples spectra. Spectrochim. Acta A 2021, 260, 119802. [Google Scholar] [CrossRef]
- Quelal-Vásconez, M.A.; Lerma-García, M.J.; Pérez-Esteve, É.; Arnau-Bonachera, A.; Barat, J.M.; Talens, P. Fast detection of cocoa shell in cocoa powders by near infrared spectroscopy and multivariate analysis. Food Control. 2019, 99, 68–72. [Google Scholar] [CrossRef]
- Dixit, Y.; Al-Sarayreh, M.; Craigie, C.R.; Reis, M.M. A global calibration model for prediction of intramuscular fat and pH in red meat using hyperspectral imaging. Meat Sci. 2021, 181, 108405. [Google Scholar] [CrossRef]
- Huang, Y.; Du, G.; Ma, Y.; Zhou, J. Predicting heavy metals in dark sun-cured tobacco by near-infrared spectroscopy modeling based on the optimized variable selections. Ind. Crop. Prod. 2021, 172, 114003. [Google Scholar] [CrossRef]
- Chen, H.; Zhao, G.; Zhang, X.; Wang, R.; Chen, J. Improving estimation precision of soil organic matter content by removing effect of soil moisture from hyperspectra. Trans. Chin. Soc. Agric. Eng. 2014, 30, 91–100. [Google Scholar] [CrossRef]
- Rodrigues, R.R.T.; Rocha, J.T.C.; Oliveira, L.M.S.L.; Dias, J.C.M.; Müller, E.I.; Castro, E.V.R.; Filgueiras, P.R. Evaluation of calibration transfer methods using the ATR-FTIR technique to predict density of crude oil. Chemometr. Intell. Lab. 2017, 166, 7–13. [Google Scholar] [CrossRef]
- Feudale, R.N.; Woody, N.A.; Tan, H.; Myles, A.J.; Brown, S.D.; Ferré, J. Transfer of multivariate calibration models: A review. Chemometr. Intell. Lab. 2002, 64, 181–192. [Google Scholar] [CrossRef]
- Liang, C.; Yuan, H.; Zhao, Z.; Song, C.; Wang, J. A new multivariate calibration model transfer method of near-infrared spectral analysis. Chemometr. Intell. Lab. 2016, 153, 51–57. [Google Scholar] [CrossRef]
- Li, H.; Liang, Y.; Xu, Q.; Cao, D. Key wavelengths screening using competitive adaptive reweighted sampling method for multivariate calibration. Anal. Chim. Acta 2009, 648, 77–84. [Google Scholar] [CrossRef]
- Liu, C.; Chu, Z.; Weng, S.; Zhu, G.; Han, K.; Zhang, Z.; Huang, L.; Zhu, Z.; Zheng, S. Fusion of electronic nose and hyperspectral imaging for mutton freshness detection using input-modified convolution neural network. Food Chem. 2022, 385, 132651. [Google Scholar] [CrossRef]
- Xu, Z.P.; Zhang, P.F.; Wang, Q.; Fan, S.; Cheng, W.M.; Liu, J.; Wang, H.P.; Liu, B.M.; Wu, Y.J. Comparative study of different wavelength selection methods in the transfer of crop kernel qualitive near-infrared models. Infrared Phys. Technol. 2022, 123, 104120. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Elmasry, G.; Sun, D.; Allen, P. Non-destructive assessment of instrumental and sensory tenderness of lamb meat using NIR hyperspectral imaging. Food Chem. 2013, 141, 389–396. [Google Scholar] [CrossRef] [PubMed]



| Samples Type | Samples Size | The Maximum/ (log10 CFU/g) | The Minimum/ (log10 CFU/g) | The Average/ (log10 CFU/g) | Standard Deviation/ (log10 CFU/g) |
|---|---|---|---|---|---|
| Calibration set | 80 | 3.8808 | 2.1347 | 3.1140 | 0.2953 |
| Prediction set | 20 | 3.7559 | 2.1047 | 3.1490 | 0.3623 |
| Samples | Pretreatment Method | Variable Filtering Method | LVs | Calibration Set | Cross-Validation | Prediction Set | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RMSEC | RMSECV | RESEP | RPD | RER | |||||||
| CS | MSC | CARS | 10 | 0.6603 | 0.1887 | 0.6650 | 0.1954 | 0.6438 | 0.2037 | 2.78 | 8.11 |
| FS | MSC | CARS | 12 | 0.6128 | 0.2005 | 0.5091 | 0.2385 | 0.3453 | 0.2713 | 2.34 | 6.08 |
| Samples | Characteristic Variables (nm) |
|---|---|
| CS | 954, 977, 994, 1000, 1027, 1030, 1040, 1057, 1067, 1117, 1120, 1213, 1237, 1243, 1247, 1260, 1310, 1327, 1685 |
| FS | 1037, 1080, 1140, 1183, 1648, 1681, 1691 |
| Samples | Pretreatment Method | Variable Filtering Method | LVs | Calibration Set | Cross-Validation | Prediction Set | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RMSEC | RMSECV | RMSEP | RPD | RER | |||||||
| CS | MSC | CARS | 8 | 0.8424 | 0.1258 | 0.7946 | 0.1653 | 0.6514 | 0.2032 | 2.67 | 8.33 |
| FS | MSC | CARS | 11 | 0.5336 | 0.2093 | 0.5217 | 0.2281 | 0.5088 | 0.2454 | 2.37 | 6.90 |
| Method | SSN | RMSEC | RMSEP | RPD | RER | ||
|---|---|---|---|---|---|---|---|
| KS | 70 | 0.8424 | 0.1258 | 0.6312 | 0.1962 | 2.71 | 8.63 |
| SPXY | 70 | 0.8424 | 0.1258 | 0.8069 | 0.1691 | 2.98 | 10.01 |
| Methods | SSN | RMSEC | RMSEP | RPD | RER | ||
|---|---|---|---|---|---|---|---|
| KS | 60 | 0.8424 | 0.1258 | 0.6519 | 0.1946 | 2.72 | 8.70 |
| SPXY | 80 | 0.8424 | 0.1258 | 0.6553 | 0.1942 | 2.72 | 8.72 |
| Based | Added | RMSEC | RMSEP | RPD | RER | ||
|---|---|---|---|---|---|---|---|
| Spectra of 80 crushed samples | 5 | 0.8802 | 0.1164 | 0.3958 | 0.5985 | 0.55 | 2.83 |
| 10 | 0.9364 | 0.0909 | 0.0057 | 0.3591 | 0.96 | 4.71 | |
| 15 | 0.5740 | 0.2308 | 0.4593 | 0.2571 | 2.36 | 6.58 | |
| 20 | 0.5889 | 0.2256 | 0.5564 | 0.2181 | 2.59 | 7.75 | |
| 25 | 0.9036 | 0.1089 | 0.4446 | 0.2991 | 2.16 | 5.65 | |
| 30 | 0.8836 | 0.1200 | 0.4347 | 0.2598 | 2.31 | 6.51 | |
| 35 | 0.9174 | 0.0986 | 0.2521 | 0.2882 | 2.67 | 5.87 | |
| 40 | 0.8140 | 0.1483 | 0.2271 | 0.2873 | 2.15 | 5.89 | |
| 45 | 0.9095 | 0.1024 | 0.1495 | 0.3252 | 2.02 | 5.20 | |
| 50 | 0.8953 | 0.1077 | 0.0363 | 0.3728 | 0.87 | 4.54 | |
| 55 | 0.8332 | 0.1371 | 0.1349 | 0.3422 | 0.94 | 4.94 | |
| 60 | 0.8174 | 0.1405 | 0.0931 | 0.3461 | 0.91 | 4.89 | |
| 65 | 0.8234 | 0.1385 | 0.0671 | 0.3541 | 0.89 | 4.78 | |
| 70 | 0.5479 | 0.2103 | 0.6285 | 0.2145 | 2.45 | 7.88 | |
| 75 | 0.4640 | 0.2247 | 0.7408 | 0.1877 | 2.63 | 9.01 | |
| 80 | 0.5401 | 0.7349 | 0.6527 | 0.2109 | 2.45 | 8.02 |
| Based | Added | RMSEC | RMSEP | RPD | RER | ||
|---|---|---|---|---|---|---|---|
| Spectra of 80 crushed samples | 5 | 0.8319 | 0.1395 | 0.7926 | 0.1528 | 2.07 | 10.72 |
| 10 | 0.8192 | 0.1476 | 0.7355 | 0.1841 | 2.79 | 9.19 | |
| 15 | 0.8010 | 0.1573 | 0.8019 | 0.1535 | 2.22 | 12.02 | |
| 20 | 0.7590 | 0.1705 | 0.7411 | 0.1725 | 2.96 | 9.80 | |
| 25 | 0.7877 | 0.1569 | 0.7360 | 0.1886 | 2.76 | 8.97 | |
| 30 | 0.8836 | 0.1200 | 0.4347 | 0.2598 | 1.31 | 6.51 | |
| 35 | 0.8301 | 0.1450 | 0.6079 | 0.2278 | 2.52 | 7.42 | |
| 40 | 0.8210 | 0.1459 | 0.6030 | 0.2210 | 2.54 | 7.65 | |
| 45 | 0.8008 | 0.1531 | 0.6266 | 0.2207 | 2.54 | 7.66 | |
| 50 | 0.8028 | 0.1499 | 0.6406 | 0.2183 | 2.53 | 7.75 | |
| 55 | 0.7468 | 0.1687 | 0.7702 | 0.1786 | 2.84 | 9.47 | |
| 60 | 0.7269 | 0.1714 | 0.7405 | 0.1900 | 2.70 | 8.90 | |
| 65 | 0.7732 | 0.1529 | 0.8174 | 0.1766 | 2.80 | 9.58 | |
| 70 | 0.7293 | 0.1652 | 0.6954 | 0.2066 | 2.52 | 8.19 | |
| 75 | 0.7300 | 0.1626 | 0.7467 | 0.1960 | 2.58 | 8.63 | |
| 80 | 0.7403 | 0.1594 | 0.8150 | 0.1692 | 2.81 | 9.99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, W.; Zhang, F.; Fu, F.; Sang, S.; Qiao, Z. Rapid Detection of Total Viable Count in Intact Beef Dishes Based on NIR Hyperspectral Hybrid Model. Sensors 2023, 23, 9584. https://doi.org/10.3390/s23239584
Wei W, Zhang F, Fu F, Sang S, Qiao Z. Rapid Detection of Total Viable Count in Intact Beef Dishes Based on NIR Hyperspectral Hybrid Model. Sensors. 2023; 23(23):9584. https://doi.org/10.3390/s23239584
Chicago/Turabian StyleWei, Wensong, Fengjuan Zhang, Fangting Fu, Shuo Sang, and Zhen Qiao. 2023. "Rapid Detection of Total Viable Count in Intact Beef Dishes Based on NIR Hyperspectral Hybrid Model" Sensors 23, no. 23: 9584. https://doi.org/10.3390/s23239584
APA StyleWei, W., Zhang, F., Fu, F., Sang, S., & Qiao, Z. (2023). Rapid Detection of Total Viable Count in Intact Beef Dishes Based on NIR Hyperspectral Hybrid Model. Sensors, 23(23), 9584. https://doi.org/10.3390/s23239584








