Cross-Parallel Transformer: Parallel ViT for Medical Image Segmentation
Abstract
:1. Introduction
- We propose an enhanced PTransUNet model by utilizing parallel ViT rather than serial ViT in the TransUNet medical image segmentation model. This model demonstrates an improvement in segmentation efficiency in comparison to the TransUNet model.
- We present an enhanced C-PTransUNet model and introduce a cross-parallel Transformer module to upgrade the parallel ViT. The MHSA part employs the Dendrite Net (DD) [20,21] layer to enhance semantic representation and remote spatial dependencies. It achieves self-crossover attention for semantic features through feature fusion. In the Feed-Forward Network section, we employ a “rude” streamlining approach. This entails the removal of the fully connected layer and using exclusively the combination of normalization and activation functions. This results in reduced computational overhead while preserving the nonlinear feature representation.
- On the public dataset Synapse [22], we experimentally evaluate the PT model and C-PT model, and the findings demonstrate that parallel ViT has superior accuracy and efficiency than sequential ViT for medical image segmentation.
2. Related Work
2.1. Vision Transformers Development
2.2. Transformer-Based Medical Image Segmentation Method
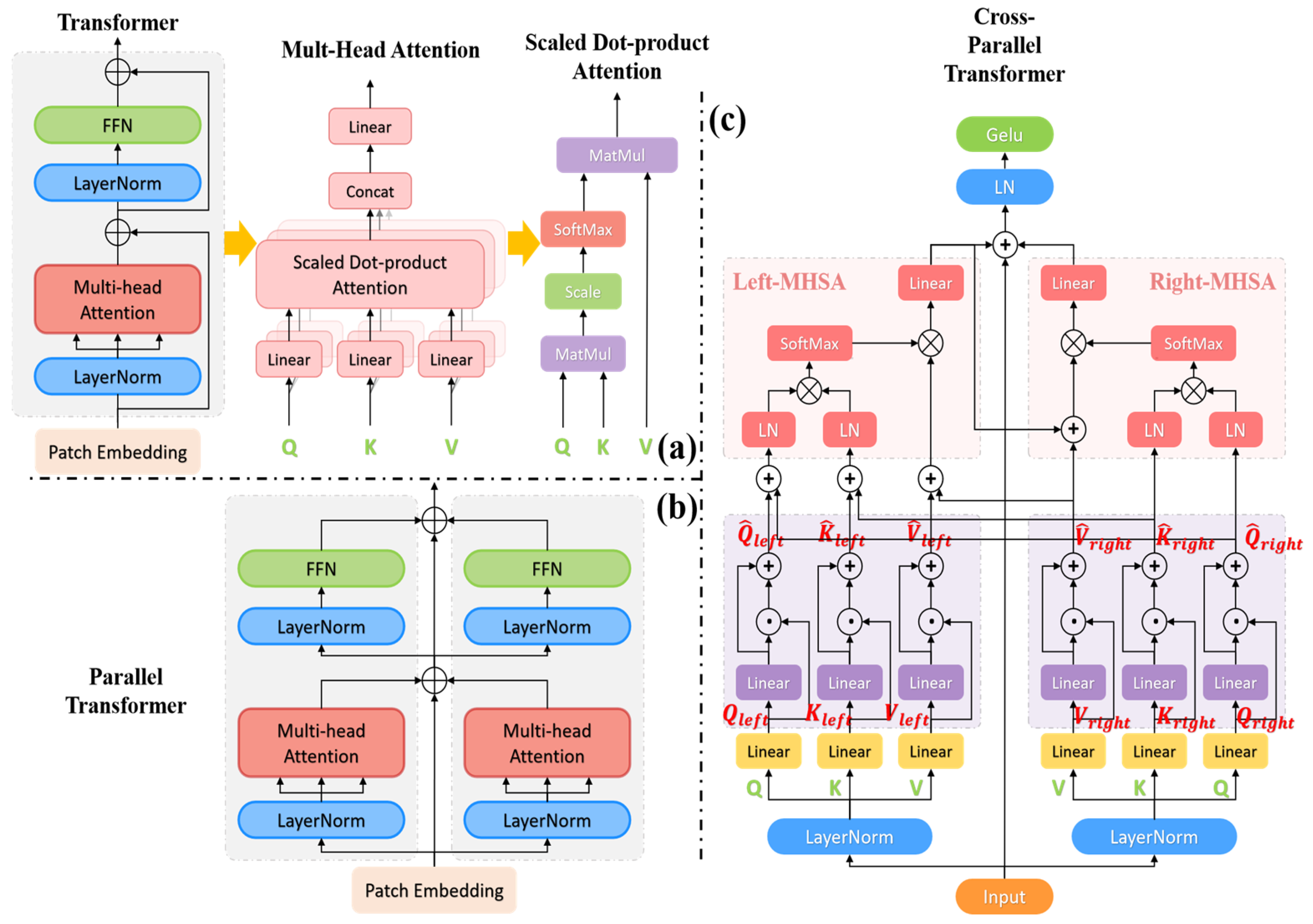
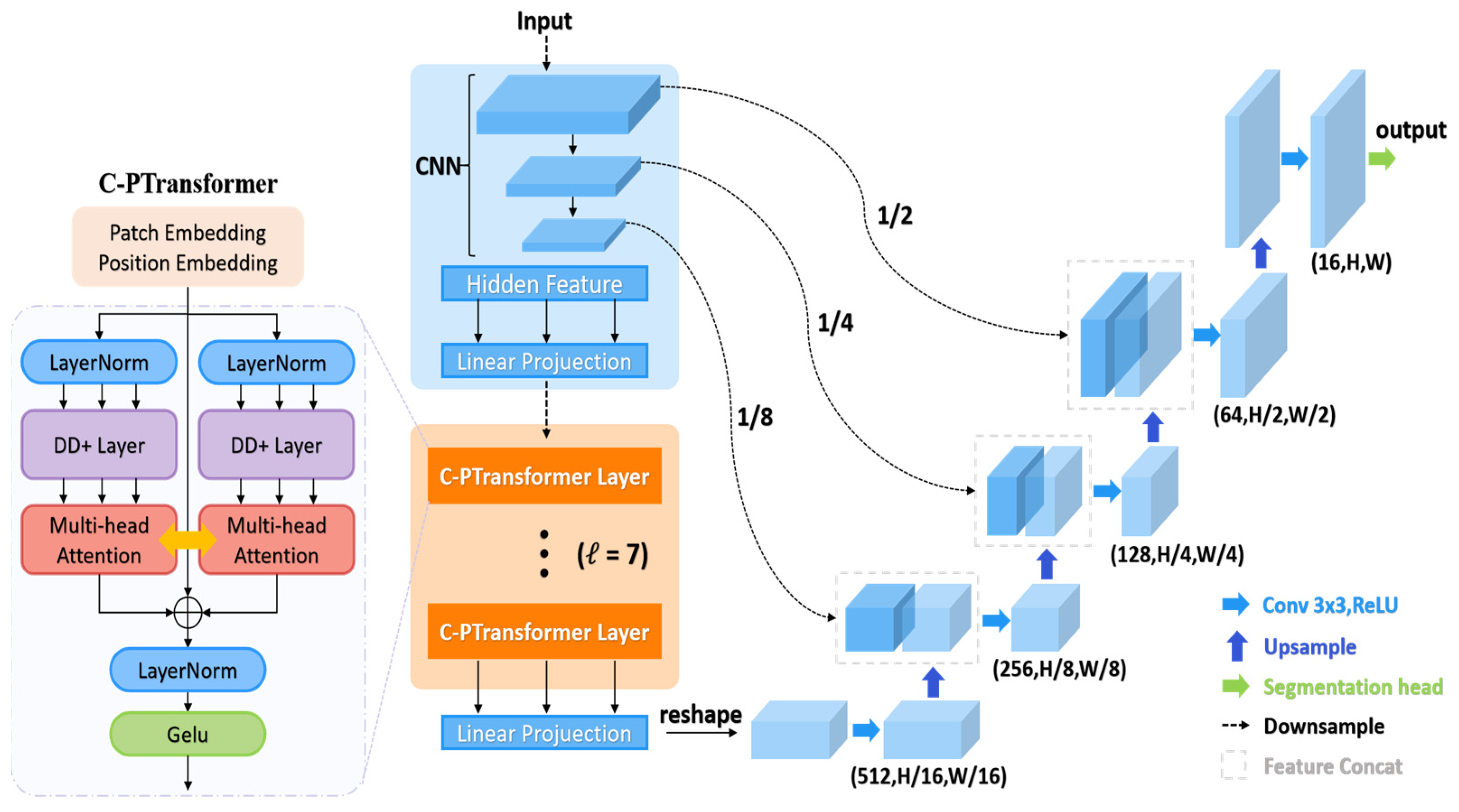
3. Methods Section
3.1. Cross-Parallel Transformer Module
3.1.1. Cross-Parallel Multi-Head Self-Attention Blocks (C-PMHSA)
3.1.2. Activation Function Block
3.2. Improvements to the TransUNet Model
4. Experiments and Results
4.1. Dataset
4.2. Implementation Details
4.3. Comparison of Baseline Models
4.3.1. Synapse Dataset Result
| Methods | Average | Size | Aorta | Gallbladder | Left Kidney | Right Kidney | Liver | Pancreas | Spleen | Stomach | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DSC↑ | HD95↓ | ||||||||||
| V-Net [44] | 68.81 | - | - | 75.34 | 51.87 | 77.10 | 80.75 | 87.84 | 40.05 | 80.56 | 56.98 |
| DARR [45] | 69.77 | - | - | 74.74 | 53.77 | 72.31 | 73.24 | 94.08 | 54.18 | 89.90 | 45.96 |
| R50+U-Net [3] | 74.68 | 36.87 | - | 84.18 | 62.84 | 79.79 | 71.29 | 93.35 | 48.23 | 84.41 | 73.92 |
| TransClaw UNet [46] | 78.09 | - | 224 | 85.87 | 61.38 | 84.83 | 79.36 | 94.28 | 57.65 | 87.74 | 73.55 |
| U-Net [3] | 78.2 | 31.96 | 224 | 88.31 | 70.2 | 79.38 | 71.57 | 93.75 | 57.53 | 86.31 | 78.52 |
| R50 VIT CUP [13] | 71.29 | 32.87 | 224 | 73.73 | 55.13 | 75.80 | 72.20 | 91.51 | 45.99 | 81.99 | 73.95 |
| CGNET [47] | 75.08 | - | 224 | 83.48 | 65.32 | 77.91 | 72.04 | 91.92 | 57.37 | 85.47 | 77.12 |
| AttUNet [48] | 75.59 | 36.97 | - | 55.92 | 63.91 | 79.20 | 72.71 | 93.56 | 49.37 | 87.19 | 74.95 |
| Swin-UNet [6] | 79.13 | 21.55 | 224 | 85.47 | 66.53 | 83.28 | 79.61 | 94.29 | 56.58 | 90.66 | 76.60 |
| TransUNet [5] | 77.48 | 31.69 | 224 | 87.23 | 63.13 | 81.87 | 77.02 | 94.08 | 55.86 | 85.08 | 75.62 |
| UCTransNet [10] | 78.23 | 26.75 | 224 | - | - | - | - | - | - | - | - |
| FFUNet [8] | 79.09 | 31.65 | 224 | 86.68 | 67.09 | 81.13 | 73.73 | 93.67 | 64.17 | 90.92 | 75.32 |
| CTC-Net [41] | 78.41 | 22.52 | 224 | 86.46 | 63.53 | 83.71 | 80.79 | 93.78 | 59.73 | 86.87 | 72.39 |
| TransDeepLab [42] | 80.16 | 21.25 | 224 | 86.04 | 69.16 | 84.08 | 79.88 | 93.53 | 61.19 | 89.00 | 78.40 |
| Ours 1 | 78.35 | 26.38 | 224 | 86.65 | 60.86 | 82.18 | 77.50 | 94.57 | 58.28 | 89.65 | 77.12 |
| Ours 2 | 80.73 | 21.15 | 224 | 88.33 | 65.99 | 83.84 | 82.27 | 94.54 | 63.36 | 88.28 | 79.27 |
| TransUNet [5] | 81.41 | 23.28 | 320 | 90.37 | 64.98 | 85.51 | 81.60 | 94.67 | 68.34 | 88.41 | 77.40 |
| Ours 1 | 81.88 | 30.17 | 320 | 89.54 | 64.90 | 84.40 | 80.18 | 95.39 | 66.27 | 91.28 | 83.12 |
| Ours 2 | 82.66 | 21.11 | 320 | 89.21 | 67.97 | 85.54 | 81.18 | 95.09 | 66.82 | 90.79 | 84.71 |
| nnUNet [43] | 82.36 | 24.74 | 512 | 90.96 | 65.57 | 81.92 | 78.36 | 95.96 | 69.36 | 91.12 | 85.60 |
| TransUNet [5] | 81.57 | 26.89 | 512 | 90.45 | 66.20 | 79.73 | 74.99 | 95.25 | 74.24 | 88.61 | 83.14 |
| Ours 1 | 82.85 | 28.12 | 512 | 91.01 | 63.85 | 84.38 | 80.6 | 95.84 | 70.19 | 91.61 | 85.33 |
| Ours 2 | 81.85 | 22.32 | 512 | 90.94 | 60.51 | 84.99 | 79.38 | 95.28 | 68.67 | 93.39 | 81.71 |
| Evaluating Indicator | Methods | Average | Aorta | Gallbladder | Left Kidney | Right Kidney | Liver | Pancreas | Spleen | Stomach |
|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy | TransUNet | 99.88 | 99.97 | 99.98 | 99.94 | 99.93 | 99.92 | 99.91 | 99.90 | 99.70 |
| Ours 1 | 99.89 | 99.97 | 99.98 | 99.94 | 99.93 | 99.74 | 99.91 | 99.93 | 99.73 | |
| Ours 2 | 99.90 | 99.97 | 99.98 | 99.95 | 99.94 | 99.75 | 99.92 | 99.92 | 99.76 | |
| F1 Score | TransUNet | 76.53 | 87.61 | 53.24 | 81.99 | 76.89 | 93.76 | 58.44 | 86.07 | 74.24 |
| Ours 1 | 77.31 | 86.65 | 52.53 | 82.18 | 77.50 | 94.57 | 58.28 | 89.65 | 77.12 | |
| Ours 2 | 79.69 | 88.33 | 57.65 | 83.84 | 82.27 | 94.54 | 63.36 | 88.28 | 79.28 | |
| Sensitivity | TransUNet | 76.53 | 88.36 | 50.49 | 87.67 | 73.00 | 95.04 | 55.59 | 90.53 | 71.56 |
| Ours 1 | 76.85 | 87.09 | 51.62 | 82.99 | 76.62 | 95.81 | 54.80 | 92.01 | 73.89 | |
| Ours 2 | 79.56 | 89.88 | 55.32 | 82.47 | 83.30 | 95.83 | 60.63 | 91.44 | 77.66 | |
| Precision | TransUNet | 80.29 | 87.19 | 62.71 | 78.58 | 83.81 | 92.60 | 73.12 | 84.51 | 79.86 |
| Ours 1 | 80.36 | 86.72 | 58.79 | 81.72 | 80.16 | 93.46 | 70.95 | 88.54 | 82.62 | |
| Ours 2 | 82.37 | 87.30 | 64.71 | 87.45 | 81.94 | 93.42 | 74.80 | 86.24 | 83.15 |
4.3.2. ACDC Dataset Result
4.4. Parallelism Experiment
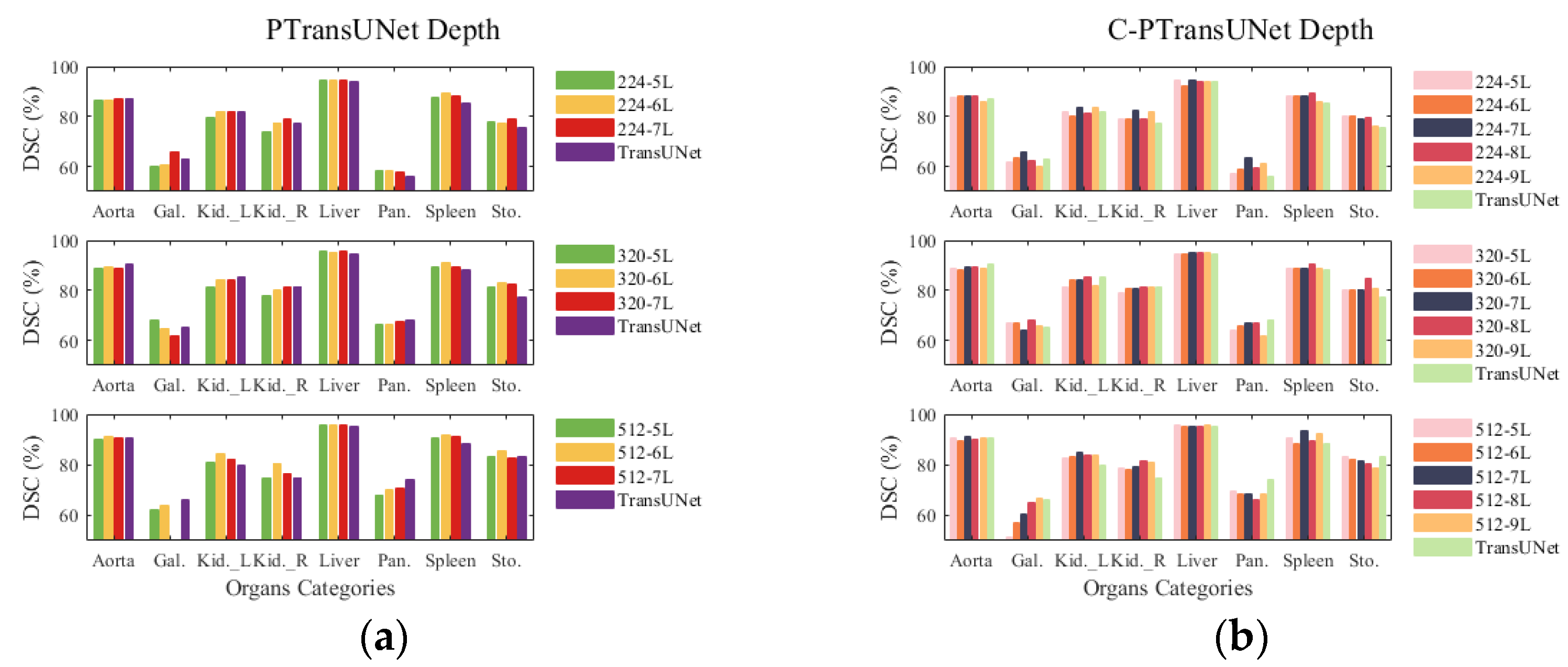
4.5. Depth Experiment
4.6. Ablation Experiment
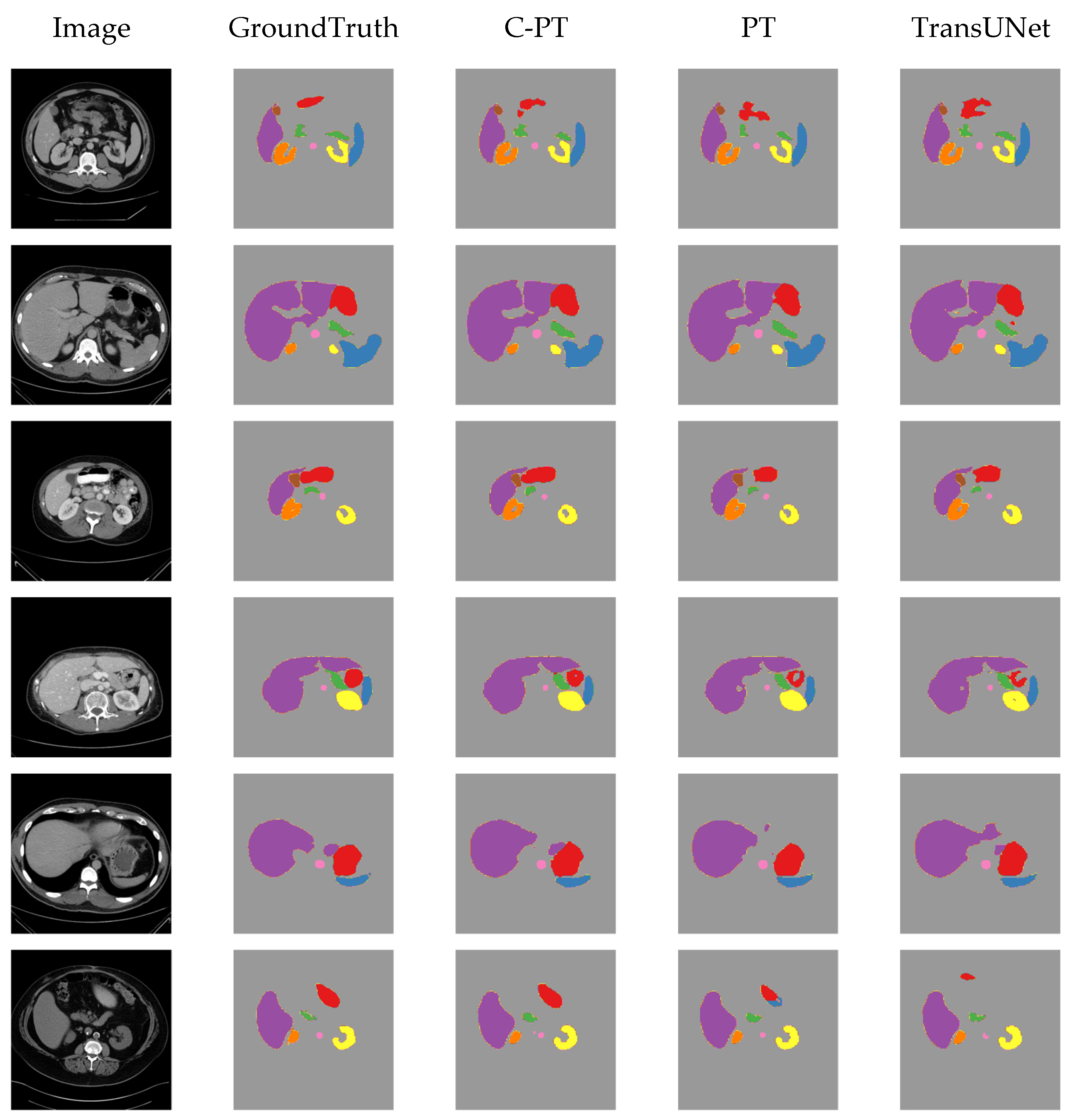
4.7. Visualising Results on Synapse Dataset
5. Conclusions
- (1)
- The PT model demonstrates superior DSC performance compared with the TransUNet baseline model while maintaining the same number of parameters and FLOPs. Additionally, the parallel ViT proves more appropriate for the baseline model than the serial ViT for feature learning at a deeper level.
- (2)
- At an input size of 224, the C-PT model decreases parameter count by 29% and FLOPs by 21.4% as compared with the baseline model, while also improving DSC accuracy by 3.25% and shortening HD edge gap by 10.54 mm compared to the baseline. The C-PT model exhibits superior segmentation performance and higher efficiency than the baseline model employed.
- (3)
- The C-PT module demonstrates improved performance and efficiency when compared to the parallel ViT module within the baseline model. This is attributed to the design of the C-PMHSA and the streamlined MLP. The MHSA block’s feature extraction capability is enhanced to ensure the overall performance of the C-PT module, while the FFN block is replaced with an activation function to reduce the number of parameters and FLOPs of the C-PT module.
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PT | PTransUNet network |
| C-PT | C-PTransUNet network |
| CNN | Convolutional neural network |
| ViT | Vision Transformer |
| MHSA | Multi-Head Self-Attention |
| FFN | Feed-Forward Network |
| DSC | Dice similarity coefficient |
| HD95 | the 95% Hausdorff Distance |
| FLOPs | Floating point operations per second |
| MLP | Multilayer Perceptron |
| LN | Layer Normalization |
| GELU | Gaussian Error Linear Unit |
References
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; pp. 205–218. [Google Scholar]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. Unetr: Transformers for 3d medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 4–8 January 2022; pp. 574–584. [Google Scholar]
- Xie, J.; Zhu, R.; Wu, Z.; Ouyang, J. FFUNet: A novel feature fusion makes strong decoder for medical image segmentation. IET Signal Process. 2022, 16, 501–514. [Google Scholar] [CrossRef]
- Zhou, H.-Y.; Guo, J.; Zhang, Y.; Yu, L.; Wang, L.; Yu, Y. nnformer: Interleaved transformer for volumetric segmentation. arXiv 2021, arXiv:2109.03201. [Google Scholar]
- Wang, H.; Cao, P.; Wang, J.; Zaiane, O.R. Uctransnet: Rethinking the skip connections in U-Net from a channel-wise perspective with transformer. In Proceedings of the AAAI Conference on Artificial Intelligence, Virtual, 22 February–2 March 2022; pp. 2441–2449. [Google Scholar]
- Gao, Y.; Zhou, M.; Metaxas, D.N. UTNet: A hybrid transformer architecture for medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; pp. 61–71. [Google Scholar]
- Peiris, H.; Hayat, M.; Chen, Z.; Egan, G.; Harandi, M. A robust volumetric transformer for accurate 3D tumor segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; pp. 162–172. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S. An image is worth 16×16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Ansari, M.Y.; Abdalla, A.; Ansari, M.Y.; Ansari, M.I.; Malluhi, B.; Mohanty, S.; Mishra, S.; Singh, S.S.; Abinahed, J.; Al-Ansari, A. Practical utility of liver segmentation methods in clinical surgeries and interventions. BMC Med. Imaging 2022, 22, 97. [Google Scholar]
- Liu, Z.; Shen, L. Medical image analysis based on transformer: A review. arXiv 2022, arXiv:2208.06643. [Google Scholar]
- Khan, S.; Naseer, M.; Hayat, M.; Zamir, S.W.; Khan, F.S.; Shah, M. Transformers in vision: A survey. ACM Comput. Surv. (CSUR) 2022, 54, 1–41. [Google Scholar] [CrossRef]
- Shamshad, F.; Khan, S.; Zamir, S.W.; Khan, M.H.; Hayat, M.; Khan, F.S.; Fu, H. Transformers in medical imaging: A survey. Med. Image Anal. 2023, 88, 102802. [Google Scholar] [CrossRef]
- Touvron, H.; Cord, M.; El-Nouby, A.; Verbeek, J.; Jégou, H. Three things everyone should know about vision transformers. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; pp. 497–515. [Google Scholar]
- Bebis, G.; Georgiopoulos, M. Feed-forward neural networks. IEEE Potentials 1994, 13, 27–31. [Google Scholar] [CrossRef]
- Liu, G.; Wang, J. Dendrite net: A white-box module for classification, regression, and system identification. IEEE Trans. Cybern. 2021, 52, 13774–13787. [Google Scholar] [CrossRef] [PubMed]
- Liu, G. It may be time to perfect the neuron of artificial neural network. TechRxiv 2023. [Google Scholar] [CrossRef]
- Landman, B.; Xu, Z.; Igelsias, J.; Styner, M.; Langerak, T.; Klein, A. Miccai multi-atlas labeling beyond the cranial vault–workshop and challenge. In Proceedings of the MICCAI Multi-Atlas Labeling Beyond Cranial Vault—Workshop Challenge, Munich, Germany, 5–9 October 2015; p. 12. [Google Scholar]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H. Training data-efficient image transformers & distillation through attention. Proc. Int. Conf. Mach. Learn. 2021, 139, 10347–10357. [Google Scholar]
- Yuan, K.; Guo, S.; Liu, Z.; Zhou, A.; Yu, F.; Wu, W. Incorporating convolution designs into visual transformers. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Virtual, 11–17 October 2021; pp. 579–588. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Virtual, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Yuan, L.; Chen, Y.; Wang, T.; Yu, W.; Shi, Y.; Jiang, Z.-H.; Tay, F.E.; Feng, J.; Yan, S. Tokens-to-token vit: Training vision transformers from scratch on imagenet. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Virtual, 11–17 October 2021; pp. 558–567. [Google Scholar]
- Wang, W.; Xie, E.; Li, X.; Fan, D.-P.; Song, K.; Liang, D.; Lu, T.; Luo, P.; Shao, L. Pyramid vision transformer: A versatile backbone for dense prediction without convolutions. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Virtual, 11–17 October 2021; pp. 568–578. [Google Scholar]
- Zhou, D.; Kang, B.; Jin, X.; Yang, L.; Lian, X.; Jiang, Z.; Hou, Q.; Feng, J. Deepvit: Towards deeper vision transformer. arXiv 2021, arXiv:2103.11886. [Google Scholar]
- Lin, T.; Wang, Y.; Liu, X.; Qiu, X. A survey of transformers. AI Open 2022, 3, 111–132. [Google Scholar] [CrossRef]
- Delalleau, O.; Bengio, Y. Shallow vs. deep sum-product networks. Adv. Neural Inf. Process. Syst. 2011, 24. [Google Scholar]
- Eldan, R.; Shamir, O. The power of depth for feedforward neural networks. In Proceedings of the Conference on Learning Theory, New York, NY, USA, 23–26 June 2016; pp. 907–940. [Google Scholar]
- Lu, Z.; Pu, H.; Wang, F.; Hu, Z.; Wang, L. The expressive power of neural networks: A view from the width. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- Zhang, Y.; Liu, H.; Hu, Q. Transfuse: Fusing transformers and cnns for medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 Septembe–1 October 2021; pp. 14–24. [Google Scholar]
- Dubey, S.R.; Singh, S.K.; Chaudhuri, B.B. Activation functions in deep learning: A comprehensive survey and benchmark. Neurocomputing 2022, 503, 92–108. [Google Scholar] [CrossRef]
- Sukhbaatar, S.; Grave, E.; Lample, G.; Jegou, H.; Joulin, A. Augmenting self-attention with persistent memory. arXiv 2019, arXiv:1907.01470. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Zhao, Y.; Wang, G.; Tang, C.; Luo, C.; Zeng, W.; Zha, Z.-J. A battle of network structures: An empirical study of cnn, transformer, and mlp. arXiv 2021, arXiv:2108.13002. [Google Scholar]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.-A.; Cetin, I.; Lekadir, K.; Camara, O.; Ballester, M.A.G. Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Zhang, S.; Xu, Y.; Wu, Z.; Wei, Z. CTC-Net: A Novel Coupled Feature-Enhanced Transformer and Inverted Convolution Network for Medical Image Segmentation. In Proceedings of the Asian Conference on Pattern Recognition, Kitakyushu, Japan, 5–8 November 2023; pp. 273–283. [Google Scholar]
- Azad, R.; Heidari, M.; Shariatnia, M.; Aghdam, E.K.; Karimijafarbigloo, S.; Adeli, E.; Merhof, D. Transdeeplab: Convolution-free transformer-based deeplab v3+ for medical image segmentation. In Proceedings of the International Workshop on PRedictive Intelligence in MEdicine, Singapore, 22 September 2022; pp. 91–102. [Google Scholar]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Fu, S.; Lu, Y.; Wang, Y.; Zhou, Y.; Shen, W.; Fishman, E.; Yuille, A. Domain adaptive relational reasoning for 3d multi-organ segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020; pp. 656–666. [Google Scholar]
- Chang, Y.; Menghan, H.; Guangtao, Z.; Xiao-Ping, Z. Transclaw u-net: Claw u-net with transformers for medical image segmentation. arXiv 2021, arXiv:2107.05188. [Google Scholar]
- Wu, T.; Tang, S.; Zhang, R.; Cao, J.; Zhang, Y. Cgnet: A light-weight context guided network for semantic segmentation. IEEE Trans. Image Process. 2020, 30, 1169–1179. [Google Scholar] [CrossRef]
- Schlemper, J.; Oktay, O.; Schaap, M.; Heinrich, M.; Kainz, B.; Glocker, B.; Rueckert, D. Attention gated networks: Learning to leverage salient regions in medical images. Med. Image Anal. 2019, 53, 197–207. [Google Scholar] [CrossRef]
- Huang, X.; Deng, Z.; Li, D.; Yuan, X. Missformer: An effective medical image segmentation transformer. arXiv 2021, arXiv:2109.07162. [Google Scholar] [CrossRef]


| Model | Sequential Transformer | Parallel Transformer |
|---|---|---|
| TransUNet | √ | |
| Swin-Unet | √ | |
| UNETR | √ | |
| PTransUNet | √ | |
| C-PTransUNet | √ |
| Methods | Average | RV | Myo | LV | ||||
|---|---|---|---|---|---|---|---|---|
| DSC↑ | HD95↓ | DSC↑ | HD95↓ | DSC↑ | HD95↓ | DSC↑ | HD95↓ | |
| R50 U-Net [3] | 87.55 | - | 87.1 | - | 80.63 | - | 94.92 | - |
| R50 Att-Unet [48] | 86.75 | - | 87.58 | - | 79.2 | - | 93.47 | - |
| VIT CUP [13] | 81.45 | - | 81.46 | - | 70.71 | - | 92.18 | - |
| R50 VIT CUP [13] | 87.57 | - | 86.07 | - | 81.88 | - | 94.75 | - |
| SwinUNet [6] | 90.00 | - | 88.55 | - | 85.62 | - | 95.83 | - |
| MISSFormer [49] | 87.90 | - | 86.36 | - | 85.75 | - | 91.59 | - |
| UNETR [7] | 87.38 | - | 88.49 | - | 82.04 | - | 91.62 | - |
| CTC-Net [41] | 90.77 | - | 90.09 | - | 85.52 | - | 96.72 | - |
| TransUNet | 89.59 | 6.91 | 88.54 | 18.11 | 86.13 | 1.36 | 94.12 | 1.28 |
| Ours 1 | 89.49 | 6.50 | 88.16 | 16.91 | 86.29 | 1.32 | 94.02 | 1.26 |
| Ours 2 | 90.44 | 5.30 | 90.02 | 13.34 | 86.51 | 1.34 | 94.80 | 1.22 |
| Methods | Average | RV | Myo | LV | ||||
| Sens↑ | Prec↑ | Sens↑ | Prec↑ | Sens↑ | Prec↑ | Sens↑ | Prec↑ | |
| TransUNet | 87.01 | 87.16 | 82.52 | 79.58 | 85.23 | 86.48 | 93.29 | 95.43 |
| Ours 1 | 86.64 | 87.19 | 81.84 | 79.21 | 85.29 | 86.72 | 92.79 | 95.66 |
| Ours 2 | 87.90 | 86.84 | 83.76 | 79.05 | 85.89 | 86.64 | 94.08 | 94.82 |
| Methods | Average | RV | Myo | LV | ||||
| Acc↑ | F1-S↑ | Acc↑ | F1-S↑ | Acc↑ | F1-S↑ | Acc↑ | F1-S↑ | |
| TransUNet | 99.80 | 86.71 | 99.79 | 80.37 | 99.73 | 85.65 | 99.89 | 94.13 |
| Ours 1 | 99.80 | 86.61 | 99.79 | 79.99 | 99.73 | 85.82 | 99.89 | 94.02 |
| Ours 2 | 99.80 | 87.08 | 99.79 | 80.88 | 99.73 | 86.04 | 99.89 | 94.32 |
| Models | Size | Parallelism | Average | Params (M) | FLOPs (G) | Train Time | ||
|---|---|---|---|---|---|---|---|---|
| Branches | Layer | DSC↑ | HD↓ | |||||
| PT/6 × 2 | 224 | 2 | 6 | 78.35 | 26.38 | 88.91 | 24.73 | 1:43:03 |
| 320 | 2 | 6 | 81.88 | 30.17 | 88.91 | 50.5 | 3:33:34 | |
| 512 | 2 | 6 | 82.85 | 28.12 | 88.91 | 129.56 | 10:23:42 | |
| PT/4 × 3 | 224 | 3 | 4 | 78.9 | 26.53 | 88.91 | 24.73 | 1:42:33 |
| 320 | 3 | 4 | 81.91 | 25.33 | 88.91 | 50.5 | 3:30:20 | |
| 512 | 3 | 4 | 81.84 | 33.25 | 88.91 | 129.56 | 10:23:45 | |
| PT/3 × 4 | 224 | 4 | 3 | 78.49 | 25.28 | 88.91 | 24.73 | 1:42:39 |
| 320 | 4 | 3 | 80.96 | 29.87 | 88.91 | 50.5 | 3:28:56 | |
| 512 | 4 | 3 | 74.39 | 27.48 | 88.91 | 129.56 | 10:28:56 | |
| Models | Size | Parallelism | Average | Params (M) | FLOPs (G) | Train time | ||
|---|---|---|---|---|---|---|---|---|
| Branches | Layer | DSC↑ | HD↓ | |||||
| C-PT/ 6 × 2 | 224 | 2 | 6 | 78.87 | 24.51 | 55.17 | 17.8 | 1:26:59 |
| 320 | 2 | 6 | 81.2 | 28.86 | 55.17 | 36.37 | 2:58:21 | |
| 512 | 2 | 6 | 80.24 | 26.07 | 55.17 | 93.37 | 8:58:56 | |
| C-PT/ 4 × 3 | 224 | 3 | 4 | 79.55 | 26.97 | 55.16 | 17.8 | 1:25:38 |
| 320 | 3 | 4 | 79.03 | 38.38 | 55.16 | 36.35 | 2:57:19 | |
| 512 | 3 | 4 | 75.12 | 18.17 | 55.16 | 93.34 | 8:58:51 | |
| C-PT/ 3 × 4 | 224 | 4 | 3 | 79.67 | 25.02 | 31.48 | 17.8 | 1:26:02 |
| 320 | 4 | 3 | 80.58 | 33.51 | 31.48 | 36.36 | 2:57:53 | |
| 512 | 4 | 3 | 82.63 | 26.11 | 31.48 | 93.37 | 8:58:54 | |
| Model | Size | Depth | Average | Params (M) | FLOPs (G) | Train Time | ||
|---|---|---|---|---|---|---|---|---|
| Branches | Layer | DSC↑ | HD↓ | |||||
| PT | 224 | 2 | 5 | 77.31 | 31.52 | 75.39 | 21.95 | 1:28:56 |
| 224 | 2 | 6 | 78.35 | 26.38 | 88.91 | 24.73 | 1:43:03 | |
| 224 | 2 | 7 | 79.18 | 27.82 | 102.43 | 27.51 | 1:51:51 | |
| PT | 320 | 2 | 5 | 81.11 | 27.63 | 75.39 | 44.82 | 3:07:48 |
| 320 | 2 | 6 | 81.88 | 30.17 | 88.91 | 50.5 | 3:33:34 | |
| 320 | 2 | 7 | 81.37 | 23.58 | 102.43 | 56.18 | 3:51:43 | |
| PT | 512 | 2 | 5 | 80.82 | 37.47 | 75.39 | 114.97 | 9:09:44 |
| 512 | 2 | 6 | 82.85 | 28.12 | 88.91 | 129.56 | 10:23:42 | |
| 512 | 2 | 7 | 73.66 | 27.44 | 102.43 | 144.14 | 11:37:46 | |
| Model | Size | Depth | Average | Params (M) | FLOPs (G) | Train Time | ||
|---|---|---|---|---|---|---|---|---|
| Branches | Layer | DSC↑ | HD↓ | |||||
| C-PT/ 6 × 2 | 224 | 2 | 5 | 78.78 | 30.36 | 47.28 | 16.18 | 1:19:27 |
| 224 | 2 | 6 | 78.87 | 24.51 | 55.17 | 17.8 | 1:26:59 | |
| 224 | 2 | 7 | 80.73 | 21.15 | 63.07 | 19.43 | 1:34:25 | |
| 224 | 2 | 8 | 79.17 | 27.52 | 70.96 | 21.05 | 1:41:10 | |
| 224 | 2 | 9 | 78.59 | 28.78 | 78.86 | 22.68 | 1:48:22 | |
| C-PT/ 4 × 3 | 320 | 2 | 5 | 80.47 | 31.62 | 47.28 | 33.04 | 2:41:18 |
| 320 | 2 | 6 | 81.2 | 28.86 | 55.17 | 36.37 | 2:58:21 | |
| 320 | 2 | 7 | 81.26 | 29.43 | 63.07 | 39.69 | 3:14:48 | |
| 320 | 2 | 8 | 82.66 | 21.11 | 70.96 | 43.01 | 3:36:37 | |
| 320 | 2 | 9 | 80.51 | 25.91 | 78.86 | 46.34 | 3:52:46 | |
| C-PT/ 3 × 4 | 512 | 2 | 5 | 80.28 | 28.27 | 47.28 | 84.82 | 8:05:24 |
| 512 | 2 | 6 | 80.24 | 26.07 | 55.17 | 93.37 | 8:58:56 | |
| 512 | 2 | 7 | 81.85 | 22.32 | 63.07 | 101.93 | 9:52:13 | |
| 512 | 2 | 8 | 81.52 | 23.21 | 70.96 | 110.48 | 10:47:44 | |
| 512 | 2 | 9 | 82.1 | 24.89 | 78.86 | 119.03 | 11:44:54 | |
| Model | Average | CP-T Block | Block Layer | Params (M) | FLOPs G) | ||
|---|---|---|---|---|---|---|---|
| DSC↑ | HD↓ | DD Layer | MLP | ||||
| C-PT/ 0DD | 77.77 | 29.93 | 0 | f | 6 | 34.89 | 13.64 |
| 77.8 | 29.03 | 0 | f | 7 | 39.41 | 14.57 | |
| 77.47 | 33.91 | 0 | f | 8 | 43.93 | 15.51 | |
| C-PT/ 1DD | 78.87 | 24.51 | +1 | f | 6 | 55.17 | 17.8 |
| 80.73 | 21.15 | +1 | f | 7 | 63.07 | 19.43 | |
| 79.17 | 27.52 | +1 | f | 8 | 70.96 | 21.05 | |
| C-PT/ 2DD | 77.29 | 28.64 | +2 | f | 6 | 75.45 | 21.96 |
| 78.69 | 28.57 | +2 | f | 7 | 86.72 | 24.28 | |
| 78.24 | 30.76 | +2 | f | 8 | 97.99 | 26.61 | |
| Model | Average | CP-T Block | Block Layer | Params (M) | FLOPs (G) | ||
|---|---|---|---|---|---|---|---|
| DSC↑ | HD↓ | DD Layer | MLP | ||||
| C-PT/ 0MLP | 77.66 | 28.23 | +1 | 0 | 6 | 55.16 | 17.8 |
| 77.48 | 35.41 | +1 | 0 | 7 | 63.06 | 19.42 | |
| 78.2 | 27.68 | +1 | 0 | 8 | 70.95 | 21.05 | |
| C-PT/ 1MLP | 77.31 | 28.88 | +1 | +1 | 6 | 82.19 | 23.35 |
| 77.82 | 27.72 | +1 | +1 | 7 | 94.59 | 25.9 | |
| 78.05 | 27.6 | +1 | +1 | 8 | 106.99 | 24.45 | |
| C-PT/ 2MLP | 78.17 | 31.38 | +1 | +2 | 6 | 109.22 | 28.9 |
| 79.08 | 28.14 | +1 | +2 | 7 | 126.13 | 32.38 | |
| 78.25 | 29.54 | +1 | +2 | 8 | 143.03 | 35.86 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Wang, Z.; Chen, L.; Xiao, H.; Yang, B. Cross-Parallel Transformer: Parallel ViT for Medical Image Segmentation. Sensors 2023, 23, 9488. https://doi.org/10.3390/s23239488
Wang D, Wang Z, Chen L, Xiao H, Yang B. Cross-Parallel Transformer: Parallel ViT for Medical Image Segmentation. Sensors. 2023; 23(23):9488. https://doi.org/10.3390/s23239488
Chicago/Turabian StyleWang, Dong, Zixiang Wang, Ling Chen, Hongfeng Xiao, and Bo Yang. 2023. "Cross-Parallel Transformer: Parallel ViT for Medical Image Segmentation" Sensors 23, no. 23: 9488. https://doi.org/10.3390/s23239488
APA StyleWang, D., Wang, Z., Chen, L., Xiao, H., & Yang, B. (2023). Cross-Parallel Transformer: Parallel ViT for Medical Image Segmentation. Sensors, 23(23), 9488. https://doi.org/10.3390/s23239488






