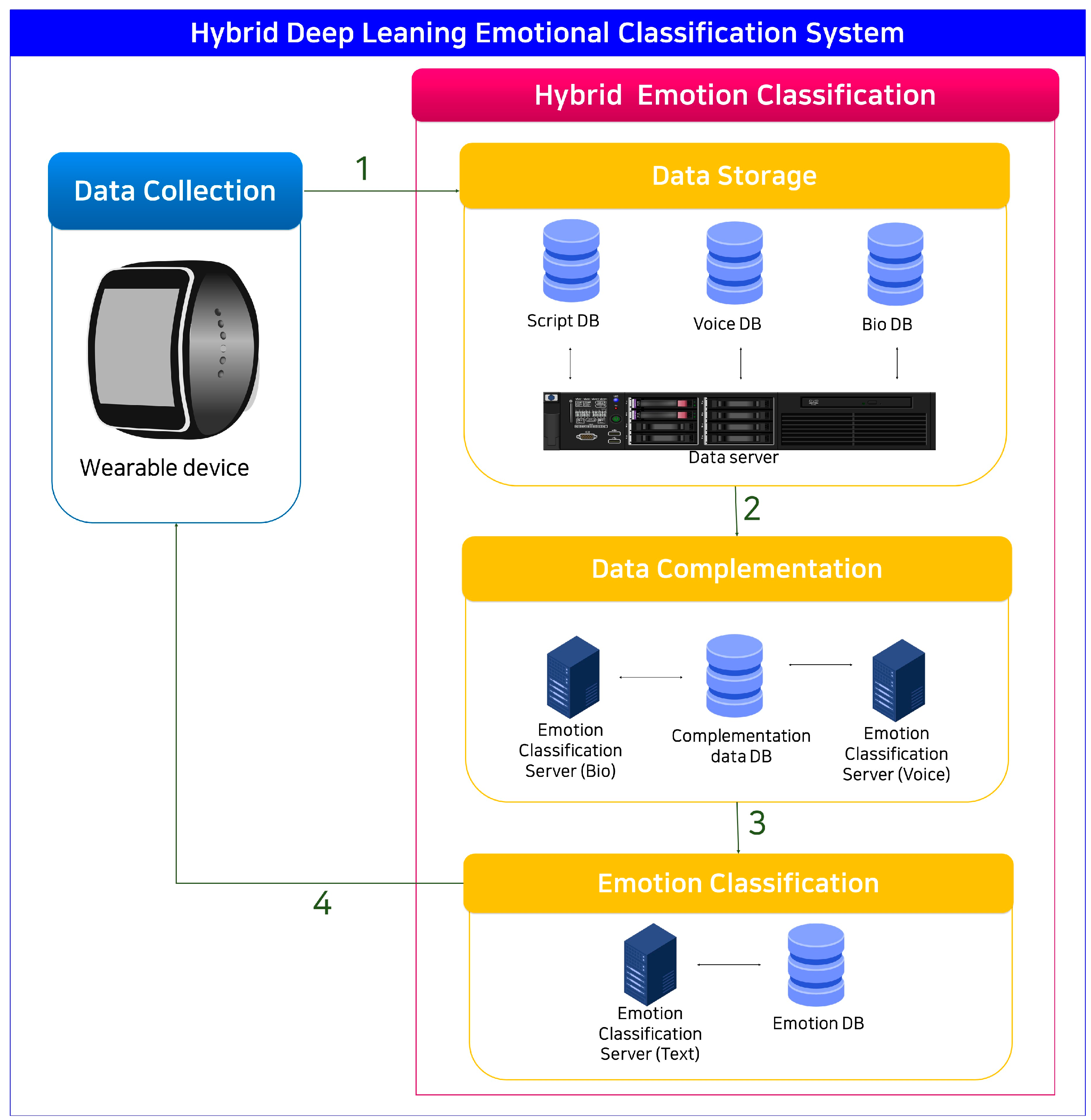
3.4.1. Predicting IBI Missing Values Using LSTM Models
Missing values in IBI data can be attributed to various factors, including weak heartbeats or external shocks during data collection. Furthermore, gaps in the periods of IBI data may arise due to differences in data acquisition durations compared to other biosignals. The challenge of managing such data with missing values is addressed in this paper; the process of imputing missing values in these data is described using Algorithm 1, with the corresponding parameters detailed in
Table 1.
In
Table 1, the “TotalTime” signifies the aggregate duration of the input data, segmented into dil units. The time span extending from the present moment to the most recent time slot containing an IBI value is denoted as “pre_time”, while the interval from the first time slot featuring an IBI value after the current time is termed “post_time”. The discrepancy between “post_time” and “pre_time” is defined as “interval_time”, and the variations in IBI values for the respective time slots are designated as “interval_bio”.
| Algorithm 1: Data_complementation |
| Input | Bio |
| Output | result_model |
| 1. | Def Data_complementation(bio): |
| 2. | Interpolated_bioes = [] |
| 3. | For time in TotalTime: |
| 4. | if post_time = pre_time: |
| 5. | Interpolated_bio = bio[post_time] |
| 6. | else: |
| 7. | Interpolated_bio = bio[pre_time]+(dil-pre_time)*(Interval_bio)/(Interval_time) |
| 8. | Interpolated_bioes.append(Interpolated_bio) |
| 9. | Return Interpolated_bioes |
| 10. | Use Algorithm 2 (Lstm model) |
| 11. | return result_model |
Lines 1 through 9 of Algorithm 1 delineate the “Data_complementation” function, which has been developed to address the issue of missing values in IBI data. This function employs linear interpolation to bridge the gaps or missing data points between the recorded measurement times. In line 7, a linear connection is established between values at “pre_time” and “post_time”, subsequently assigning values corresponding to the unit time of the absent data.
Following this, the outcomes generated by the “Data_complementation” function are harnessed to predict short-term IBI for the entire measurement duration, as depicted in lines 10 to 11. In this instance, an LSTM model is employed, which adheres to a structure akin to that presented in Algorithm 2 and makes use of the parameters delineated in
Table 2.
The values denoted as “Metrics_x” in
Table 2 are derived through the utilization of weights (W_x) and a bias (b_x) for the computation of a weighted sum. This summation process involves taking the previous time’s hidden state and the current input value, represented as “dfm”, and transforming these components into a matrix.
| Algorithm 2: LSTM model |
| Input | dfm |
| Output | Pred_res |
| 1. | Initialization weights and biases |
| 2. | Metrics_x = (W_x*[hst-1, dfm[t]] + b_x) |
| 3. | for s from 1 to seq: |
| 4. | Forget Gate: = |
| 5. | Input Gate: = |
| 6. | Cell State: cst = fgt*cst-1 + igt |
| 7. | Output Gate: ogt = = |
| 8. | pred_res = |
| 9. | return pred_res |
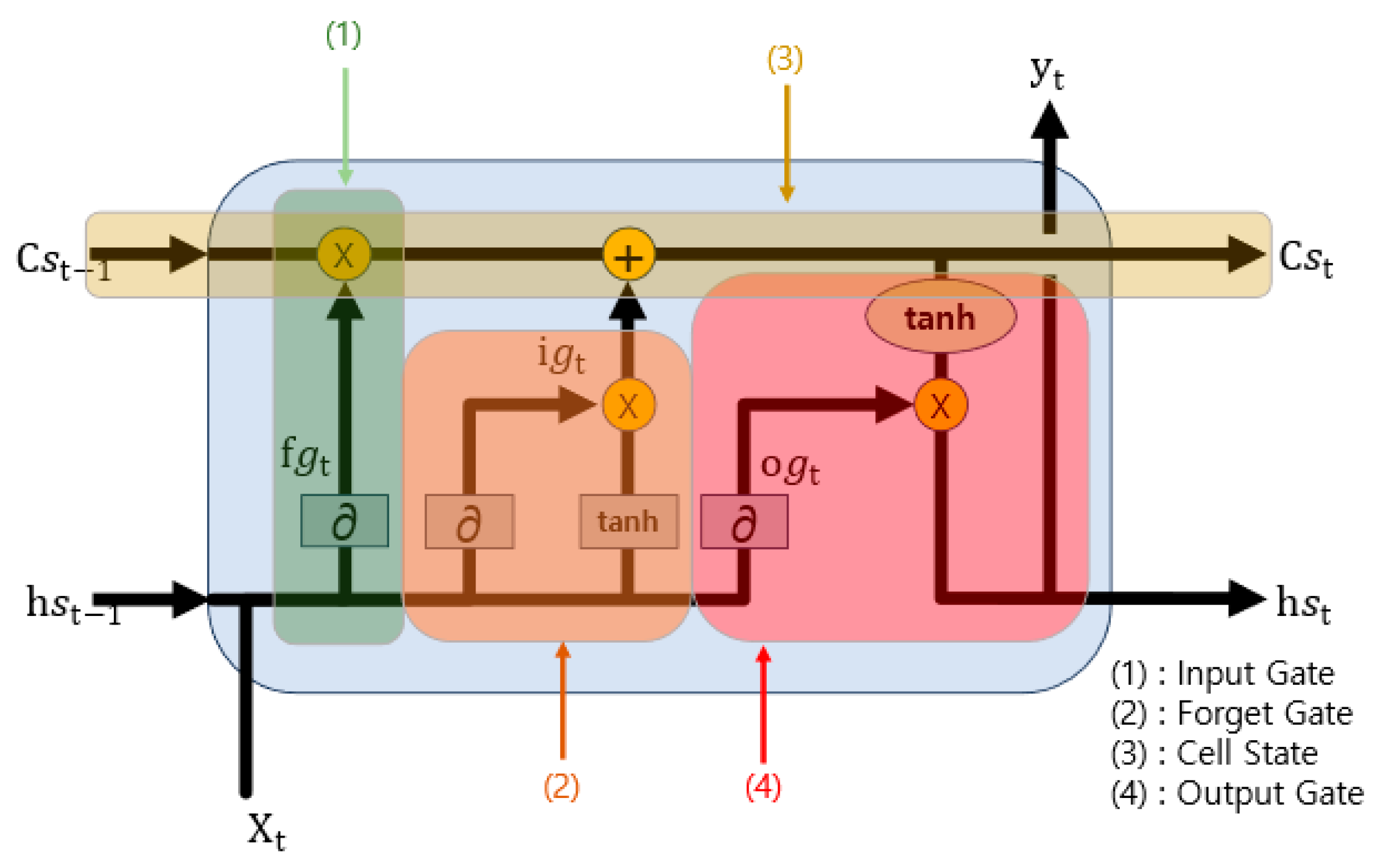
The LSTM model utilized for the prediction of missing values adheres to Algorithm 2 and is visually represented in
Figure 3. Algorithm 2 iterates a specified number of times, denoted as “seq”. In line 4, the forget gate, which is akin to element (1) in
Figure 3, plays a crucial role. It employs a sigmoid function to assess the importance and, based on this evaluation, determines how much of the previous state to consider. Line 5 corresponds to the input gate, which is like element (2) in
Figure 3. This gate employs a sigmoid function to determine the proportion of the input to include and subsequently updates the cell state in line 6 by amalgamating the input data that have been processed through the hyperbolic tangent (tanh) function. The resultant cell gate is analogous to element (3) in
Figure 3. Line 7 pertains to the output gate, which is responsible for generating the final output, equivalent to element (4) in
Figure 3. This gate, after passing through the Softmax function, produces the ultimate output. This algorithm is implemented as a model with a structure closely resembling that described in
Table 3, specifically designed for the purpose of imputing missing values. The process of missing value imputation is visually depicted in
Figure 4,
Figure 5 and
Figure 6.
Table 3 delineates the configuration of the LSTM model employed in Algorithm 1. This model comprises a solitary LSTM layer utilized for prediction. Following this, it traverses a dropout layer, implemented to mitigate the risk of overfitting. Subsequently, the data are processed through a Softmax layer, culminating in the generation of the ultimate output.
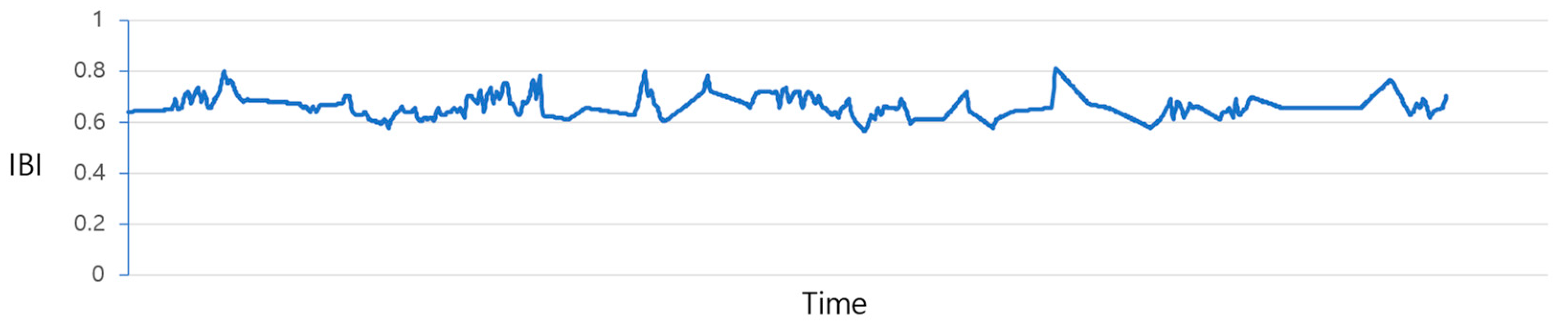
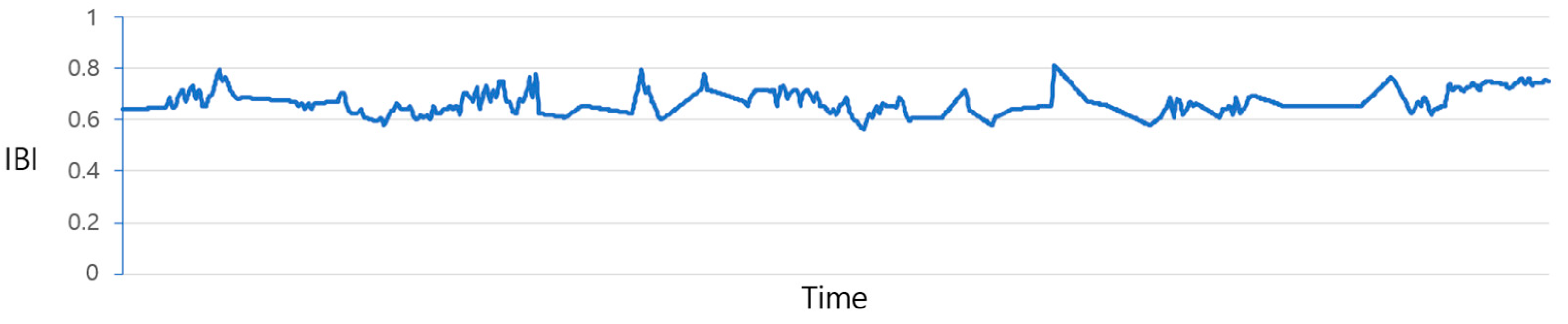
Figure 4 illustrates the plot of IBI values before the prediction process. Due to the irregular data measurement intervals, as previously discussed, there exist gaps where values are missing. The process of filling in these gaps is illustrated in
Figure 5 and
Figure 6. The data from
Figure 4 are adjusted utilizing the “Data_complementation” function, as defined in line 1 of Algorithm 1. This transformation aligns the intervals between data points with those of other biodata and rectifies missing values stemming from variations in time intervals.
Figure 5 depicts the graph of the data after undergoing this adjustment. Nevertheless, there are still time intervals without IBI values, even after this procedure.
In such instances, applying the same missing value imputation method shown in
Figure 6 becomes challenging because of the insufficient data available to adequately supplement the missing values. Consequently, predictions are generated by employing the LSTM model outlined in line 10 of Algorithm 1.
Figure 6 represents the distribution of the results obtained through LSTM predictions.
3.4.3. Mel Spectrogram CNN
The Mel spectrogram [
28] is a technique used to analyze audio data by applying the Fourier transform [
29] to them. These data are then converted to the Mel scale [
30], considering the pitch variations in human speech. To reduce information loss, audio data undergo this Mel scale transformation and are further subject to a logarithmic transformation, resulting in a log Mel spectrogram [
31].
Table 4 below lists a detailed explanation of the key parameters in Algorithm 3.
The STFT function in
Table 4 performs the short-time Fourier transform [
32], which is calculated on the original audio data using the parameters n_fft and hop_len, where n_fft stands for the frame length, and hop_len indicates how much the frames overlap. This function analyzes the audio signal in the frequency domain to generate the spectrogram, stft_sp. Algorithm 3 below represents the process of converting speech files into log Mel spectrograms. The input for Algorithm 3 includes wavs, n_fft, h_len, and n_mels, and the output is padded_mels.
| Algorithm 3: Make Mel scale spectrogram and pad from speech files |
| Input | waves, num_fft, hop_len, num_mels |
| Output | log_mels |
| 1. | for each wav in waves: |
| 2. | stft_sp = STFT(wav, num_fft, hop_len) |
| 3. | convert stft_sp to mel_sp using the mel_filters with num_mels |
| 4. | log_mel = 10*log10stft_sp’s power |
| 5. | add log_mel to log_mels |
| 6. | find the longest column length in the log_mels |
| 7. | pad column with zeros to the longest column length |
| 8. | return log_mels |
To conduct STFT(1), the audio file is first split into frames of a consistent length determined by num_fft. These frames have intervals set using hop_len to ensure they overlap. Then, a Hamming window function (2) is applied to each frame. This function [
33] helps maintain continuity, enhances specific frequency domains, and diminishes others.
Lines 1 to 5 explain how audio files are transformed into log Mel spectrograms. In line 2, the STFT function is applied to the original signal x along with parameters num_fft, hop_len, and num_mels, performing a local Fourier transformation. For this, num_mels sets the number of Mel filters, which controls how finely the frequency domain is divided in the Mel scale. In line 3, mel_filters represents the Mel filter bank [
34]. This filter bank is visualized as triangular shapes in the frequency domain. These triangular filters encapsulate specific frequency ranges and are employed to extract energy within corresponding segments of the frequency spectrum. The shape and size of the Mel filter bank are determined based on the sampling rate of the input data and the desired frequency range representation in the Mel scale. So, in lines 2 to 3, the STFT function produces the spectrogram stft_sp, and the Mel spectrogram mel_sp, which captures human auditory characteristics, is acquired by employing a Mel filter bank. In line 4, the power of the Mel spectrogram mel_sp is obtained using Equation (3), and then it is transformed into the log_mel representation in Decibel units [
35].
The log Mel spectrogram data from the audio file are represented as a 2D matrix. Notably, n_mels, the number of Mel filters, determines the fixed number of rows, while the number of columns varies depending on the length of the original audio data. As explained in lines 6 to 7, zero padding is carried out, where any empty spaces are filled with zeros.
The input data with a fixed size are trained with a CNN-based ResNet-50 model, as shown in
Table 5, along with
Figure 7 and
Figure 8. The ResNet-50 used here reduces the size of feature maps and increases their numbers as the layers deepen. Moreover, the model structure in
Figure 7 and
Figure 8 incorporates shortcut connections, which serve to prevent overfitting and gradient vanishing as the layers grow deeper. This design also leads to faster training and superior performance compared to earlier versions of ResNet models.
The model for training audio data consists of a CNN-based ResNet-50 [
36] architecture, which includes convolution blocks, identity blocks, pooling layers, global average pooling (GAP) layers [
37], and fully connected (dense) layers for classification, as shown in
Table 5.
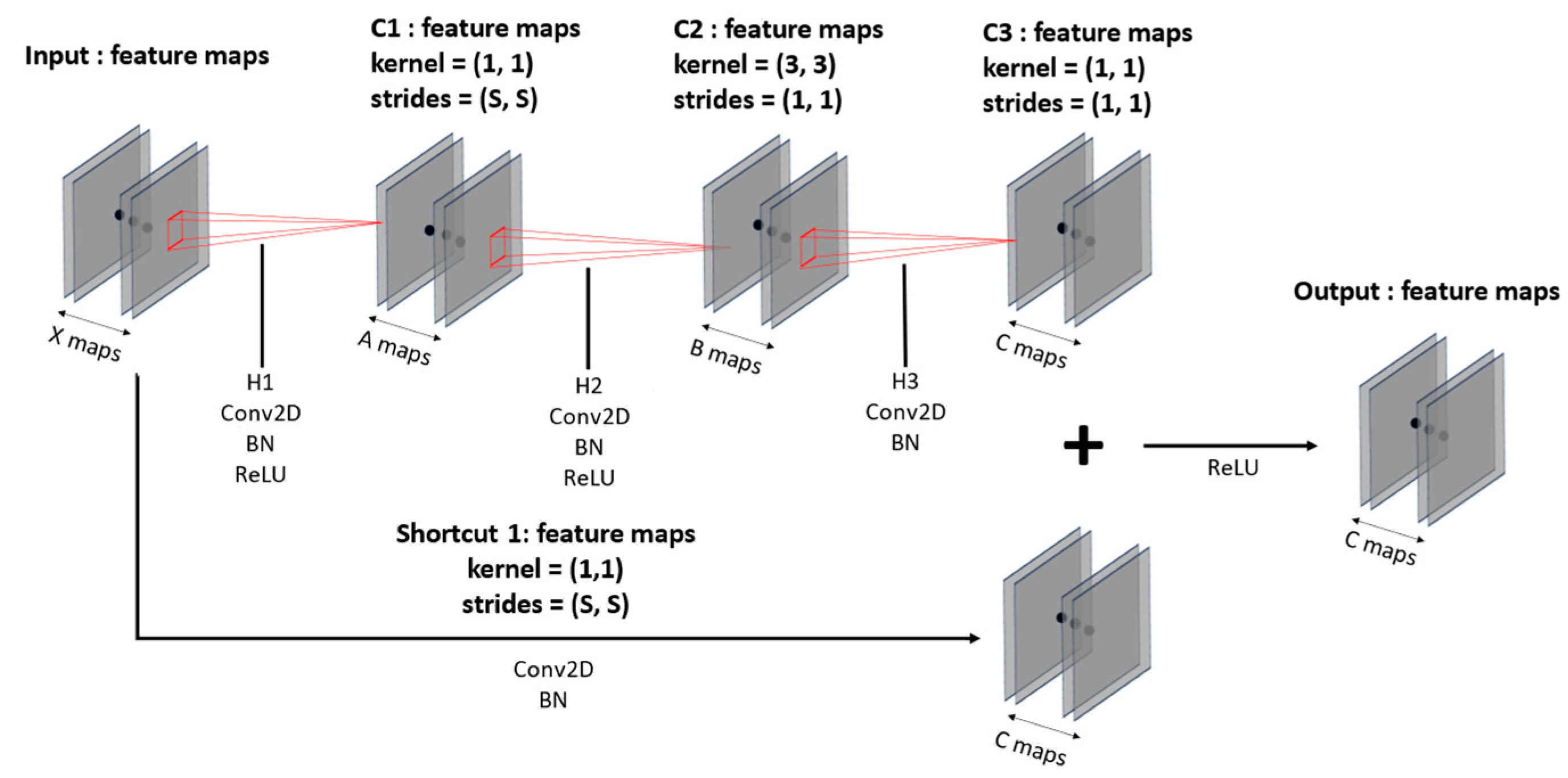
Figure 7 illustrates the convolution block and
Figure 8 shows the identity block. Both blocks compress the number of input feature maps in the initial convolution layers and then expand it back to the initial number of a 1 × 1 kernel; the second layer employs a 3 × 3 kernel; and the third layer returns to a 1 × 1 kernel, forming a deep neural network. This kernel configuration, shown in
Figure 7 and
Figure 8, allows for the construction of deeper neural networks with reduced training time compared to the original ResNet. The convolution blocks can flexibly set strides to increase or decrease the size of feature maps, and identity blocks leverage the fact that input and output sizes remain the same to construct deep layers. Batch normalization [
38] is applied to each layer following the convolution layers to normalize the distribution of input data, enhancing training stability and speed. Additionally, a GAP layer is used to flatten 3D data into 1D data and reduce spatial information loss. With the utilization of these spatial features, the model learns and comprehends spatial information more effectively.
The audio data are transformed into spectrograms and pass through ResNet-50, as described in
Table 5. Throughout this process, the data that initially had a single feature map exhibit a reduction in feature map size as they are processed through the deeper layers, leading to faster processing. However, the number of feature maps increases, allowing the model to learn more detailed spatial information. Because of these characteristics, the model can reduce input data size while maintaining high accuracy on the dataset. Furthermore, it addresses issues that arise with deepening layers, such as overfitting and gradient vanishing, through shortcut connections.
Table 6 presents the values used when transforming audio data into log Mel spectrograms. The value for the padding size used in this context corresponds to the max_length obtained in Algorithm 3.
The CNN model is trained on the entire dataset, which was partitioned with an equal distribution of participants and divided into training, validation, and test sets at an 8:1:1 ratio. About 13,000 voice files were extracted from KEMDy20 conversation situations and were divided after preprocessing. The training process involves randomly extracting 20 samples per class from the training dataset and repeating this process three times, for a total of 1600 iterations. Under the specified conditions in
Table 6, the trained model achieved an accuracy of 0.772.
3.4.4. Cross-LSTM and CNN
To incorporate the results from
Section 3.4.2 and
Section 3.4.3 [
39] into the scripts by adding sentences that convey emotions, Algorithm 4 below is employed.
Table 7 below outlines the parameters of Algorithm 4. The input for Algorithm 3 includes P
lstm and P
cnn, and the output is emo_scripts.
| Algorithm 4: Cross-LSTM and CNN (CLC) |
| Input | Plstm, Pcnn, scripts |
| Output | emo_scripts |
| 1. | pre_emo = [ ] |
| 2. | for each plstm, pcnn in ZIP(Plstm, Pcnn): |
| 3. | if plstm and pcnn are same: |
| 4. | add plstm to pre_emo |
| 5. | else: |
| 6. | add the higher-scoring result between plstm and pcnn to pre_emo |
| 7. | convert the emotions in pre_emo into emotional sentences |
| 8. | emo_scripts = scripts + pre_emo |
| 9. | return emo_scripts |
In line 1, a list is created to compare and store the prediction results of two models P
lstm and P
cnn. In lines 2 to 6, identical emotion predictions from P
lstm and P
cnn are directly appended to the pre_emo list. For different emotion predictions, the model selects the emotion with the higher score between p
lstm and p
cnn and stores it in pre_emo. Finally, in lines 7 to 9, sentences are generated from the emotion classification results and then appended to the original script. The emotional expression sentences are based on
Table 8 below, and the inclusion of these emotional expression sentences in the script serves to complement the model’s emotional classification performance.
Using the threshold values in CLC to derive results is significant. This is because it takes into account the results predicted using different types of data. Disregarding this aspect and immediately aggregating results significantly reduce the reliability of the modified data, leading to a decrease in the final model’s performance. For these reasons, to reflect the characteristics of different data, each result is compared, and, in order to enhance the meaningful performance of BERT, cross-validation is performed to calculate the threshold values as follows: First, the entire dataset is divided into 10 equal portions. For these 10 divided datasets, if the results obtained with CLC improve BERT’s performance, training is stopped, and accuracy is assessed. Finally, an average of accuracy is calculated and used. The value used in this paper’s CLC was 0.75.
In the experiments, sentences expressing emotions intuitively, as presented in
Table 8, were integrated. Depending on the specific model and its application, other sentence forms may be utilized. These sentences were predominantly tailored for emotion classification. The emotion data predicted with CLC exhibited an average emotion prediction accuracy of 0.926 when compared to the entire set of emotion labels.
3.4.5. BERT’s Input Complement
In this paper, the learning approach is modified by implementing BERT’s dynamic masking technique and adopting a pretrained roBERTa model with an increased batch size and a more extensive training dataset. This choice aligns with the strengths of roBERTa models [
40] for emotion classification compared to various BERT-based models. To assess the performance of a Korean-specific model using Korean as one of the languages, the KLUE/roBERTa-base model, pretrained with the KLUE dataset, is employed.
In the HDECS approach, usable data are initially separated from challenging biodata and audio data. Subsequently, complementary data for the script are generated by applying emotion classification algorithms customized for biodata and audio data. The audio data are subjected to visualization using the Mel spectrogram algorithm, and emotion classification is executed using a CNN-based ResNet model. Regarding biodata, any missing values in IBI are addressed using the IBI prediction LSTM model (IPL). Subsequently, the emotion classification LSTM model (EPL) is used for emotion classification. The outcomes from both models are compared to engage in the cross-LSTM and CNN (CLC) process to generate emotion-related scripts. These scripts are appended to the existing scripts and combined with the KLUE/roBERTa model to enhance model performance, as depicted in
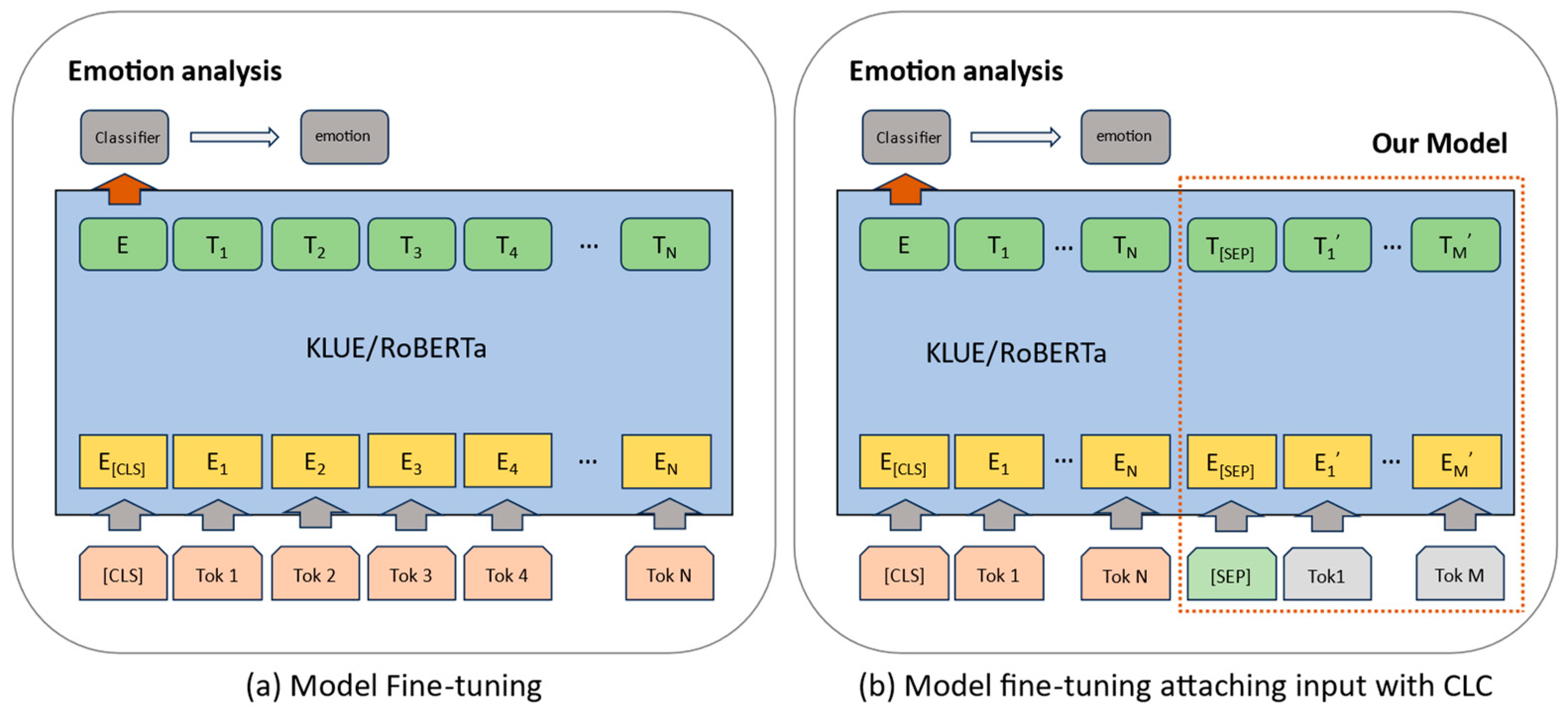
Figure 9.
Figure 9a indicates the fine-tuning of emotion-labeled scripts on the KLUE/roBERTa model. In contrast,
Figure 9b, as proposed in this paper, illustrates the process through which emotion-related scripts are generated using CLC and added to the existing scripts. Subsequently, fine-tuning is carried out.
The traditional BERT model processes parsed script data, commencing with the [CLS] token to designate the beginning of a sentence. It computes the vector values of the embeddings for each token and produces the token with the highest score in the model. These tokens are concatenated to construct sentences, and the resulting sentences undergo a classification process to predict the corresponding emotion for each sentiment.
To facilitate sentiment analysis with the KLUE/roBERTa model, fine-tuning with emotion-labeled text data is required, followed by transfer learning. For transfer learning, scripts must be tokenized using the byte-pair encoding (BPE) to conform to KLUE/roBERTa’s input specifications. In
Figure 9b, the process commences with the [CLS] token, followed by listing tokenized subwords and inserting [SEP] tokens for sentence concatenation. Subsequently, CLC generates the newly added scripts, tokenizes them into subwords using BPE, and appends them. The input is then padded to the maximum token length of 70 tokens, and emotions (neutral, happy, angry, surprised, sad, disgust, and fear) are one-hot encoded to serve as labels, forming the input for the model.