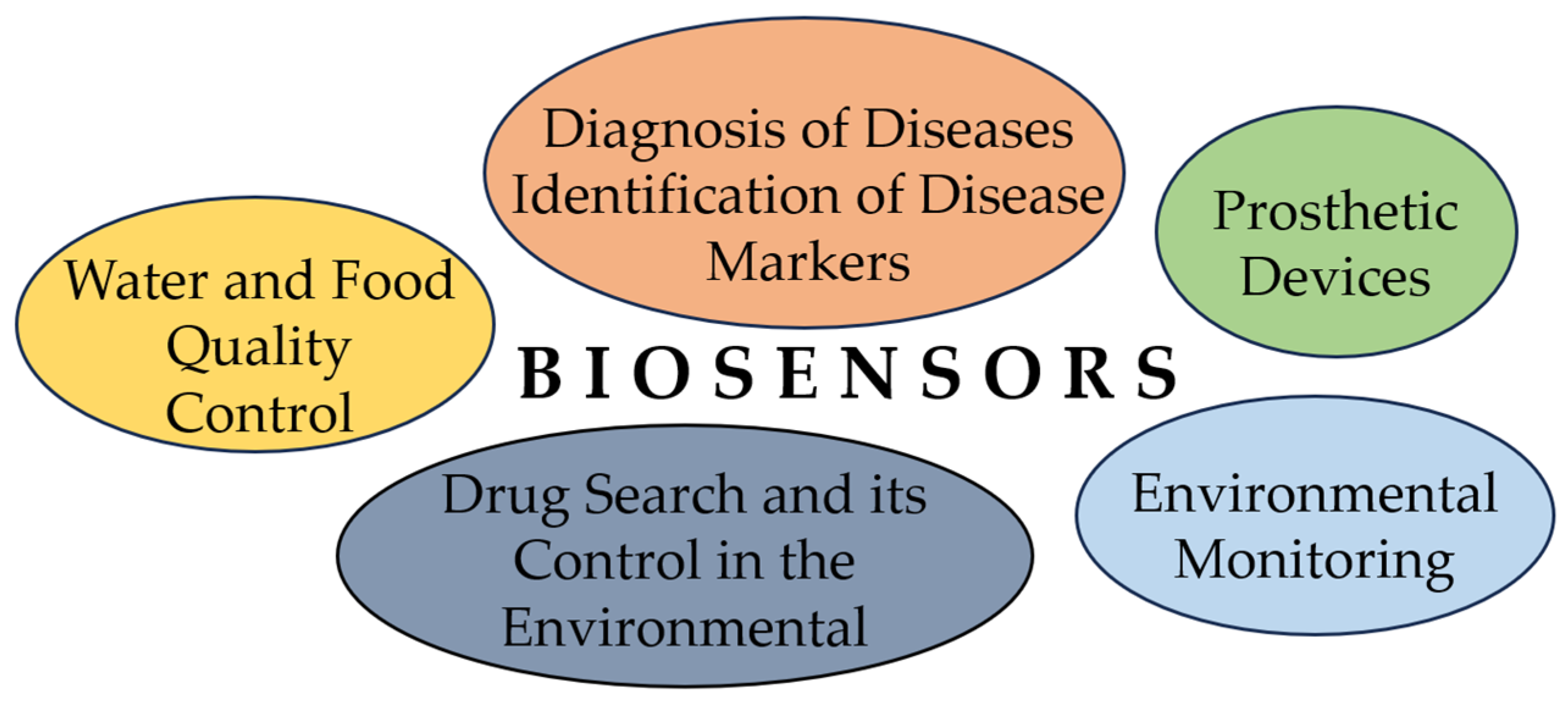
Optical Sensors for Bacterial Detection
Abstract
:1. Introduction
2. Classical Bacterial Detection Methods and Approaches
- -
- Enzyme-linked immunosorbent assay.
- -
- Immunomagnetic separation, an analytic tool used to detect bacteria and based on the capture of target bacteria with antibody-coated magnetic beads. It is combined with various detection methods such as PCR [10].
- -
- Methods based on fluorophore-labeled secondary antibodies [11].
- -
- Enzyme-labeled antibodies [12].
- -
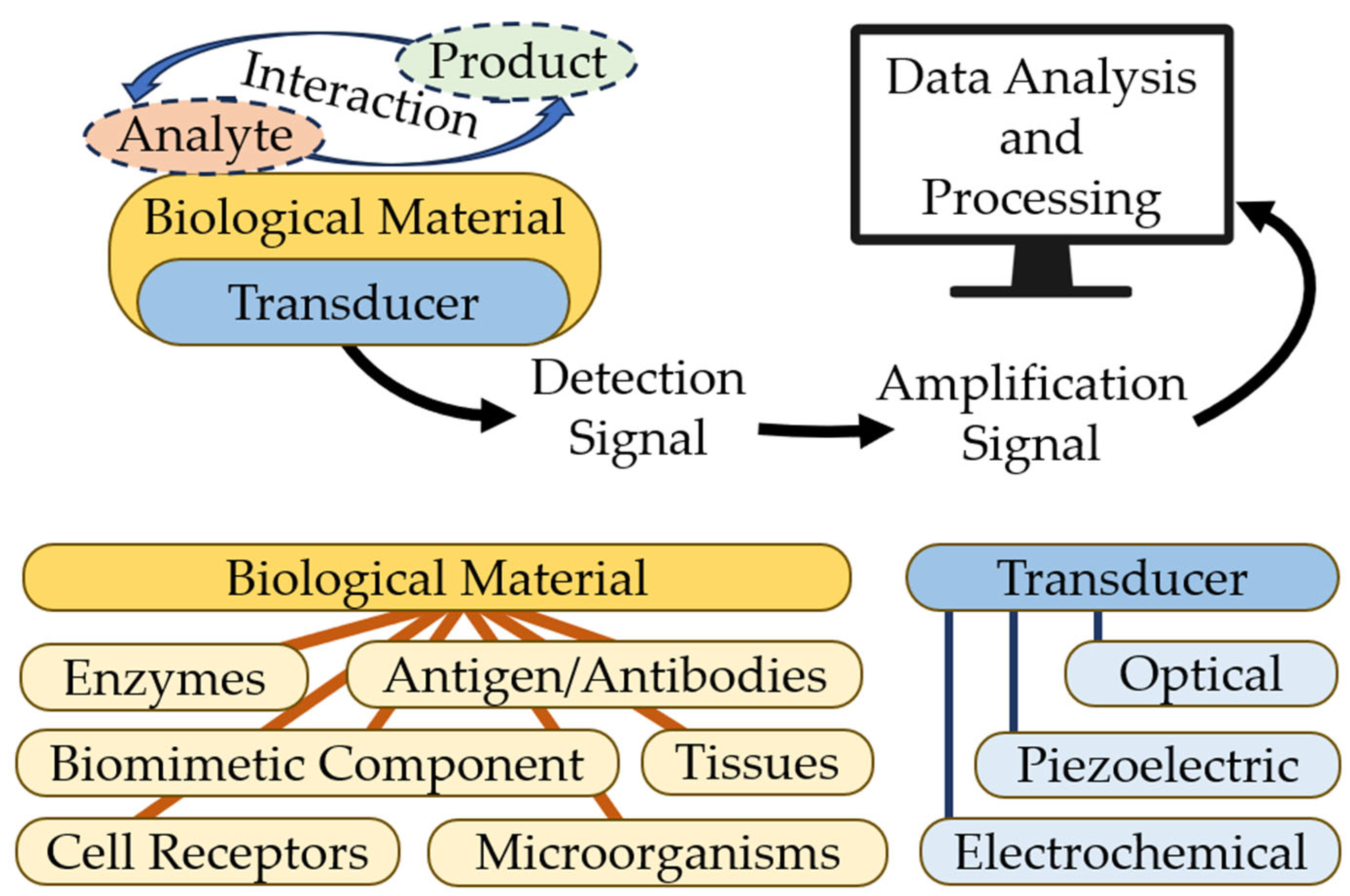
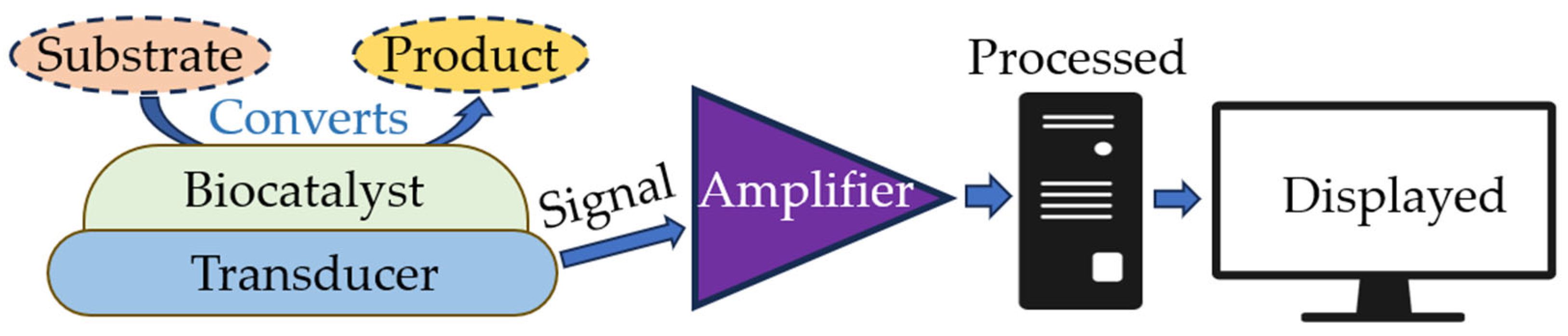
3. Biosensors: Operating Principle and Classification
- -
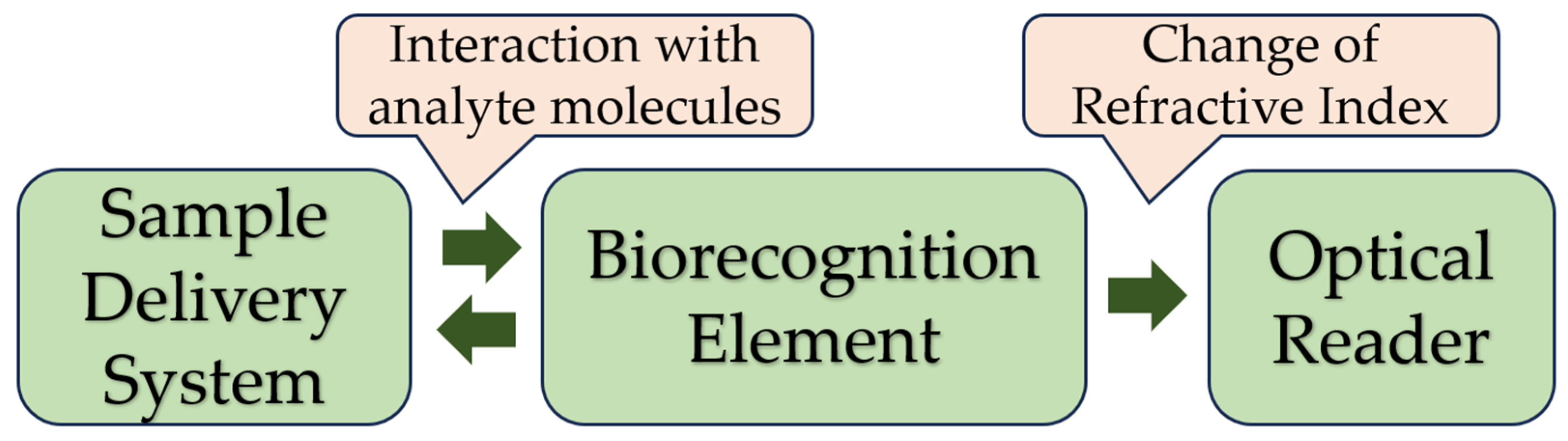
- Bioselective element (biological material, (e.g., tissues, microorganisms, organelles, cellular receptors, enzymes, antibodies, and nucleic acids), material of biological origin, or a biomimic material). The sensing element can be made using bioengineering.
- -
- An optical, piezoelectric, or electrochemical transducer, which converts the signal resulting from the interaction of the analyte with the bioselective element into a signal that is easier to measure.
- -
- Associated electronics, primarily responsible for displaying results in a user-friendly manner [18]. Figure 2 shows the basic scheme for a biosensor. Modern technologies in infectious diagnostics and epidemiology have made label-free biosensors increasingly widespread. This allows intermolecular interactions and cellular reactions to be screened, yielding details of the selectivity of bacterial exotoxins, the specificity of antibacterial agents, the antigen–antibody interaction, the kinetics of inflammation, and immunological and serological responses [19,20]. This type of biosensor requires only one recognition element, which simplifies the analysis and reduces its duration and reagent costs. The modern generation of label-free biosensors enables the real-time measurement of the products of biomolecular reactions, continuous recording of data, and kinetic monitoring of the recognition of the ligand–receptor interaction [3,21].
4. Optical Sensor Systems for Bacterial Detection
- -
- Absorption (the ability of a substance to absorb optical radiation).
- -
- Reflection (when a stream of light falls on the interface between two media, part of its radiation is reflected back).
- -
- Luminescence (the glow of a substance that occurs after it absorbs excitation energy and is excess radiation, as compared with the thermal radiation of the body at a given temperature).
- -
- Photoluminescence (the emission of photons by a substance that occurs when the substance is excited by electromagnetic radiation in the ultraviolet, visible, and infrared wavelength ranges). Photoluminescence is characterized by absorption and luminescence spectra, polarization, energy yield (the ratio of the energy emitted by a substance in the form of luminescence to the absorbed energy), quantum yield (the ratio of the number of emitted quanta to the number of absorbed ones), and kinetics.
4.1. Colorimetric Sensor System for Bacterial Detection
4.2. Fluorescence Sensor Systems for Bacterial Analysis
- (1)
- Fluorescence imaging to identify the bacteria;
- (2)
- Enhancement or changing of the method’s characteristics when targets are combined;
- (3)
- Quenching of fluorescent signals through energy receptors, such as quenching agents [35].
- -
- Use of appropriate fluorescent materials;
- -
- Use of DNA, bacteria, and metabolites as the response targets;
4.3. Chemiluminescence
4.4. Sensors Based on the Surface Plasmon Resonance
4.5. Fiber Optic Sensors
4.6. Optical Sensors Based on Ionophores and Planar Biosensors
4.7. Photonic Crystal Biosensors
5. Optical Sensor Systems Based on Measurement of Orientational Effects
5.1. Optical Sensor Systems Based on Measurements of Bacterial Electrical Characteristics
5.1.1. Electrorotational Sensors
5.1.2. Electro-Optical Sensors with a Fixed Direction of the Electric Field Vector
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- GBD 2015 Mortality and Causes of Death Collaborators. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1459–1544. [Google Scholar] [CrossRef] [PubMed]
- GBD 2019 Diseases and Injuries Collaborators. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef] [PubMed]
- Dudak, F.C.; Boyacı, I.H. Rapid and label-free bacteria detection by surface plasmon resonance (SPR) biosensors. Biotechnol. J. 2009, 4, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Zourob, M.; Elwary, S.; Turner, A. (Eds.) Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems; Springer Science + Business Media, LLC: Berlin, Germany, 2008. [Google Scholar]
- Wang, P.; Sun, H.; Yang, W.; Fang, Y. Optical methods for label-free detection of bacteria. Biosensors 2022, 12, 1171. [Google Scholar] [CrossRef] [PubMed]
- Kaura, B.; Kumar, S.; Kaushik, B.K. Trends, challenges, and advances in optical sensing for pathogenic bacteria detection (PathoBactD). Biosens. Bioelectron. X 2023, 14, 100352. [Google Scholar] [CrossRef]
- Manafi, M. Fluorogenic and chromogenic enzyme substrates in culture media and identification tests. Int. J. Food Microbiol. 1996, 31, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Rantsiou, K.; Alessandria, V.; Urso, R.; Dolci, P.; Cocolin, L. Detection, quantification, and vitality of Listeria monocytogenes in food as determined by quantitative PCR. Int. J. Food Microbiol. 2008, 121, 99–105. [Google Scholar] [CrossRef]
- Hobson, N.S.; Tothill, I.; Turner, A.P.F. Microbial detection. Biosens. Bioelectron. 1996, 11, 455–477. [Google Scholar] [CrossRef]
- Docherty, L.; Adams, M.R.; Patel, P.; McFadden, J. The magnetic immunopolymerase chain reaction assay for the detection of Camplyobacter in milk and poultry. Lett. Appl. Microbiol. 1996, 37, 288–292. [Google Scholar] [CrossRef]
- Yu, L.S.L.; Reed, S.A.; Golden, M.H. Time-resolved fluorescence immunoassay (TRFIA) for the detection of Escherichia coli O157:H7 in apple cider. J. Microbiol. Meth. 2002, 49, 63–68. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y. Detection of Escherichia coli O157:H7 using immunomagnetic separation and absorbance measurement. J. Microbiol. Meth. 2002, 51, 369–377. [Google Scholar] [CrossRef]
- Su, X.L.; Li, Y. Quantum dot biolabeling coupled with immunomagnetic separation for detection of Escherichia coli O157:H7. Anal. Chem. 2004, 76, 4806–4810. [Google Scholar] [CrossRef] [PubMed]
- Dudak, F.C.; Boyacı, I.H. Enumeration of immunomagnetically captured Escherichia coli in water samples using quantum dot-labeled antibodies. J. Rapid Methods Autom. Microbiol. 2008, 16, 122–131. [Google Scholar] [CrossRef]
- Lazcka, O.; Del Campo, F.J.; Munoz, F.X. Pathogen detection: A perspective of traditional methods and biosensors. Biosens. Bioelectron. 2007, 22, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, B.V.; Cordeiro, T.A.R.; e Freitas, G.R.O.; Ferreira, L.F.; Franco, D.L. Biosensors for the detection of respiratory viruses: A review. Talanta Open 2020, 2, 100007. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Dkhar, D.S.; Chandra, P.; Azad, U.P. Nanobiosensors design using 2D materials: Implementation in infectious and fatal disease diagnosis. Biosensors 2023, 13, 166. [Google Scholar] [CrossRef] [PubMed]
- Purohit, B.; Vernekar, P.R.; Shetti, N.P.; Chandra, P. Biosensor nanoengineering: Design, operation, and implementation for biomolecular analysis. Sens. Int. 2020, 1, 100040. [Google Scholar] [CrossRef]
- Liu, X.; Marrakchi, M.; Xu, D.; Dong, H.; Andreescu, S. Biosensors based on modularly designed synthetic peptides for recognition, detection and live/dead differentiation of pathogenic bacteria. Biosens. Bioelectron. 2016, 80, 9–16. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, H.; Hu, Z.; Yu, G.; Yang, D.; Zhao, J. Label and label-free based surface-enhanced Raman scattering for pathogen bacteria detection: A review. Biosens. Bioelectron. 2017, 94, 131–140. [Google Scholar] [CrossRef]
- Fan, X.; White, I.M.; Shopova, S.I.; Zhu, H.; Suter, J.D.; Sun, Y. Sensitive optical biosensors for unlabeled targets: A review. Anal. Chim. Acta 2008, 620, 8–26. [Google Scholar] [CrossRef]
- Wu, X.; Xu, C.; Tripp, R.A.; Huang, Y.W.; Zhao, Y. Detection and differentiation of foodborne pathogenic bacteria in mung bean sprouts using field deployable label-free SERS devices. Analyst 2013, 138, 3005–3012. [Google Scholar] [CrossRef] [PubMed]
- Marks, H.; Schechinger, M.; Garza, J.; Locke, A.; Coté, G. Surface enhanced Raman spectroscopy (SERS) for in vitro diagnostic testing at the point of care. Nanophotonics 2017, 6, 681–701. [Google Scholar] [CrossRef]
- Kahraman, M.; Mullen, E.R.; Korkmaz, A.; Wachsmann-Hogiu, S. Fundamentals and applications of SERS-based bioanalytical sensing. Nanophotonics 2017, 6, 831–852. [Google Scholar] [CrossRef]
- Wu, R.; Ma, Y.; Pan, J.; Lee, S.H.; Liu, J.; Zhu, H.; Gu, R.; Shea, K.J.; Pan, G. Efficient capture, rapid killing and ultrasensitive detection of bacteria by a nano-decorated multi-functional electrode sensor. Biosens. Bioelectron. 2018, 101, 52–59. [Google Scholar] [CrossRef]
- Kimuda, S.G.; Biraro, I.A.; Bagaya, B.S.; Raynes, J.G.; Cose, S. Characterising antibody avidity in individuals of varied Mycobacterium tuberculosis infection status using surface plasmon resonance. PLoS ONE 2018, 13, e0205102. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.X.; Sun, Y.; Li, X.; Wang, L.; Xu, Y.; He, L.L.; Li, G.L. Recent advances in dual recognition based surface enhanced Raman scattering for pathogenic bacteria detection: A review. Anal. Chim. Acta 2021, 1157, 338279. [Google Scholar] [CrossRef] [PubMed]
- Andryukov, B.G.; Lyapun, I.N.; Matosova, E.V.; Somova, L.M. Biosensor technologies in medicine: From detection of biochemical markers to research into molecular targets (Review). Mod. Technol. Med. 2020, 12, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Qi, P.; Zhang, D. A simple, rapid and cost-effective colorimetric assay based on the 4-mercaptophenylboronic acid functionalized silver nanoparticles for bacteria monitoring. Sens. Actuators B Chem 2018, 260, 983–989. [Google Scholar] [CrossRef]
- Hong, B.; Li, Y.; Wang, W.; Ma, Y.; Wang, J. Separation and colorimetric detection of Escherichia coli by phage tail fiber protein combined with nano-magnetic beads. Mikrochim. Acta 2023, 190, 202. [Google Scholar] [CrossRef]
- You, S.M.; Jeong, K.B.; Luo, K.; Park, J.S.; Park, J.W.; Kim, Y.R. Paper-based colorimetric detection of pathogenic bacteria in food through magnetic separation and enzyme-mediated signal amplification on paper disc. Anal. Chim. Acta 2021, 1151, 338252. [Google Scholar] [CrossRef]
- Valeur, B.; Berberan-Santos, M.N. Molecular Fluorescence Principles and Applications; Wiley: Weinheim, Germany, 2012; ISBN 9783527328376. [Google Scholar] [CrossRef]
- Fluorescence in Biological Research. Available online: https://ru.wikipedia.org/wiki/%D0%A4%D0%BB%D1%83%D0%BE%D1%80%D0%B5%D1%81%D1%86%D0%B5%D0%BD%D1%86%D0%B8%D1%8F_%D0%B2_%D0%B1%D0%B8%D0%BE%D0%BB%D0%BE%D0%B3%D0%B8%D1%87%D0%B5%D1%81%D0%BA%D0%B8%D1%85_%D0%B8%D1%81%D1%81%D0%BB%D0%B5%D0%B4%D0%BE%D0%B2%D0%B0%D0%BD%D0%B8%D1%8F%D1%85 (accessed on 19 November 2023).
- Chen, L.Y.; Hwang, E.; Zhang, J. Fluorescent nanobiosensors for sensing glucose. Sensors 2018, 18, 1440. [Google Scholar] [CrossRef]
- Fernando, K.A.S.; Sahu, S.; Liu, Y.M.; Lewis, W.K.; Guliants, E.A.; Jafariyan, A.; Wang, P.; Bunker, C.E.; Sun, Y.P. Carbon quantum dots and applications in photocatalytic energy conversion. ACS Appl. Mater. Infer. 2015, 7, 8363–8376. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Lv, F.; Qi, R.; Senthilkumar, T.; Zhao, H.; Chen, Y.; Li, L.; Wang, S. Förster resonance energy transfer mediated rapid and synergistic discrimination of bacteria over fungi using a cationic conjugated glycopolymer. ACS Appl. Bio Mater. 2020, 3, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.H.; Zhang, J.; Liu, J.L.; Zhang, Y. Recent progress of rare-earth doped upconversion nanoparticles: Synthesis, optimization, and applications. Adv. Sci. 2019, 6, 1901358. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, M.; Li, X.; Fan, Y.; Li, J.; Lu, K.; Wen, H.; Ren, J. Recent advances of fluorescent sensors for bacteria detection-A review. Talanta 2023, 254, 124133. [Google Scholar] [CrossRef] [PubMed]
- Kourti, D.; Angelopoulou, M.; Petrou, P.; Kakabakos, S. Optical immunosensors for bacteria detection in food matrices. Chemosensors 2023, 11, 430. [Google Scholar] [CrossRef]
- Pligovka, A.; Lazavenka, A.; Turavets, U.; Hoha, A.; Salerno, M. Two-level 3D column-like nanofilms with hexagonally–packed tantalum fabricated via anodizing of Al/Nb and Al/Ta layers—A potential nano-optical biosensor. Materials 2023, 16, 993. [Google Scholar] [CrossRef] [PubMed]
- Zuo, W.; Liang, L.; Ye, F.; Zhao, S. An integrated platform for label-free fluorescence detection and inactivation of bacteria based on boric acid functionalized Zr-MOF. Sens. Actuators B Chem. 2021, 345, 130345. [Google Scholar] [CrossRef]
- Sun, X.; Wang, Y.; Zhang, L.; Liu, S.; Zhang, M.; Wang, J.; Ning, B.; Peng, Y.; He, J.; Hu, Y.; et al. CRISPR-Cas9 triggered two-step isothermal amplification method for E. coli O157:H7 detection based on a metal–organic framework platform. Anal. Chem. 2020, 92, 3032–3041. [Google Scholar] [CrossRef]
- Gupta, A.; Garg, M.; Singh, S.; Deep, A.; Sharma, A.L. Highly sensitive optical detection of Escherichia coli using terbium-based metal–organic framework. ACS Appl. Mater. Interfaces 2020, 12, 48198–48205. [Google Scholar] [CrossRef]
- Jenie, S.N.A.; Kusumastuti, Y.; Krismastuti, F.S.H.; Untoro, Y.M.; Dewi, R.T.; Udin, L.Z.; Artanti, N. Rapid fluorescence quenching detection of Escherichia coli using natural silica-based nanoparticles. Sensors 2021, 21, 881. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Yang, X.; Zhang, B.; Cheng, S.; Han, H.; Jina, Q.; Wang, C.; Xiao, R. Sensitive detection of Escherichia coli O157:H7 and Salmonella typhimurium in food samples using two-channel fluorescence lateral flow assay with liquid Si@quantum dot. Food Chem. 2021, 363, 130400. [Google Scholar] [CrossRef]
- Nandi, S.; Ritenberga, M.; Jelinek, R. Bacterial detection with amphiphilic carbon dots. Analyst 2015, 140, 4232. [Google Scholar] [CrossRef] [PubMed]
- Bruce, J.A.; Clapper, J.C. Conjugation of carboxylated graphene quantum dots with cecropin P1 for bacterial biosensing applications. ACS Omega 2020, 5, 26583–26591. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Yang, D.; Yin, X. Rapid detection of three common bacteria based on fluorescence spectroscopy. Sensors 2022, 22, 1168. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Zheng, L.; Wang, S.; Huang, F.; Liu, Y.; Jiang, H.; Lin, J. A microfluidic biosensor for rapid and automatic detection of Salmonella using metal-organic framework and Raspberry Pi. Biosens. Bioelectron. 2021, 178, 113020. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Huang, X.; Li, Z.; Shi, J.; Zhai, X.; Hu, X.; Nini, L.; Zou, X. Rapid and highly sensitive detection of Salmonella typhimurium in lettuce by using magnetic fluorescent nanoparticles. Anal. Methods 2020, 12, 5861–5868. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.; Qiao, Y.; Jiang, N.; Li, C.; Zhu, X.; Li, W.; Cai, Q. Water-soluble ZnCuInSe quantum dots for bacterial classification, detection, and imaging. Anal. Bioanal. Chem. 2020, 412, 8379–8389. [Google Scholar] [CrossRef]
- Sur, V.P.; Mazumdar, A.; Ashrafi, A.; Mukherjee, A.; Milosavljevic, V.; Michalkova, H.; Kopel, P.; Richtera, L.; Moulick, A. A novel biocompatible titanium–gadolinium quantum dot as a bacterial detecting agent with high antibacterial activity. Nanomaterials 2020, 10, 778. [Google Scholar] [CrossRef]
- Ouyang, Q.; Yang, Y.; Ali, S.; Wang, L.; Li, H.; Chen, Q. Upconversion nanoparticles-based FRET system for sensitive detection of Staphylococcus aureus. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 255, 119734. [Google Scholar] [CrossRef]
- Nißler, R.; Bader, O.; Dohmen, M.; Walter, S.G.; Noll, C.; Selvaggio, G.; Groß, U.; Kruss, S. Remote near infrared identification of pathogens with multiplexed nanosensors. Nat. Commun. 2020, 11, 5995. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhong, Y.; Li, W.; Wang, W.; Li, C.; Wang, A.; Yan, H.; Wan, Y.; Li, J. Fluorescent and opt-electric recording bacterial identification device for ultrasensitive and specific detection of microbials. ACS Sens. 2021, 6, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.A.; Park, S.Y.; Cha, Y.; Gopala, L.; Lee, M.H. Strategies of detecting bacteria using fluorescence-based dyes. Front. Chem. 2021, 9, 743923. [Google Scholar] [CrossRef] [PubMed]
- Kabiraz, M.P.; Majumdar, P.R.; Mahmud, M.M.C.; Bhowmik, S.; Ali, A. Conventional and advanced detection techniques of foodborne pathogens: A comprehensive review. Heliyon 2023, 9, e15482. [Google Scholar] [CrossRef] [PubMed]
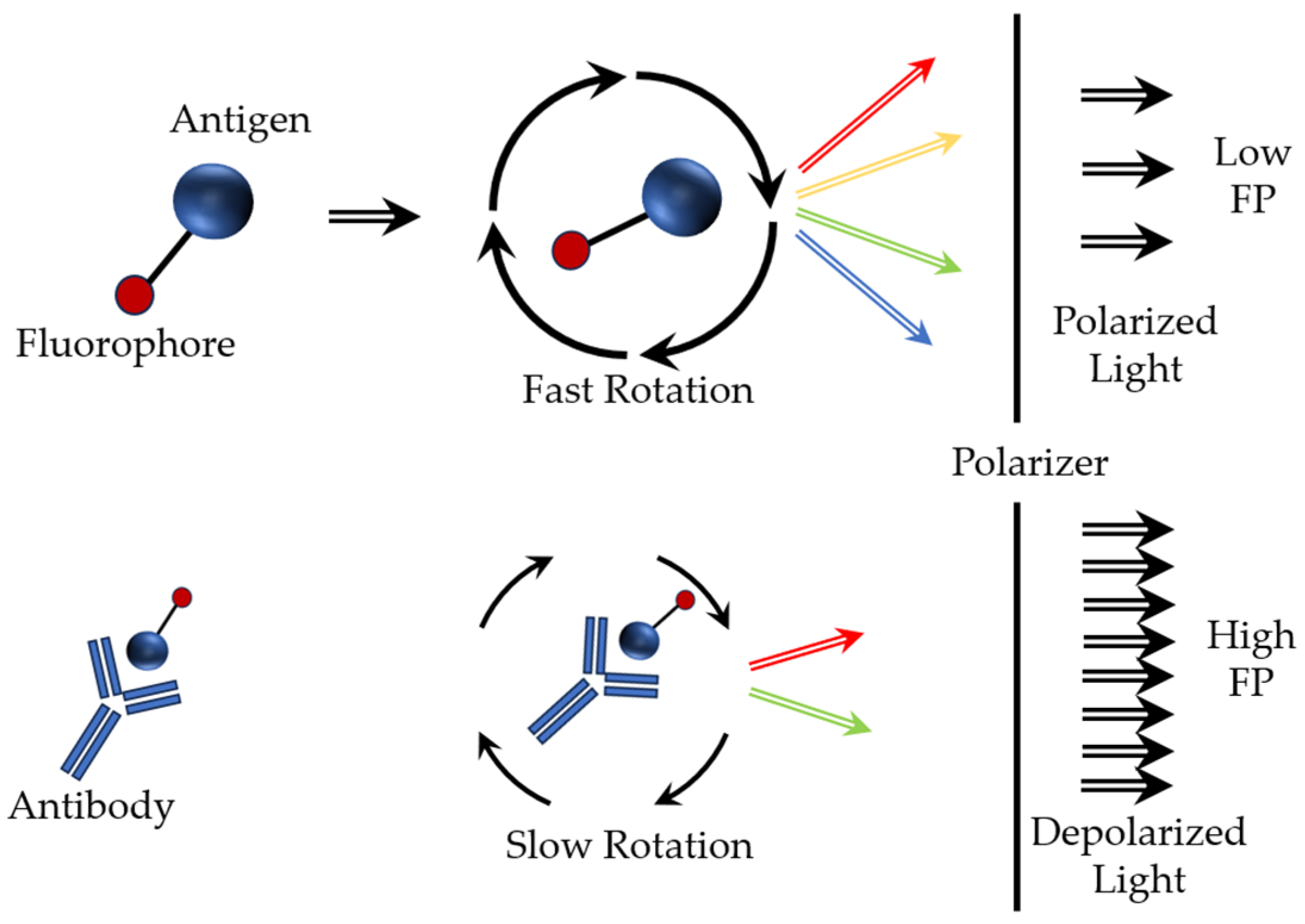
- Zhang, H.; Yang, S.; De Ruyck, K.; Beloglazova, N.; Eremin, S.A.; De Saeger, S.; Zhang, S.; Shen, J.; Wang, Z. Fluorescence polarization assays for chemical contaminants in food and environmental analyses. Trends Anal. Chem. 2019, 114, 293–313. [Google Scholar] [CrossRef]
- Hendrickson, O.D.; Taranova, N.A.; Zherdev, A.V.; Dzantiev, B.B.; Eremin, S.A. Fluorescence polarization-based bioassays: New horizons. Sensors 2020, 20, 7132. [Google Scholar] [CrossRef] [PubMed]
- Javed, R.; Narang, D.; Kaur, P.; Chandra, M.; Filia, G.; Singh, S.T. A fluorescence polarization assay using recombinant protein ESAT-6 for the detection of antibodies against pathogenic. Mycobacterium bovis in bovine. Iran. J. Vet. Res. Shiraz Univ. 2022, 23, 204–209. [Google Scholar] [CrossRef]
- Sotolongo-Rodríguez, D.; Gomez-Flores, R.; Navarro-Soto, M.C.; Arellano-Reynoso, B.; Tamez-Guerra, P.; Ramírez-Pfeiffer, C. Evaluation of the fluorescence polarization assay for the diagnosis of brucellosis in goat milk. Vet. Sci. 2022, 9, 303. [Google Scholar] [CrossRef]
- Sentry 300. The Most Accurate and Advanced Fluorescence Polarization Reader. Available online: https://ellielab.com/sentry-300/ (accessed on 19 November 2023).
- Roda, A.; Mirasoli, M.; Michelini, E.; Di Fusco, M.; Zangheri, M.; Cevenini, L.; Simoni, P. Progress in chemical luminescence-based biosensors: A critical review. Biosens. Bioelectron. 2016, 76, 164–179. [Google Scholar] [CrossRef]
- Calabretta, M.M.; Lopreside, A.; Montali, L.; Zangheri, M.; Evangelisti, L.; D’Elia, M.; Michelini, E. Portable light detectors for bioluminescence biosensing applications: A comprehensive review from the analytical chemist’s perspective. Anal. Chim. Acta 2022, 1200, 339583. [Google Scholar] [CrossRef]
- Anderson, W.A.; Dippel, A.B.; Maiden, M.M.; Waters, C.M.; Hammond, M.C. Chemiluminescent sensors for quantitation of the bacterial second messenger cyclic di-GMP. Methods Enzymol. 2020, 640, 83–104. [Google Scholar] [CrossRef] [PubMed]
- Pour, S.R.S.; Calabria, D.; Emamiamin, A.; Lazzarini, E.; Pace, A.; Guardigli, M.; Zangheri, M.; Mirasoli, M. Electrochemical vs. optical biosensors for point-of-care applications: A critical Review. Chemosensors 2023, 11, 546. [Google Scholar] [CrossRef]
- Shen, J.; Zhou, T.; Huang, R. Recent advances in electrochemiluminescence sensors for pathogenic bacteria detection. Micromachines 2019, 10, 532. [Google Scholar] [CrossRef] [PubMed]
- Deisingh, A.K.; Thompson, M. Biosensors for the detection of bacteria. Can. J. Microbiol. 2004, 50, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Utkin, D.V.; Ossina, N.A.; Kouklev, V.E.; Erokhin, P.S.; Scherbakova, S.A.; Kutyrev, V.V. Biosensors: Current state and prospects of applying in laboratory diagnostics of particularly dangerous infectious diseases. Probl. Part. Danger. Infect. 2009, 102, 11–14. [Google Scholar] [CrossRef]
- Homola, J.; Vaisocherová, H.; Dostálek, J.; Piliarik, M. Multi-analyte surface plasmon resonance biosensing. Methods 2005, 37, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Piliarik, M.; Vaisocherová, H.; Homola, J. Surface plasmon resonance biosensing. In Methods in Molecular Biology: Biosensors and Biodetection; Rasooly, A., Herold, K.E., Eds.; Humana Press: Totowa, NJ, USA, 2009; Volume 503, pp. 65–88. [Google Scholar] [CrossRef]
- Homola, J. (Ed.) Surface Plasmon Resonance Based Sensors; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar] [CrossRef]
- Raether, H. Surface-Plasmons on Smooth and Rough Surfaces and on Gratings; Springer Tracts in Modern Physics; Springer: Berlin/Heidelberg, Germany, 1988; Volume 111, pp. 1–133. [Google Scholar]
- Lahiri, B.; Khokhar, A.Z.; De La Rue, R.M.; McMeekin, S.G.; Johnson, N.P. Asymmetric split ring resonators for optical sensing of organic materials. Opt. Express 2009, 17, 1107. [Google Scholar] [CrossRef]
- De Angelis, F.; Patrini, M.; Das, G.; Maksymov, I.; Galli, M.; Businaro, L.; Andreani, L.C.; Di Fabrizio, E. A hybrid plasmonic–photonic nanodevice for label-free detection of a few molecules. Nano Lett. 2008, 8, 2321–2327. [Google Scholar] [CrossRef]
- Xavier, J.; Vincent, S.; Meder, F.; Vollmer, F. Advances in optoplasmonic sensors—Combining optical nano/microcavities and photonic crystals with plasmonic nanostructures and nanoparticles. Nanophotonics 2018, 7, 1–38. [Google Scholar] [CrossRef]
- Yalçin, A.; Popat, K.C.; Aldridge, J.C.; Desai, T.A.; Hryniewicz, J.; Chbouki, N.; Little, B.E.; King, O.; Van, V.; Chu, S.; et al. Optical sensing of biomolecules using microring resonators. IEEE J. Sel. Top. Quantum Electron. 2006, 12, 148–154. [Google Scholar] [CrossRef]
- Xu, D.X.; Densmore, A.; Delâge, A.; Waldron, P.; McKinnon, R.; Janz, S.; Lapointe, J.; Lopinski, G.; Mischki, T.; Post, E.; et al. Folded cavity SOI microring sensors for high sensitivity and real time measurement of biomolecular binding. Opt. Express 2008, 16, 15137–15148. [Google Scholar] [CrossRef]
- Densmore, A.; Vachon, M.; Xu, D.-X.; Janz, S.; Ma, R.; Li, Y.-H.; Lopinski, G.; Delâge, A.; Lapointe, J.; Luebbert, C.C.; et al. Silicon photonic wire biosensor array for multiplexed real-time and label-free molecular detection. Opt. Lett. 2009, 34, 3598–3600. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Long, K.D.; Lu, M.; Chaudhery, V.; Yu, H.; Choi, J.S.; Polans, J.; Zhuo, Y.; Harley, B.A.C.; Cunningham, B.T. Photonic crystal enhanced microscopy for imaging of live cell adhesion. Analyst 2013, 138, 5886–5894. [Google Scholar] [CrossRef]
- Triggs, G.J.; Fischer, M.; Stellinga, D.; Scullion, M.G.; Evans, G.J.O.; Krauss, T.F. Spatial resolution and refractive index contrast of resonant photonic crystal surfaces for biosensing. IEEE Photonics J. 2015, 7, 6801810. [Google Scholar] [CrossRef] [PubMed]
- Nazirizadeh, Y.; Reverey, J.; Geyer, U.; Lemmer, U.; Selhuber-Unkel, C.; Gerken, M. Material-based three-dimensional imaging with nanostructured surfaces. Appl. Phys. Lett. 2013, 102, 011116. [Google Scholar] [CrossRef]
- Wu, H.-Y.; Choi, C.J.; Cunningham, B.T. Plasmonic nanogap-enhanced Raman scattering using a resonant nanodome array. Small 2012, 8, 2878–2885. [Google Scholar] [CrossRef]
- Yazdi, S.H.; White, I.M. A nanoporous optofluidic microsystem for highly sensitive and repeatable surface enhanced Raman spectroscopy detection. Biomicrofluidics 2012, 6, 14105–141059. [Google Scholar] [CrossRef] [PubMed]
- Kundu, J.; Le, F.; Nordlander, P.; Halas, N.J. Surface enhanced infrared absorption (SEIRA) spectroscopy on nanoshell aggregate substrates. Chem. Phys. Lett. 2008, 452, 115–119. [Google Scholar] [CrossRef]
- Bukasov, R.; Shumaker-Parry, J.S. Silver nanocrescents with infrared plasmonic properties as tunable substrates for surface enhanced infrared absorption spectroscopy. Anal. Chem. 2009, 81, 4531–4535. [Google Scholar] [CrossRef]
- Tittl, A.; Leitis, A.; Liu, M.; Yesilkoy, F.; Choi, D.-Y.; Neshev, D.N.; Kivshar, Y.S.; Altug, H. Imaging-based molecular barcoding with pixelated dielectric metasurfaces. Science 2018, 360, 1105–1109. [Google Scholar] [CrossRef]
- Mathias, P.C.; Wu, H.-Y.; Cunningham, B.T. Employing two distinct photonic crystal resonances to improve fluorescence enhancement. Appl. Phys. Lett. 2009, 95, 021111. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-H.; Cho, Y.-W.; Kim, T.-H. Recent advances in surface plasmon resonance sensors for sensitive optical detection of pathogens. Biosensors 2022, 12, 180. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Irudayaraj, J.; Ryan, T. A mixed self-assembled monolayer-based surface plasmon immunosensor for detection of E. coli O157:H7. Biosens. Bioelectron. 2006, 21, 998–1006. [Google Scholar] [CrossRef] [PubMed]
- Homola, J.; Dostálek, J.; Chen, S.F.; Rasooly, A.; Jiang, S.Y.; and Yee, S.S. Spectral surface plasmon resonance biosensor for detection of staphylococcal enterotoxin B in milk. Int. J. Food Microbiol. 2002, 75, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, M.; Nomura, Y.; Zhang, W.; Sakino, M.; Lee, K.H.; Ikebukuro, K.; Karube, I. Simple and rapid detection method using surface plasmon resonance for dioxins, polychlorinated biphenylx and atrazine. Anal. Chim. Acta 2001, 434, 223–230. [Google Scholar] [CrossRef]
- Mazumdar, S.D.; Hartmann, M.; Kampfer, P.; Keusgen, M. Rapid method for detection of Salmonella in milk by surface plasmon resonance (SPR). Biosens. Bioelectron. 2007, 22, 2040–2046. [Google Scholar] [CrossRef]
- Skottrup, P.; Nicolaisen, M.; Justesen, A.F. Rapid determination of Phytophthora infestans sporangia using a surface plasmon resonance immunosensor. J. Microbiol. Meth. 2007, 68, 507–515. [Google Scholar] [CrossRef]
- Skottrup, P.; Hearty, S.; Frøkiaer, H.; Leonard, P.; Hejgaard, J.; O’Kennedy, R.; Nicolaisen, M.; Justesen, A.F. Detection of fungal spores using a generic surface plasmon resonance immunoassay. Biosens. Bioelectron. 2007, 22, 2724–2729. [Google Scholar] [CrossRef]
- Handa, H.; Gurczynski, S.; Jackson, M.P.; Auner, G.; Walker, J.; Mao, G. Recognition of Salmonella typhimurium by immobilized phage P22 monolayers. Surf. Sci. 2008, 602, 1392–1400. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Sorokulova, I.B.; Vodyanoy, V.J.; Simonian, A.L. Lytic phage as a specific and selective probe for detection of Staphylococcus aureus—A surface plasmon resonance spectroscopic study. Biosens. Bioelectron. 2007, 22, 948–955. [Google Scholar] [CrossRef]
- Nanduri, V.; Bhunia, A.K.; Tu, S.I.; Paoli, G.C.; Brewster, J.D. SPR biosensor for the detection of L. monocytogenes using phage-displayed antibody. Biosens. Bioelectron. 2007, 23, 248–252. [Google Scholar] [CrossRef]
- Idil, N.; Bakhshpour, M.; Perçin, I.; Mattiasson, B. Whole cell recognition of Staphylococcus aureus using biomimetic SPR sensors. Biosensors 2021, 11, 140. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Rushworth, J.V.; Hirst, N.A.; Millner, P.A. Biosensors for whole-cell bacterial detection. Clin. Microbiol. Rev. 2014, 27, 631–646. [Google Scholar] [CrossRef] [PubMed]
- David, S.; Polonschii, C.; Luculescu, C.; Gheorghiu, M.; Gáspár, S.; Gheorghiu, E. Magneto-plasmonic biosensor with enhanced analytical response and stability. Biosens. Bioelectron. 2015, 63, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Uniyal, A.; Srivastava, G.; Pal, A.; Taya, S.; Muduli, A. Recent advances in optical biosensors for sensing applications: A review. Plasmonics 2023, 18, 735–750. [Google Scholar] [CrossRef]
- Chen, K.-L.; Yang, Z.-Y.; and Lin, C.-W. A magneto-optical biochip for rapid assay based on the Cotton–Mouton effect of γ-Fe2O3@ Au core/shell nanoparticles. J. Nanobiotechnol. 2021, 19, 301. [Google Scholar] [CrossRef] [PubMed]
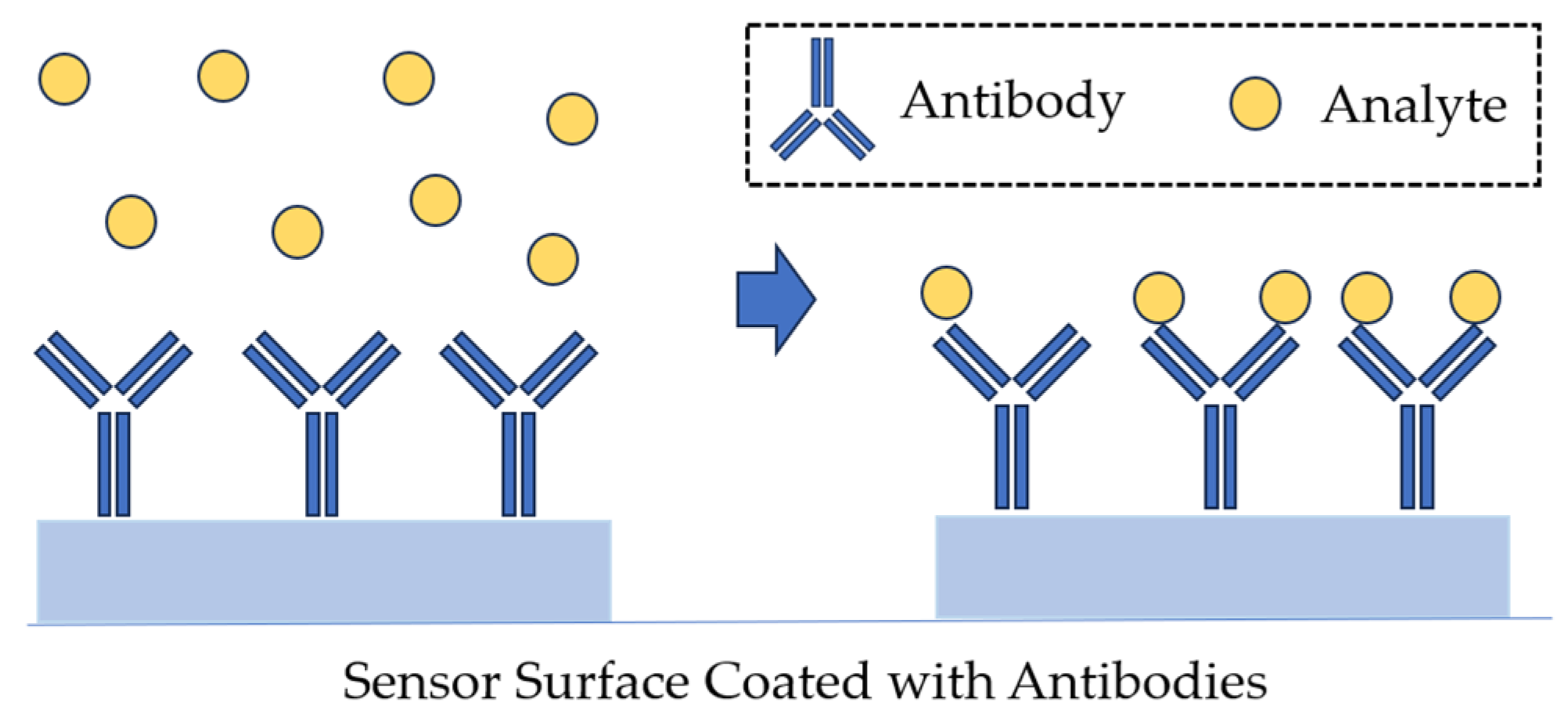
- Banada, P.P.; Bhunia, A.K. Antibodies and immunoassays for detection of bacterial pathogens. In Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems; Zourob, M., Elwary, S., Turner, A., Eds.; Springer Science + Business Media, LLC: Berlin, Germany, 2008; Chapter 21. [Google Scholar]
- Anderson, G.P.; King, K.D.; Gaffney, K.L.; Johnson, L.H. Multi-analyte interrogation using the fiber optic biosensor. Biosens. Bioelectron. 2000, 14, 771–777. [Google Scholar] [CrossRef]
- Geng, T.; Morgan, M.T.; Bhunia, A.K. Detection of low levels of Listeria monocytogenes cells by using a fiber-optic immunosensor. Appl. Environ. Microbiol. 2004, 70, 6138–6146. [Google Scholar] [CrossRef]
- Geng, T.; Uknalis, J.; Tu, S.I.; Bhunia, A.K. Fiber-optic biosensor employing Alexa-fluor conjugated antibody for detection of Escherichia coli O157: H7 from ground beef in four hours. Sensors 2006, 6, 796–807. [Google Scholar] [CrossRef]
- Nanduri, V.; Kim, G.; Morgan, M.T.; Ess, D.; Hahm, B.K.; Kothapalli, A.; Valadez, A.; Geng, T.; Bhunia, A.K. Antibody immobilization on waveguides using a flow-through system shows improved Listeria monocytogenes detection in an automated fiber optic biosensor: RAPTOR™. Sensors 2006, 6, 808–822. [Google Scholar] [CrossRef]
- Taitt, C.R.; Anderson, G.P.; Ligler, S. Evanescent wave fluorescence biosensors. Biosens. Bioelectron. 2005, 20, 2470–2487. [Google Scholar] [CrossRef]
- Kaushik, S.; Tiwari, U.; Nilima, P.S.; Das, B.; Sinha, R.K. Label-free detection of Escherichia coli bacteria by cascaded chirped long period gratings immunosensor. Rev. Sci. Instrum. 2019, 90, 025003. [Google Scholar] [CrossRef]
- Janik, M.; Brzozowska, E.; Czyszczoń, P.; Celebańska, A.; Koba, M.; Gamian, A.; Bock, W.J.; Śmietana, M. Optical fiber aptasensor for label-free bacteria detection in small volumes. Sens. Actuator. B Chem. 2021, 330, 129316. [Google Scholar] [CrossRef]
- Li, H.C.; Leng, Y.K.; Liao, Y.C.; Liu, B.; Luo, W.; Liu, J.; Shi, J.L.; Yuan, J.; Xu, H.Y.; Xiong, Y.H.; et al. Tapered microfiber MZI biosensor for highly sensitive detection of Staphylococcus aureus. IEEE Sens. J. 2022, 22, 5531–5539. [Google Scholar] [CrossRef]
- Anderson, G.P.; Breslin, K.A.; Ligler, F.S. Assay development for a portable fiberoptic biosensor. ASAIO J. 1996, 42, 942–946. [Google Scholar] [CrossRef]
- Anderson, G.P.; Jacoby, M.A.; Ligler, F.S.; King, K.D. Effectiveness of protein A for antibody immobilization for a fiber optical biosensor. Biosens. Bioelectron. 1997, 12, 329–336. [Google Scholar] [CrossRef]
- Cao, L.K.; Anderson, G.P.; Ligler, F.S.; Ezzell, J. Detection of Yersinia pestis fraction 1 antigen with a fiber optic biosensor. J Clin. Microbiol. 1995, 33, 336–341. [Google Scholar] [CrossRef]
- Anderson, G.P.; King, K.D.; Cao, L.K.; Jacoby, M.; Ligler, F.S.; Ezzell, J. Quantifying serum antiplague antibody with a fiber-optic biosensor. Clin. Diagn. Lab. Immunol. 1998, 5, 609–612. [Google Scholar] [CrossRef]
- Lim, D.V.; Simpson, J.M.; Kearns, E.A.; Kramer, M.F. Current and developing technologies for monitoring agents of bioterrorism and biowarfare. Clin. Microbiol. Rev. 2005, 18, 583–607. [Google Scholar] [CrossRef]
- Leskinen, S.D.; Lim, D.V. Rapid ultrafiltration concentration and biosensor detection of Enterococci from large volumes of Florida recreational water. Appl. Environ. Microbiol. 2008, 74, 4792–4798. [Google Scholar] [CrossRef]
- 121 Kishen, A.; John, M.S.; Lim, C.S.; Asundi, A. A fiber optic biosensor (FOBS) to monitor mutans streptococci in human saliva. Biosens. Bioelectron. 2003, 18, 1371–1378. [Google Scholar] [CrossRef]
- Kramer, M.F.; Lim, D.V. A rapid and automated fiber optic-based biosensor assay for the detection of Salmonella in spent irrigation water used in the sprouting of sprout seeds. J. Food Prot. 2004, 67, 46–52. [Google Scholar] [CrossRef]
- Cui, J.; Zhou, M.; Li, Y.; Liang, Z.; Li, Y.; Yu, L.; Liu, Y.; Liang, Y.; Chen, L.; Yang, C. A new optical fiber probe-based quantum dots immunofluorescence biosensors in the detection of Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2021, 11, 665241. [Google Scholar] [CrossRef]
- Tripathi, S.M.; Bock, W.J.; Mikulic, P.; Chinnappan, R.; Ng, A.; Tolba, M.; Zourob, M. Long period grating based biosensor for the detection of Escherichia coli bacteria. Biosens. Bioelectron. 2012, 35, 308–312. [Google Scholar] [CrossRef]
- Horváth, R.; Pedersen, H.C.; Skivesen, N.; Selmeczi, D.; Larsen, N.B. Optical waveguide sensor for on-line monitoring of bacteria. Opt. Lett. 2003, 28, 1233–1235. [Google Scholar] [CrossRef]
- Cooper, I.R.; Meikle, S.T.; Standen, G.; Hanlon, G.W.; Santin, M. The rapid and specific real-time detection of Legionella pneumophila in water samples using optical waveguide lightmode spectroscopy. J. Microbiol. Methods 2009, 78, 40–44. [Google Scholar] [CrossRef]
- Mathesz, A.; Valkai, S.; Újvárosy, A.; Aekbote, B.; Sipos, O.; Stercz, B.; Kocsis, B.; Szabó, D.; Dér, A. Integrated optical biosensor for rapid detection of bacteria. Optofluid Microfluid. Nanofluid. 2015, 2, 15. [Google Scholar] [CrossRef]
- Angelopoulou, M.; Petrou, P.S.; Makarona, E.; Haasnoot, W.; Moser, I.; Jobst, G.; Goustouridis, D.; Lees, M.; Kalatzi, K.; Raptis, I.; et al. Ultrafast multiplexed-allergen detection through advanced fluidic design and monolithic interferometric silicon chips. Anal. Chem. 2018, 90, 9559–9567. [Google Scholar] [CrossRef]
- Petrovszki, D.; Valkai, S.; Gora, E.; Tanner, M.; Bányai, A.; Fürjes, P.; Dér, A. An integrated electro-optical biosensor system for rapid, low-cost detection of bacteria. Microelectron. Eng. 2021, 239–240, 111523. [Google Scholar] [CrossRef]
- Li, M.; Singh, R.; Wang, Y.; Marques, C.; Zhang, B.; Kumar, S. Advances in novel nanomaterial-based optical fiber biosensors—A review. Biosensors 2022, 12, 843. [Google Scholar] [CrossRef]
- Zhang, H.; Zhou, X.; Li, X.; Gong, P.; Zhang, Y.; Zhao, Y. Recent advancements of LSPR fiber-optic biosensing: Combination methods, structure, and prospects. Biosensors 2023, 13, 405. [Google Scholar] [CrossRef]
- Elsherif, M.; Salih, A.E.; Muñoz, M.G.; Alam, F.; AlQattan, B.; Antonysamy, D.S.; Zaki, M.F.; Yetisen, A.K.; Park, S.; Wilkinson, T.D.; et al. Optical fiber sensors: Working principle, applications, and limitations. Adv. Photonics Res. 2022, 3, 210037. [Google Scholar] [CrossRef]
- Wang, X.; Meier, R.; Wolfbeis, O. Fluorescent pH-sensitive nanoparticles in an agarose matrix for imaging of bacterial growth and metabolism. Chem. Int. Ed. 2013, 52, 406. [Google Scholar] [CrossRef] [PubMed]
- Mistlberger, G.; Crespo, G.A.; Bakker, E. Ionophore-based optical sensors. Annu. Rev. Anal. Chem. 2014, 7, 483–512. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.D.; Wolfbeis, O.S. Fiber-optic chemical sensors and biosensors (2008–2012). Anal. Chem. 2013, 85, 487–508. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Bakker, E. Ion selective optodes: From the bulk to the nanoscale. Anal. Bioanal. Chem. 2015, 407, 3899–3910. [Google Scholar] [CrossRef] [PubMed]
- Staal, M.; Borisov, S.; Rickelt, L.; Klimant, I.; KuÈhl, M. Ultrabright planar optodes for luminescence life-time based microscopic imaging of O2 dynamics in biofilms. J. Microbiol. Methods 2011, 85, 67–74. [Google Scholar] [CrossRef]
- Bamsey, M.; Berinstain, A.; Dixon, M. Development of a potassium-selective optode for hydroponic nutrient solution monitoring. Anal. Chim. Acta 2012, 737, 72–82. [Google Scholar] [CrossRef]
- Tiefenthaler, K.; Lukosz, W. Integrated optical switches and glass sensor. Opt. Letter. 1984, 10, 137–139. [Google Scholar] [CrossRef]
- Singh, V.; Kumar, D. Theoretical modeling of a metal-clad planar waveguide based biosensors for the detection of Pseudomonas-like bacteria. Prog. Electromagn. Res. M 2009, 6, 167–184. [Google Scholar] [CrossRef]
- Kozma, P.; Kehl, F.; Ehrentreich, E.; Stamm, C.; Bier, F.F. Integrated planar optical waveguide interferometer biosensors: A comparative review. Biosens. Bioelectron. 2014, 58, 287–307. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Leng, Y.K.; Liu, B.; Liu, J.; Wan, S.P.; Wu, T.; Yuan, J.; Shao, L.; Gu, G.; Fu, Y.Q.; et al. Ultrahigh-sensitivity label-free optical fiber biosensor based on a tapered singlemode- no core-single mode coupler for Staphylococcus aureus detection. Sens. Actuators B 2020, 320, 128283. [Google Scholar] [CrossRef]
- Zaytseva, N.; Miller, W.; Goral, V.; Hepburn, J.; Fang, Y. Microfluidic resonant waveguide grating biosensor system for whole cell sensing. Appl. Phys. Lett. 2011, 98, 1–4. [Google Scholar] [CrossRef]
- Xu, J.; Suarez, D.; Gottfried, D.S. Detection of avian influenza virus using an interferometric biosensor. Anal. Bioanal. Chem. 2007, 389, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Antiochia, R.; Bollella, P.; Favero, G.; Mazzei, F. Nanotechnology based surface plasmon resonance affinity biosensors for in vitro diagnostics. Int. J. Anal. Chem. 2016, 2016, 2981931. [Google Scholar] [CrossRef] [PubMed]
- New Biosensor Designed to Detect Toxins and More Portable Device Can Differentiate among Bacteria, Viruses, and Other Biothreats. Available online: https://discover.lanl.gov/news/0422-pegasus-biosensor/ (accessed on 19 November 2023).
- De La Rue, R.; Herzig, H.P.; Gerken, M. (Eds.) Biomedical Optical Sensors: Differentiators for Winning Technologies; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar] [CrossRef]
- Threm, D.; Nazirizadeh, Y.; Gerken, M. Photonic crystal biosensors towards on-chip integration. J. Biophotonics 2012, 5, 601–616. [Google Scholar] [CrossRef]
- Taya, S.A. Highly sensitive nano-sensor based on a binary photonic crystal for the detection of Mycobacterium tuberculosis bacteria. J. Mater. Sci. Mater Electron. 2021, 32, 28406–28416. [Google Scholar] [CrossRef]
- Inana, H.; Poyraza, M.; Incia, F.; Lifsona, M.A.; Badaya, M.; Cunningham, B.T.; Demirci, U. Photonic crystals: Emerging biosensors and their promise for point-of-care applications. Chem. Soc. Rev. 2017, 46, 366–388. [Google Scholar] [CrossRef]
- Gowdhami, D.; Balaji, V.R.; Murugan, M.; Robinson, S. Gopalkrishna Hegde Photonic crystal based biosensors: An overview. ISSS J. Micro Smart Syst. 2022, 11, 147–167. [Google Scholar] [CrossRef]
- Zhao, X.; Lu, M. (Eds.) Nanophotonics in Biomedical Engineering; Springer Nature Singapore Pte Ltd.: Singapore, 2021; ISBN 978-981-15-6136-8. [Google Scholar] [CrossRef]
- Munson, B.R.; Young, D.F.; Okiishi, T.H. Fundamentals of Fluid Mechanics; John Wiley and Sons: New York, NY, USA, 2002; 568p. [Google Scholar]
- Huang, H.; Yang, X.; Krafczyk, M.; Xi-Yun, L. Rotation of spheroidical paricles in couette flow. J. Fluid Mech. 2012, 692, 369–394. [Google Scholar] [CrossRef]
- Bottcher, G.P.F. Theory of Electrical Polarizability; Academic Press: New York, NY, USA, 1982; Volume 1, p. 480. [Google Scholar]
- Bunin, V.D. Cell Suspension and Structures: Electrooptical Analysis. In Encyclopedia of Surface and Colloid Science; Marcel Dekker: New York, NY, USA, 2002; pp. 1–13. [Google Scholar]
- Stoylov, S.P. Colloid Electro-Optics: Theory and Applications; Academic Press: London, UK, 1991. [Google Scholar]
- Fernandez, R.E.; Rohani, A.; Farmehini, V.; Swami, N.S. Review: Microbial analysis in dielectrophoretic microfluidic systems. Anal. Chim. Acta 2017, 966, 11–33. [Google Scholar] [CrossRef]
- Gimsa, J.; Prüger, B.; Eppmann, P.; Donath, E. Electrorotation of particles measured by dynamic light scattering—A new dielectric spectroscopy technique. Colloids Surf. A Physicochem. Eng. Asp. 1995, 98, 243–249. [Google Scholar] [CrossRef]
- Gimsa, J.; Müller, T.; Schnelle, T.; Fuhr, G. Dielectric spectroscopy of single human erythrocytes at physiological ionic strength: Dispersion of the cytoplasm. Biophys. J. 1996, 71, 495–506. [Google Scholar] [CrossRef]
- Maier, H. Electrorotation of colloidal particles and cells depends on surface charge. Biophys. J. 1997, 73, 1617–1626. [Google Scholar] [CrossRef]
- Gimsa, J.; Wachner, D. A unified resistor-capacitor model for impedance, dielectrophoresis, electrorotation, and induced transmembrane potential. Biophys. J. 1998, 75, 1107–1116. [Google Scholar] [CrossRef]
- Georgieva, R.; Neu, B.; Shilov, V.M.; Knippel, E.; Budde, A.; Latza, R.; Donath, E.; Kiesewetter, H.; Bäumler, H. Low frequency electrorotation of fixed red blood cells. Biophys. J. 1998, 74, 2114–2120. [Google Scholar] [CrossRef]
- Kanevskiy, M.V.; Shardin, V.V.; Bunin, V.D.; Guliy, O.I. Electrophysical sensor systems for in vitro monitoring of bacterial metabolic activity. Biosens. Bioelectron. X 2022, 11, 100179. [Google Scholar] [CrossRef]
- Bahrieh, G.; Erdem, M.; Özgür, E.; Gündüz, U.; Külah, H. Assessment of effects of multi drug resistance on dielectric properties of K562 leukemic cells using electrorotation. RSC Adv. 2014, 4, 44879–44887. [Google Scholar] [CrossRef]
- Huang, L.; Tu, L.; Zeng, X.; Mi, L.; Li, X.; Wang, W. Study of a microfluidic chip integrating single cell trap and 3D stable rotation manipulation. Micromachines 2016, 7, 141. [Google Scholar] [CrossRef]
- Guliy, O.I.; Bunin, V.D.; Korzhenevich, V.I.; Ignatov, O.V. Electro-optical assays for immunoindication of microbial cells. Curr. Immunol. Rev. 2017, 13, 153–162. [Google Scholar] [CrossRef]
- Guliy, O.I.; Bunin, V.D. Electrooptical analysis as sensing system for detection and diagnostics bacterial cells. In Biointerface Engineering: Prospects in Medical Diagnostics and Drug Delivery; Chandra, P., Pandey, L.M., Eds.; Springer Nature Singapore Pte Ltd.: Singapore, 2020; Chapter 11; pp. 233–254. [Google Scholar] [CrossRef]
- Bunin, V.D.; Ignatov, O.V.; Guliy, O.I.; Voloshin, A.G.; Dykman, L.A.; O’Neil, D.; Ivnitski, D. Studies of Listeria monocytogenes-antibody binding using electro-orientation. Biosens. Bioelectron. 2004, 19, 1759–1761. [Google Scholar] [CrossRef] [PubMed]
- Guliy, O.I.; Matora, L.Y.; Burygin, G.L.; Dykman, L.A.; Ostudin, N.A.; Bunin, V.D.; Ignatov, V.V.; Ignatov, O.V. Electrophysical characteristics of Azospirillum brasilense Sp245 during interaction with antibodies to various cell-surface epitopes. Anal. Biochem. 2007, 370, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Guliy, O.I.; Velichko, N.S.; Fedonenko, Y.P.; Bunin, V.D. Use of an electro-optical sensor and phage-displayed miniantibodies for immunodetection of Herbaspirillum. Talanta 2019, 202, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Bunin, V.D.; Angersbach, A.K.; Mehta, S.K.; Guliy, O.I. Use of an optical sensor to directly monitor the metabolic activity of growing bacteria. Talanta 2021, 234, 122590. [Google Scholar] [CrossRef] [PubMed]
- Biosensors Market Size, Global Forecast, Growth Drivers, Opportunities 2030. Available online: https://www.marketsandmarkets.com/Market-Reports/biosensors-market-798.html (accessed on 10 November 2023).
- Herrera-Domínguez, M.; Morales-Luna, G.; Mahlknecht, J.; Cheng, Q.; Aguilar-Hernández, I.; Ornelas-Soto, N. Optical biosensors and their applications for the detection of water pollutants. Biosensors 2023, 13, 370. [Google Scholar] [CrossRef] [PubMed]
- Di Nonno, S.; Ulber, R. Smartphone-based optical analysis systems. Analyst 2021, 146, 2749–2768. [Google Scholar] [CrossRef]
- Veselova, I.A.; Makedonskaya, M.I.; Eremina, O.E.; Shekhovtsova, T.N. Fluorometric and SERS sensor systems for diagnostics and monitoring of catecholamine-dependent diseases. In Macro, Micro and Nano-Biosensors: Potential Applications and Possible Limitations; Rai, M., Reshetilov, A., Plekhanova, Y., Ingle, A.P., Eds.; Springer: Cham, Switzerland, 2021; Chapter 8; pp. 133–160. [Google Scholar] [CrossRef]
- Hayman, R.B. Fiber optic biosensors for bacterial detection. In Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems; Zourob, M., Elwary, S., Turner, A., Eds.; Springer Science + Business Media, LLC: Berlin, Germany, 2008; Chapter 7; pp. 125–137. [Google Scholar]


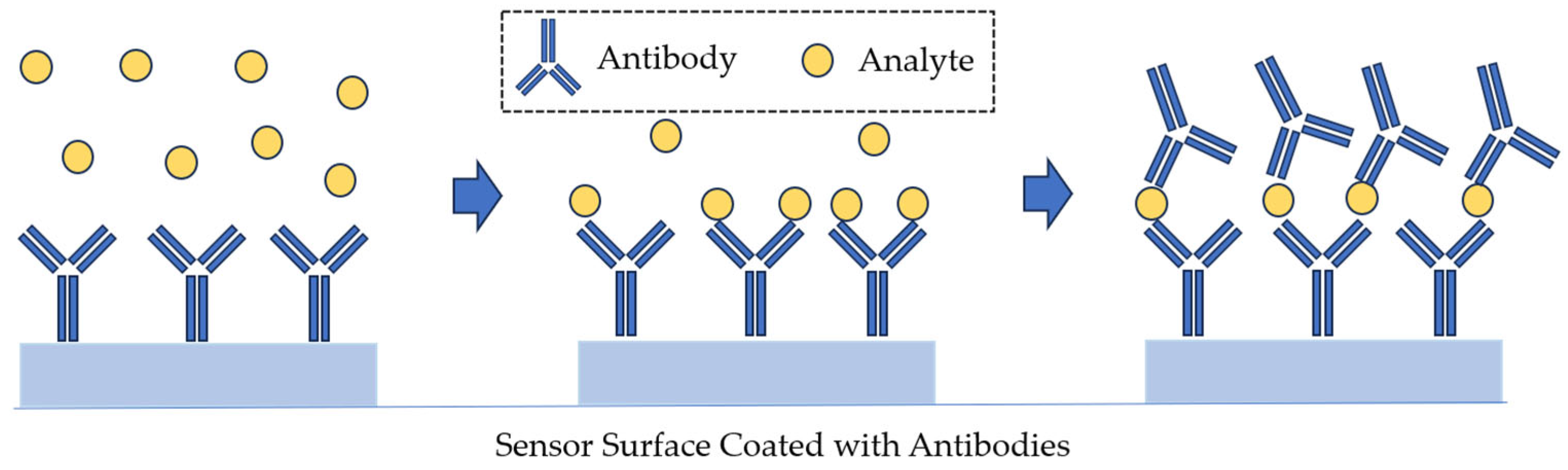
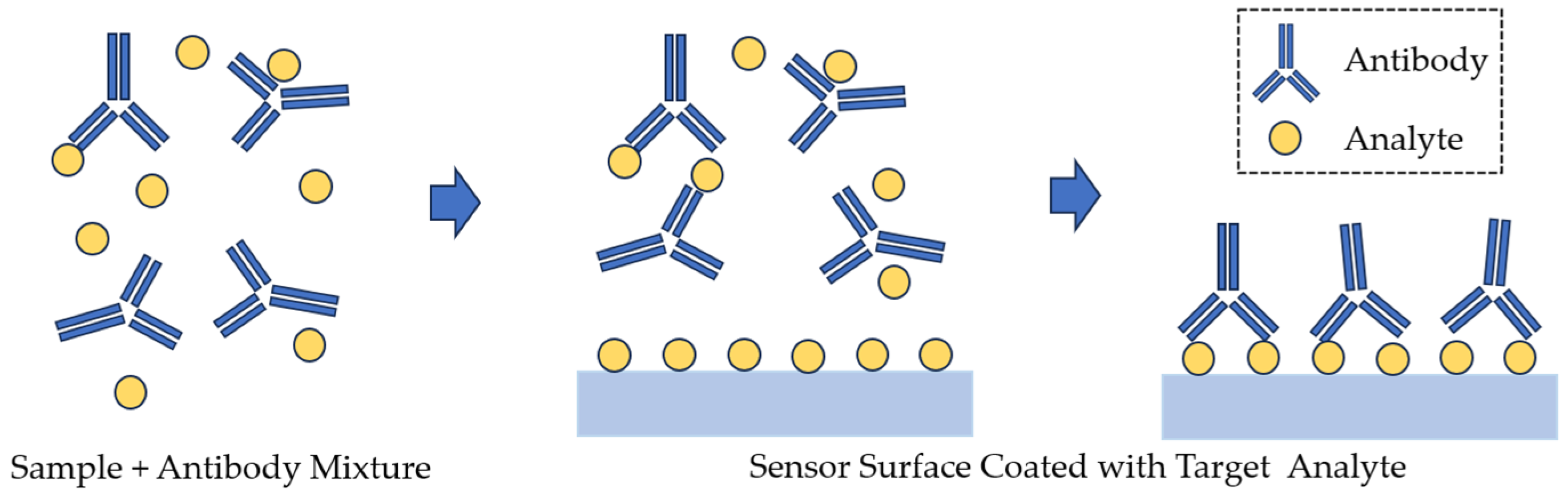

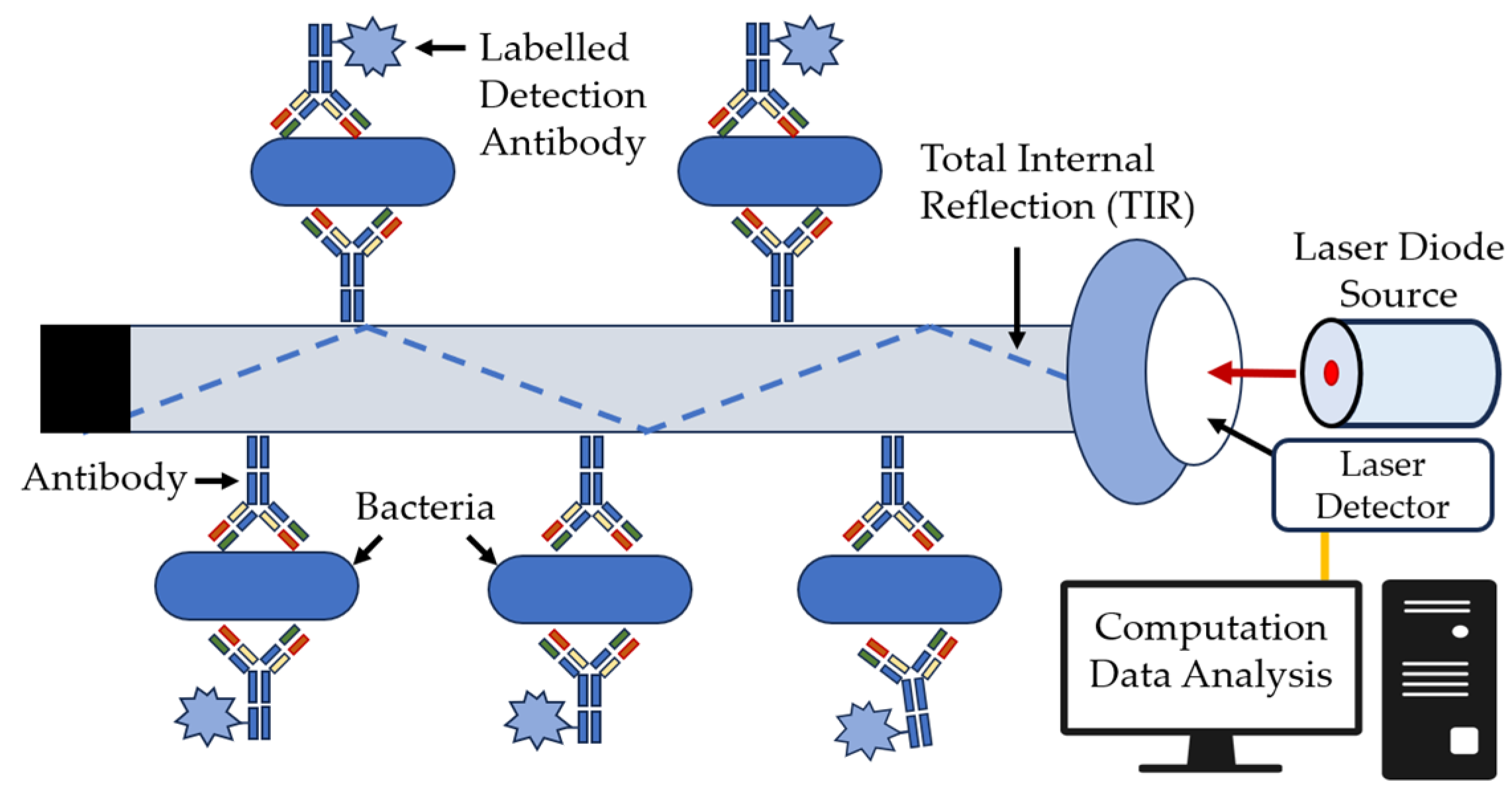

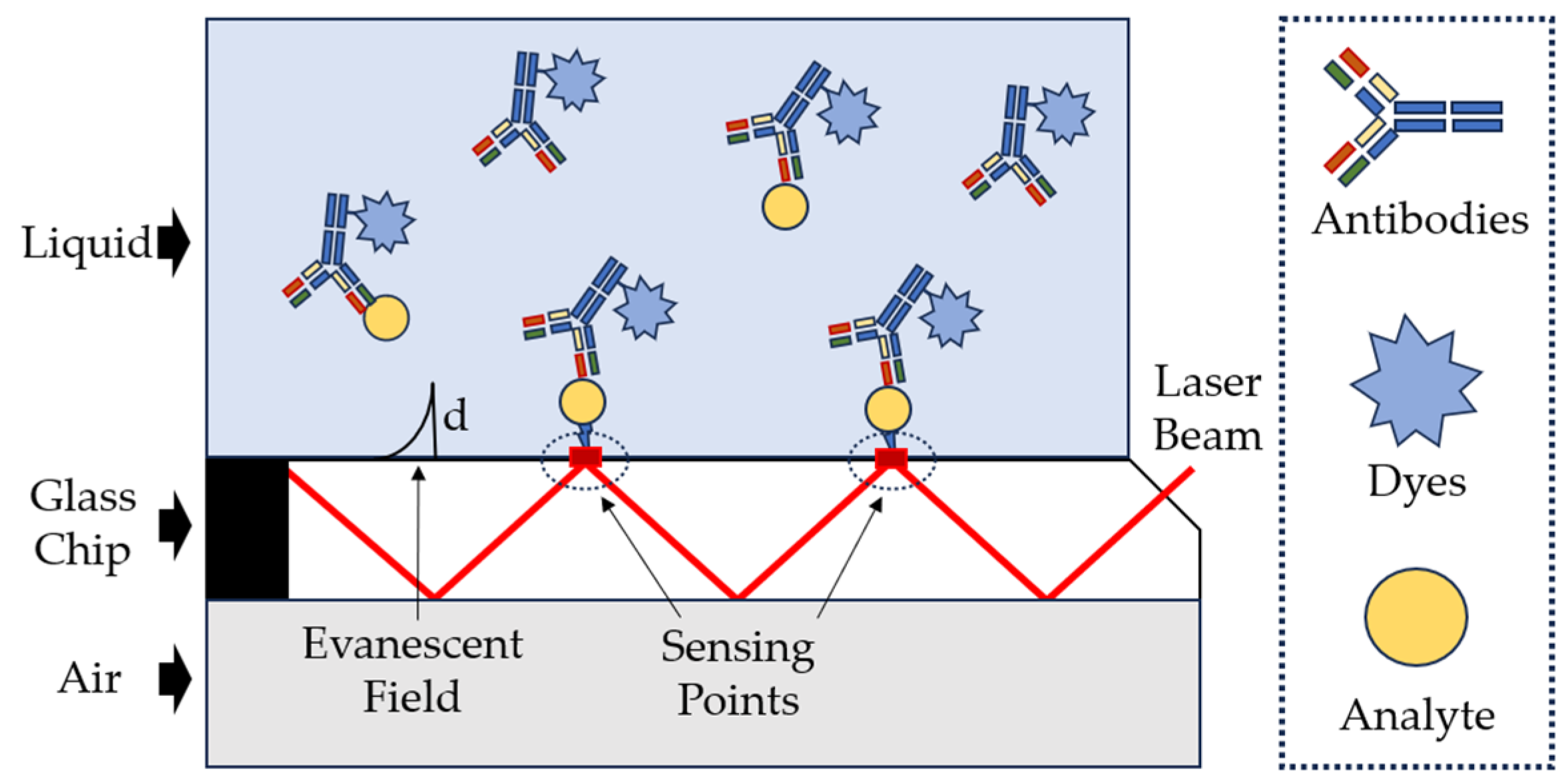
| Technique | Principle | Advantage | Disadvantage |
|---|---|---|---|
| Colorimetry | Determination of the substance concentration by the color intensity of solutions (by the absorption of light by solutions). |
|
|
| Fluorescence | Short-term absorption of a light quantum by a fluorophore (a substance capable of fluorescing), followed by rapid emission of another quantum (which has properties different from the original one). |
|
|
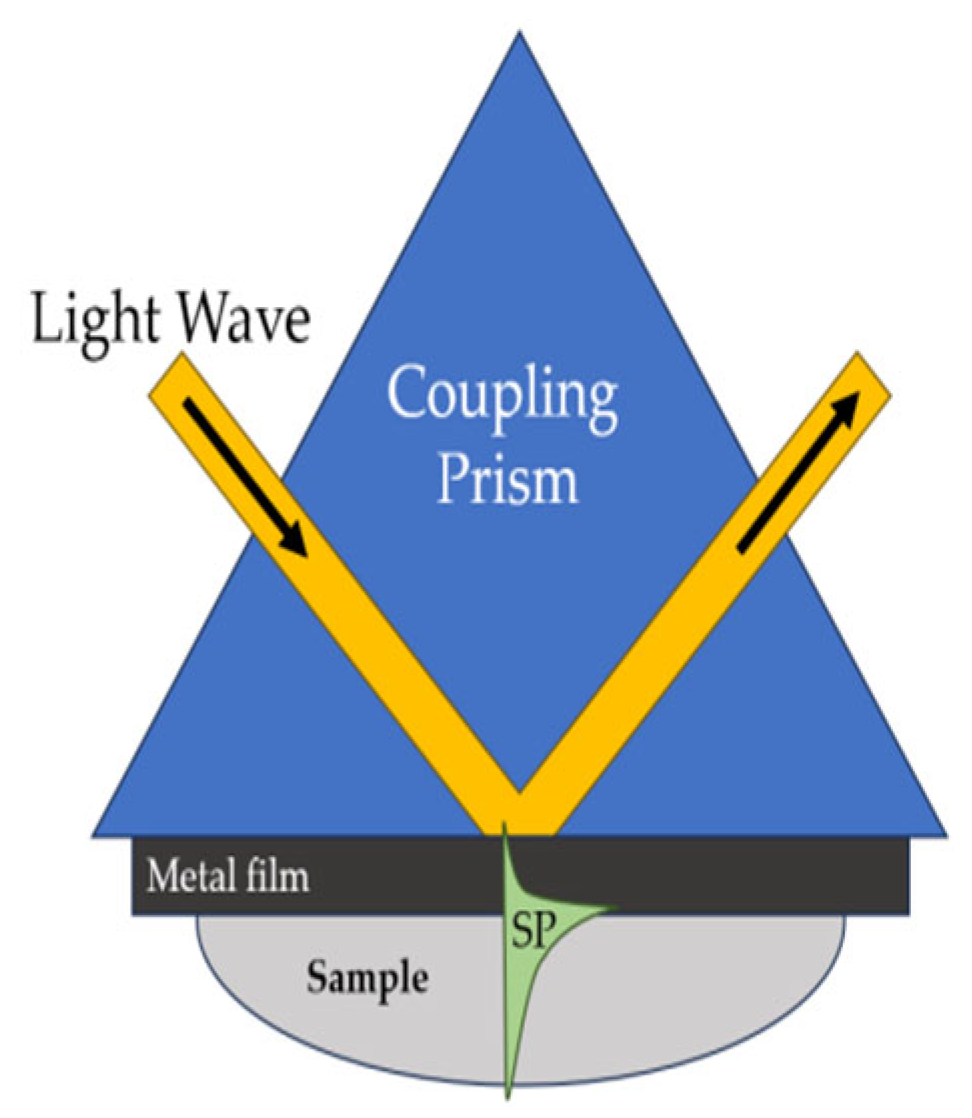
| Surface plasmon resonance (SPR) | An optical phenomenon that can be used to monitor interactions between biomolecules in their natural state in real time. It is based on changes in the direction of propagation of a light flux through an optical fiber or a triangular prism coated with a thin metal film |
|
|
| Surface-enhanced Raman scattering (SERS) | The molecule being measured is adsorbed on the surface of a rough nanometallic material, and the Raman signal from the material being measured is enhanced. |
|
|
| Fiber Optic Sensors | Use of the property of total internal reflection of the wind when it passes through the waveguide and creates a boundary of evanescent waves on the surface of the waveguide. |
|
|
| Optical Sensors Based on Ionophores | A combination of measurements on the basis of vanishing field sensing and optical phase difference. The interference signal produced by interfering fields is detected at the sensor output, and the signal is related to the analyte concentration. |
|
|
| Photonic Crystal Biosensors | The operating principle of photonic crystal waveguides is based on the detection and identification of biological objects by using the spectra of light passing through a hollow core filled with the material under study in the wavelength range 200–1100 nm. |
|
|
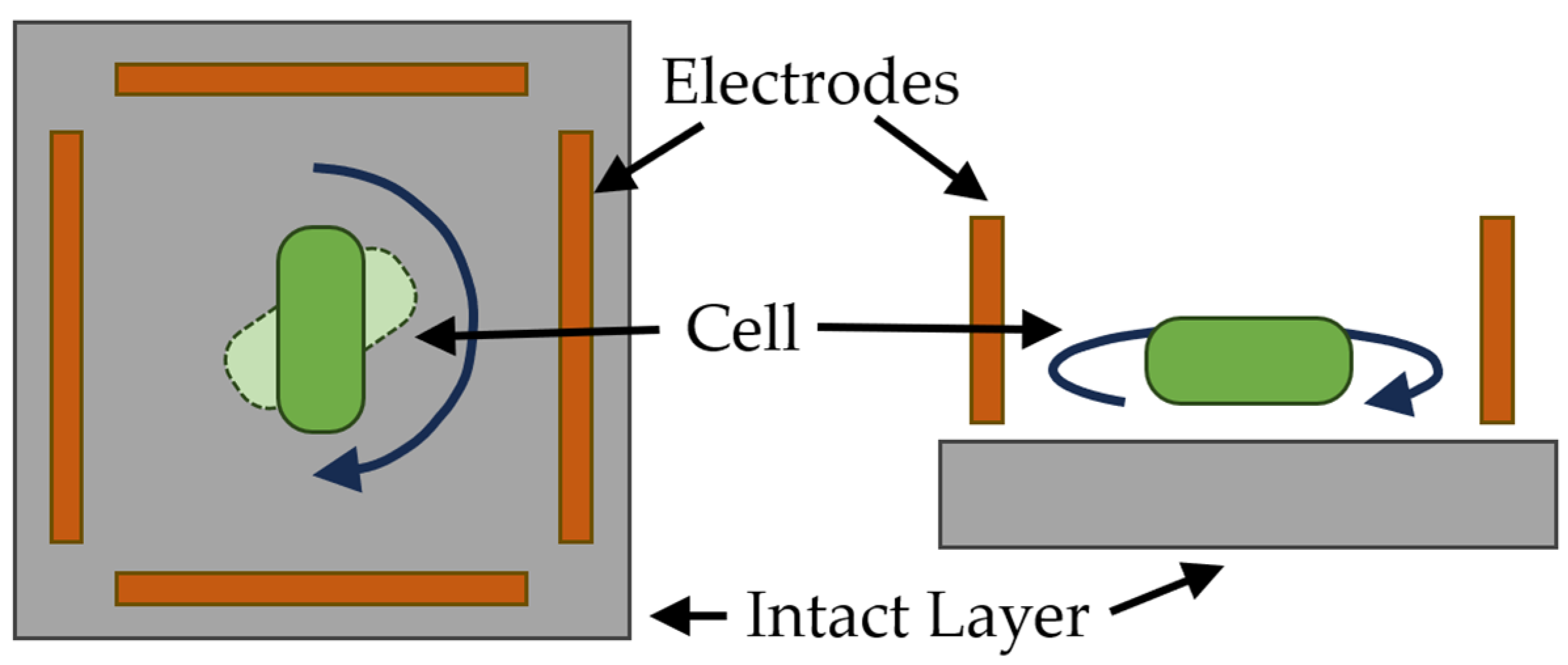
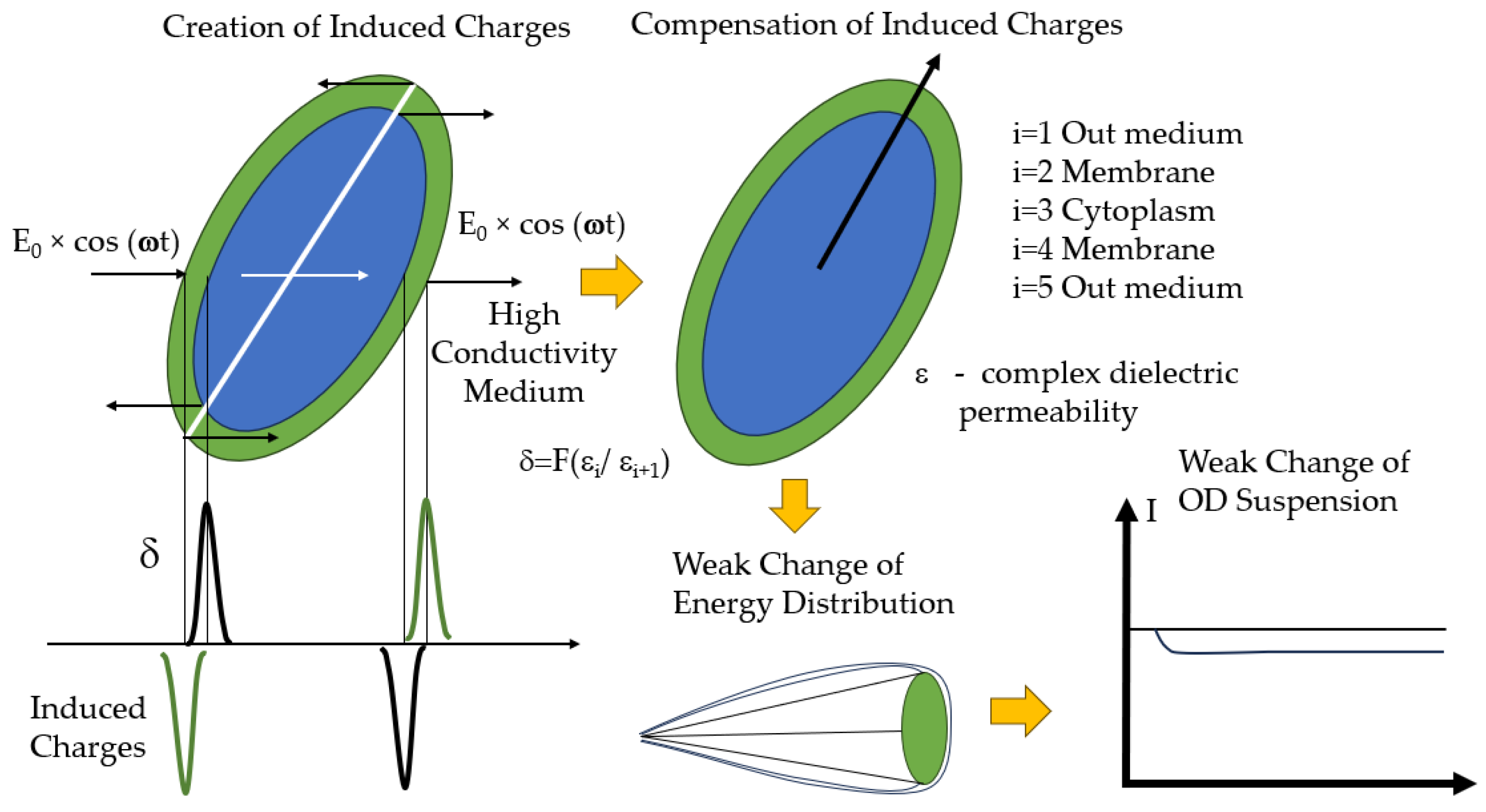
| Optical sensor systems based on measurement of orientational effects | The probing effect of an electrical field causes electrical charges to appear at the boundaries of contact between cellular structures. Their magnitude and sign depend on the complex dielectric properties of the adjacent cellular structures. |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guliy, O.I.; Karavaeva, O.A.; Smirnov, A.V.; Eremin, S.A.; Bunin, V.D. Optical Sensors for Bacterial Detection. Sensors 2023, 23, 9391. https://doi.org/10.3390/s23239391
Guliy OI, Karavaeva OA, Smirnov AV, Eremin SA, Bunin VD. Optical Sensors for Bacterial Detection. Sensors. 2023; 23(23):9391. https://doi.org/10.3390/s23239391
Chicago/Turabian StyleGuliy, Olga I., Olga A. Karavaeva, Andrey V. Smirnov, Sergei A. Eremin, and Viktor D. Bunin. 2023. "Optical Sensors for Bacterial Detection" Sensors 23, no. 23: 9391. https://doi.org/10.3390/s23239391
APA StyleGuliy, O. I., Karavaeva, O. A., Smirnov, A. V., Eremin, S. A., & Bunin, V. D. (2023). Optical Sensors for Bacterial Detection. Sensors, 23(23), 9391. https://doi.org/10.3390/s23239391






