1. Introduction
Recent advancements in artificial intelligence (AI) and computational technologies have played a pivotal role in shaping the concept of smart healthcare systems [
1]. These cutting-edge systems leverage the capabilities of AI algorithms to swiftly and accurately process medical data, empowering healthcare providers to make well-informed decisions and provide personalized patient care. The primary objective of these systems is to make healthcare more accessible and cost-effective, particularly in response to the challenges posed by aging populations and a shortage of medical professionals. Moreover, by offering remote healthcare services, they bridge the gap in medical care access for remote and underserved rural areas. The significance of healthcare systems is underscored by substantial investments, such as the estimated
$22 billion in telemedicine services in the USA [
2]. At the core of these smart healthcare systems lies machine learning (ML)-based remote medical diagnosis, which can both identify existing diseases and predict and prevent future ones. One compelling example of their importance is the ML system developed by Google, capable of detecting malignant tumors through the analysis of mammograms [
3].
Among other ML techniques, SVM stands out for its high accuracy and efficiency in the context of medical applications, which frequently involve small datasets [
4,
5,
6]. Like other ML methods, SVM follows a two-phase process: training and testing. During the training phase, healthcare providers employ historical medical data from patients to create an SVM model. However, due to resource constraints and the need for uninterrupted diagnosis services, healthcare providers often outsource this model to a third-party cloud server operated by an independent entity. In the testing phase, users like doctors and patients submit medical data, including vital signs and symptoms, to the cloud server. The cloud server utilizes this data to evaluate the model and subsequently provides the classification or diagnosis to the users [
7].
While this cloud-based approach enables medical diagnosis services for individuals and healthcare centers, the inherent trust issues associated with third-party cloud servers necessitate safeguarding patient data confidentiality and the intellectual property of the diagnosis model. Therefore, it is imperative to explore a methodology that allows the cloud server to receive and evaluate data without learning the data, diagnosis, or model parameters. This can be achieved through encryption of both the model and the data, enabling the cloud server to use the encrypted model and encrypted data to compute the diagnosis in ciphertext format, decipherable solely by the user who submitted the data.
In the existing literature, only a limited number of studies have endeavored to address the issues of privacy and safeguarding the intellectual property of models in SVM-based medical diagnosis systems [
8,
9,
10,
11,
12]. However, these studies have notable shortcomings. Some, such as those in [
8,
9,
10], primarily focus on privacy concerns but do not consider hiding the model’s parameters. Conversely, others, like [
11,
12], enable the server to learn the classification (or diagnosis) results which creates a vulnerability that can be used to breach the privacy of the users. Furthermore, existing approaches predominantly rely on cryptosystems that demand substantial communication and computational resources such as multi-party computations and Pailler cryptography. Other approaches also involve users in the computation of classifications, which is not practical given that users typically possess limited computational capabilities. Among these studies, the works in [
12,
13] are the most pertinent to our research.
The approach presented in [
12] exhibits various drawbacks. It relies on an Okamoto–Uchiyama (OU) cryptosystem [
12] based on Paillier cryptography [
14], necessitating extensive computational resources. Additionally, as the number of classes increases, the computational demands on model users escalate significantly. Furthermore, their proposed method mandates the active involvement of the model owner in every classification operation, which detracts from the advantages of outsourcing the model to a cloud server, as it requires continuous online interaction and allocation of computational resources. Another limitation is that to update the model’s parameters, ref. [
12] necessitates transmitting updated encrypted model parameters to each model user, resulting in a substantial communication burden. While the study presented in [
13] has tackled several limitations, including safeguarding the model’s intellectual property, preserving user data privacy, and concealing classification results, it does so by relying on a setup involving two non-colluding servers. Each server has the capability of performing specific computations but possesses limited knowledge. However, implementing such a dual-server setup can be expensive and may pose practical challenges. Additionally, the proposed approach incurs substantial communication and computational resources, particularly as more classes (i.e., diseases) are considered by the model, owing to its utilization of multi-party computation and Paillier cryptography.
This paper endeavors to overcome the aforementioned limitations by introducing an efficient multi-class SVM-based medical diagnosis system that preserves privacy and safeguards model intellectual property. In contrast to existing methods reliant on inefficient cryptography, our approach centers on an inner product encryption cryptosystem, characterized by efficient computation and communication. Our proposed method ensures the privacy of the medical health of users by not allowing anyone to obtain their data and diagnosis results. Additionally, our approach aims to protect the intellectual property by not allowing anyone to obtain the plaintext parameters of the model. Nevertheless, the server can still employ the ciphertext of the users’ data and encrypted model parameters to obtain a masked classification (or diagnosis). In our approach, we have modified the inner product encryption cryptosystem introduced by Kim et al. [
15] to enhance its efficiency within our context. Specifically, the cryptosystem outlined in [
15] was originally designed for a scenario involving a single encryptor and a single decryptor, where the encryptor serves as the model owner, and the decryptor acts as a model user. However, this cryptosystem necessitates outsourcing an encrypted model to the cloud server for each user and sharing a key between each user and the model owner, a process that is notably inefficient and unscalable. To address this, we have modified the cryptosystem in [
15] to make it suitable for a single-encryptor and multiple-decryptors setting. Consequently, only one encrypted model needs to be outsourced to the cloud, which can then be utilized by all users efficiently. To further optimize the efficiency of our approach, we diverge from the conventional privacy-preserving SVM schemes, as seen in many existing works [
11,
16,
17,
18], which are designed primarily for binary classification tasks. Instead, we develop a multi-class SVM classifier capable of diagnosing multiple diseases that share similar symptoms. Our method employs multiple binary classifiers rather than a single multi-class classifier. This necessitates encrypting and outsourcing each individual model to the cloud server, resulting in multiple computations required by the server to evaluate these models. While our paper primarily concentrates on medical diagnosis, the methodology we propose is versatile and applicable to various other domains, including the detection of electricity theft and the coordination of electric vehicle charging.
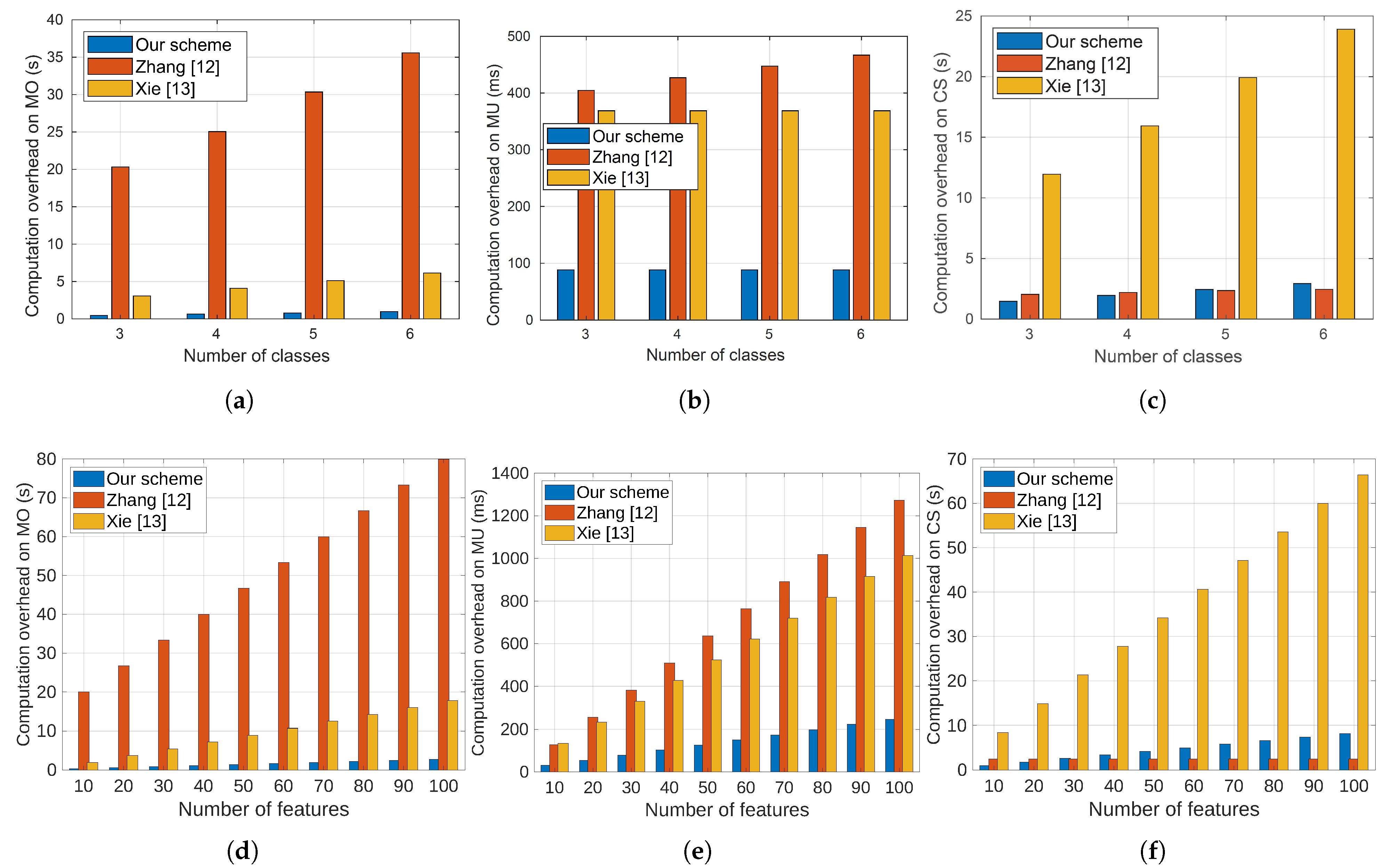
A real medical dataset in [
19] is used to train our SVM diagnosis model for dermatology diseases. This dataset is extensively utilized in the literature [
12,
20] and widely regarded as a benchmark in the field of medical applications. The dataset has six classes (skin diseases), including pityriasis rubra pilaris, psoriasis, lichen planus, pityriasis rosea, chronic dermatitis, and seboreic dermatitis. The dataset has 34 features, including age, family history, and several symptoms. To assess our proposal, extensive evaluations are conducted, and the classification accuracy and computation and computation overhead are the main metrics measured. The main contributions of this paper can be summarized as follows:
A privacy-preserving and lightweight SVM medical diagnosis scheme has been proposed by modifying an inner product encryption cryptosystem.
The results of our analysis demonstrate that our proposed scheme successfully fulfills our security and privacy objectives, including preserving the privacy of the patient’s health status and protecting the model’s parameters.
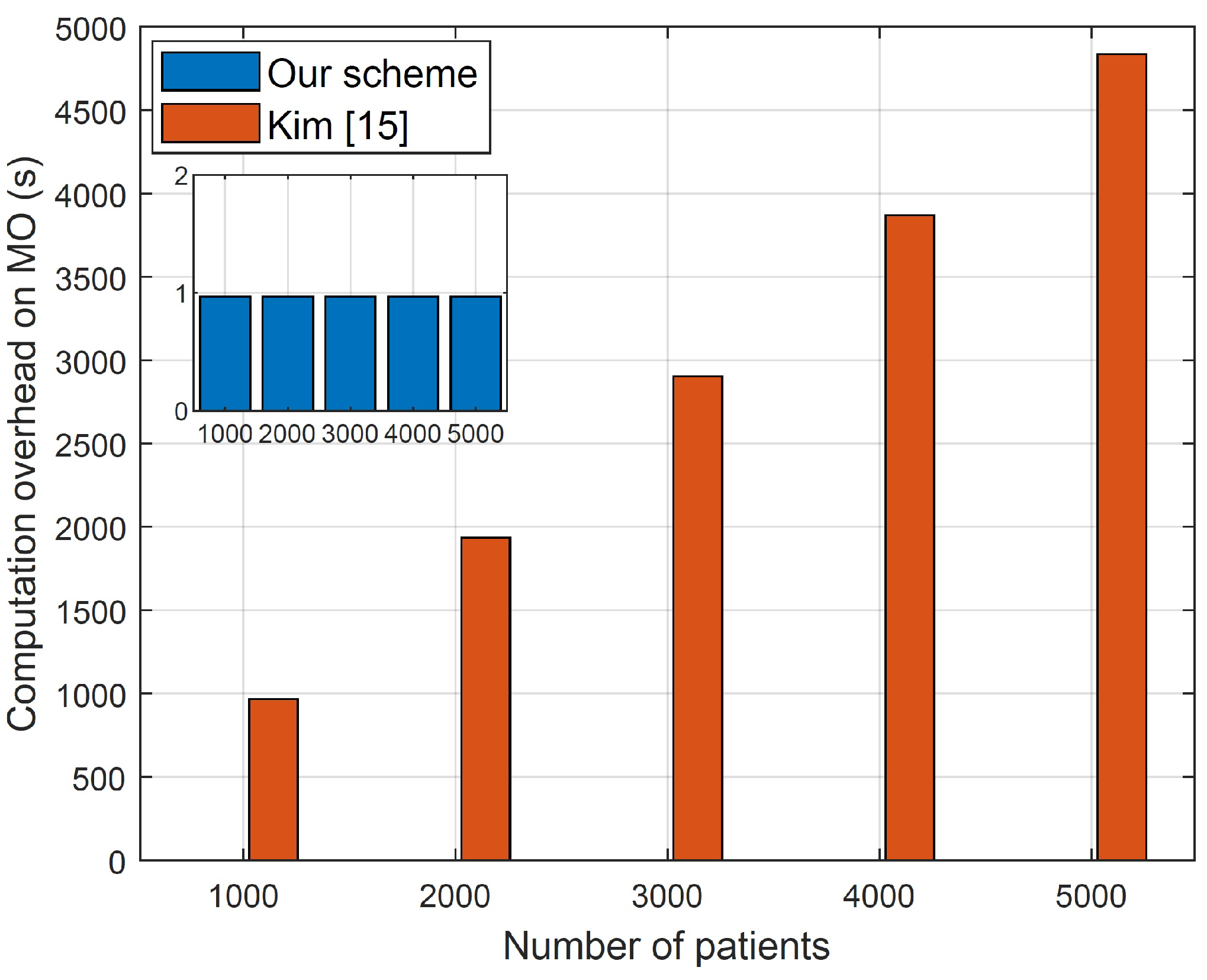
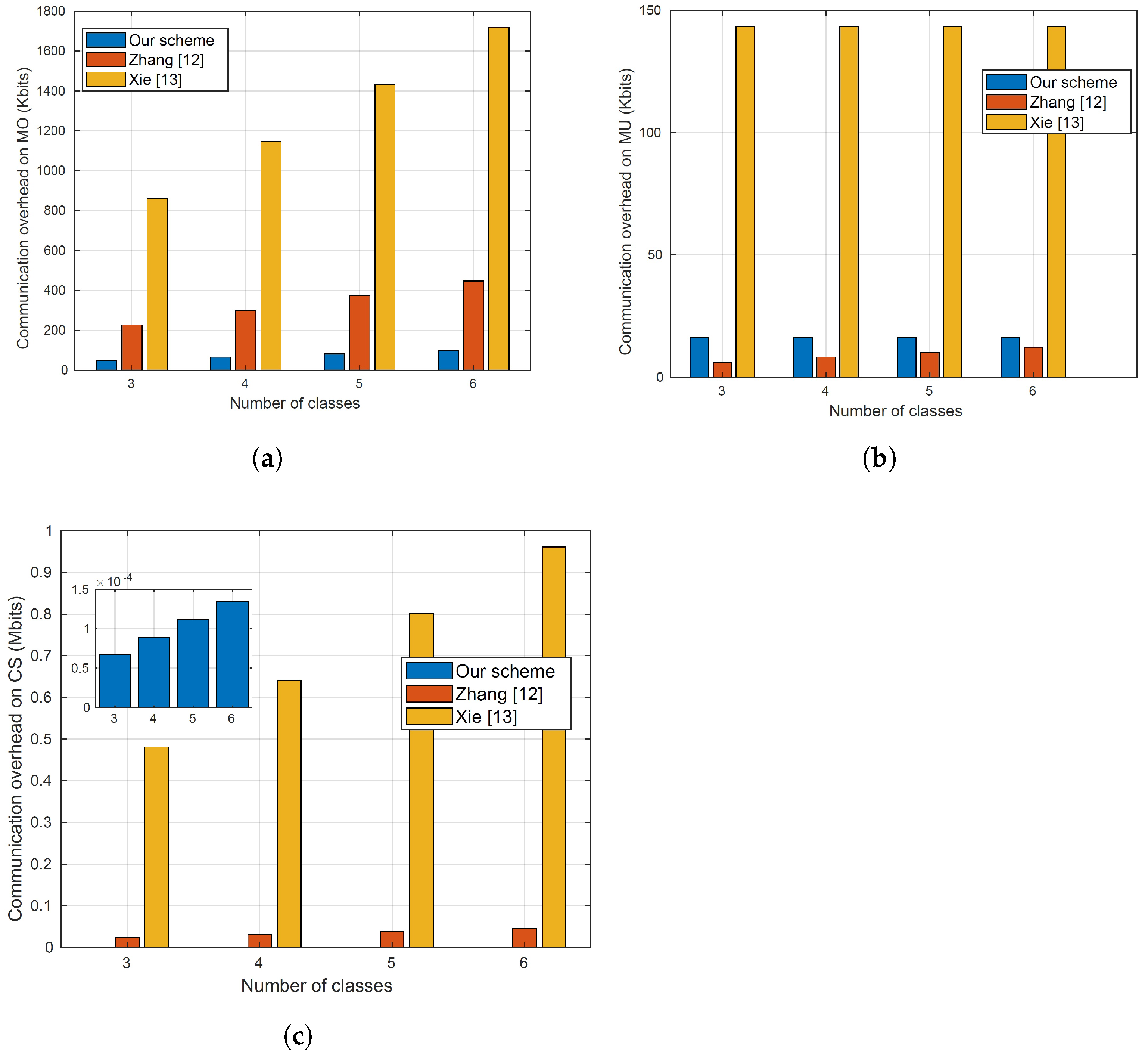
Our evaluations indicate that our proposal outperforms the most relevant approaches in the overhead while maintaining high classification accuracy.
This paper is derived from the PhD thesis of the first author [
21]. The structure of the remaining sections in this paper is as follows. The main entities considered in our system and the main messages exchanged among them, in addition to the objectives of the attackers, are discussed in
Section 2.
Section 3 offers an overview of the fundamental concepts used in our paper.
Section 4 provides an in-depth examination of our proposed scheme. We assess the capability of our scheme to preserve privacy in
Section 5.
Section 6 presents the results of experiments conducted to assess communication/computation overhead and model accuracy.
Section 7 offers insights into related works. Lastly, our conclusions are summarized in
Section 8.
3. Preliminaries
In this section, we provide a concise overview of the bilinear pairing, support vector machine, and a function-hiding inner-product encryption scheme.
3.1. Bilinear Pairing
Let
m denote the prime order of the cyclic groups
and
, with the generator of
being
V. We assume that
and
possess a non-degenerate and efficiently computed bilinear pairing map (
):
satisfying the following properties:
, for all and any , where represents a finite field of order m.
3.2. Support Vector Machine (SVM)
SVMs have garnered extensive recognition and application within the field of medical classification, primarily owing to their remarkable accuracy [
27,
28,
29]. Various notable applications within the medical domain have harnessed SVM’s potential, demonstrating its efficacy. These encompass diverse tasks, such as the classification of electrocardiogram signals [
30], aiding in clinical diagnosis [
31,
32], identifying individuals on the autism spectrum [
33], and detecting cervical cancer cells [
34], among numerous others. SVM’s versatility and precision make it a valuable tool for addressing critical medical challenges.
3.2.1. Linear SVM
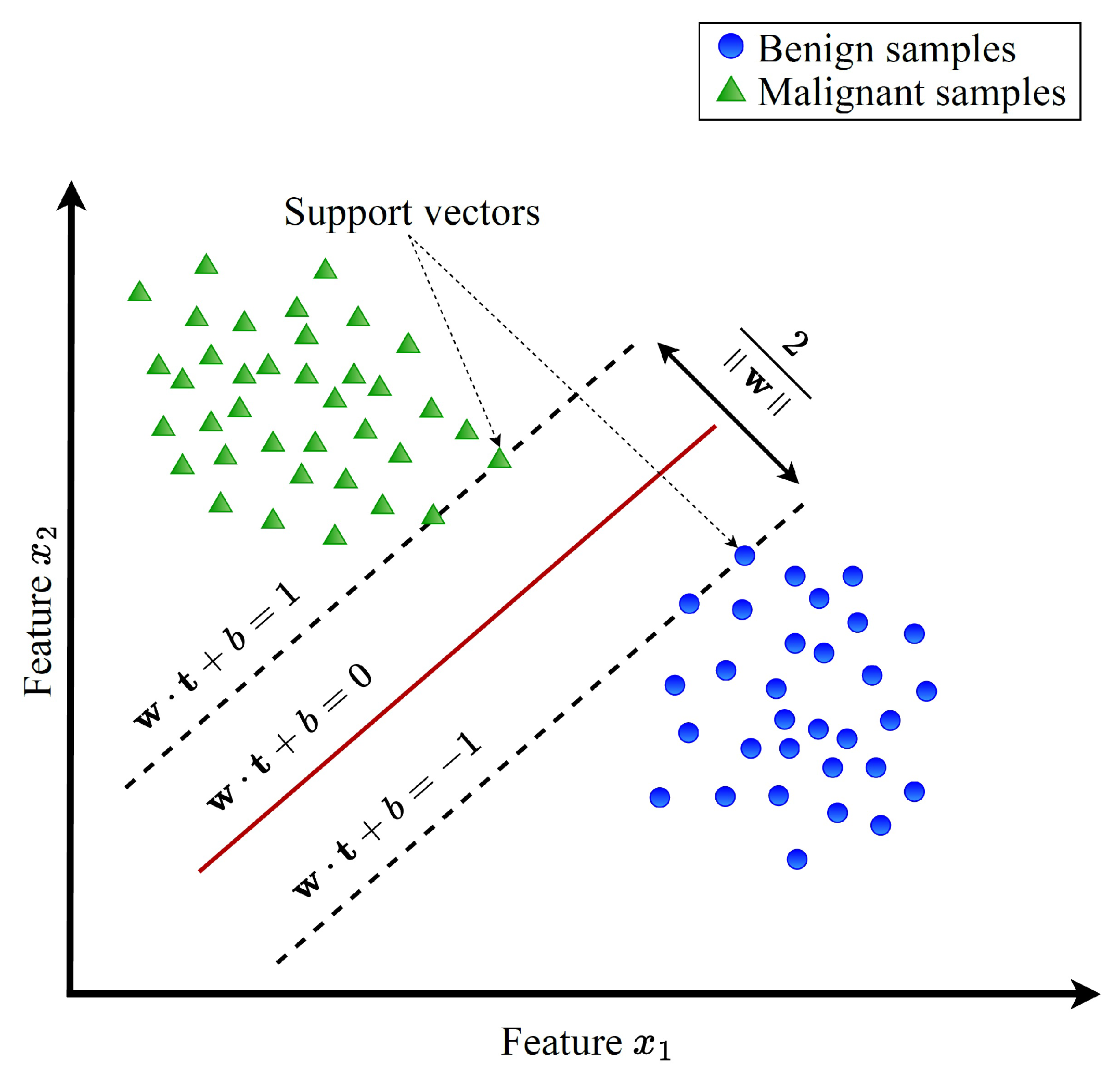
Linear support vector machine (LSVM) is a powerful tool primarily used for binary classification tasks. An illustration of the linear SVM is shown in
Figure 2. In this figure, the blue circles are used to represent the benign data while the green triangles are used to represent the malicious data. As depicted in
Figure 2, the core objective of LSVM is to discern an optimal hyperplane within the feature space, capable of effectively segregating the given set of training samples into two distinct classes. This hyperplane is essentially defined by a
decision function. Let us suppose we have a training dataset comprising
m samples, denoted as
, where each
belongs to the real-valued vector space
, representing the
jth sample with
n elements. The corresponding
is a label assigned to each sample, taking values from the set
. Typically,
corresponds to one class, while
signifies the other class.
The predictions and classifications for new, unseen data can be made after completing the training phase using the decision function defined in Equation (
1):
Here,
,
, and
b denote the weight vector, the unlabeled sample’s vector of features, and the bias, respectively.
S represents the set of support vectors
,
, and
stands for the Lagrange multiplier of the support vector, the support vector, and its associated class label, respectively. The classification of an unlabeled sample
t can be determined by using Equation (
2) to compute
, and then, the classification is benign when is negative; otherwise, it is malicious.
3.2.2. Multi-Classification SVM (MCSVM)
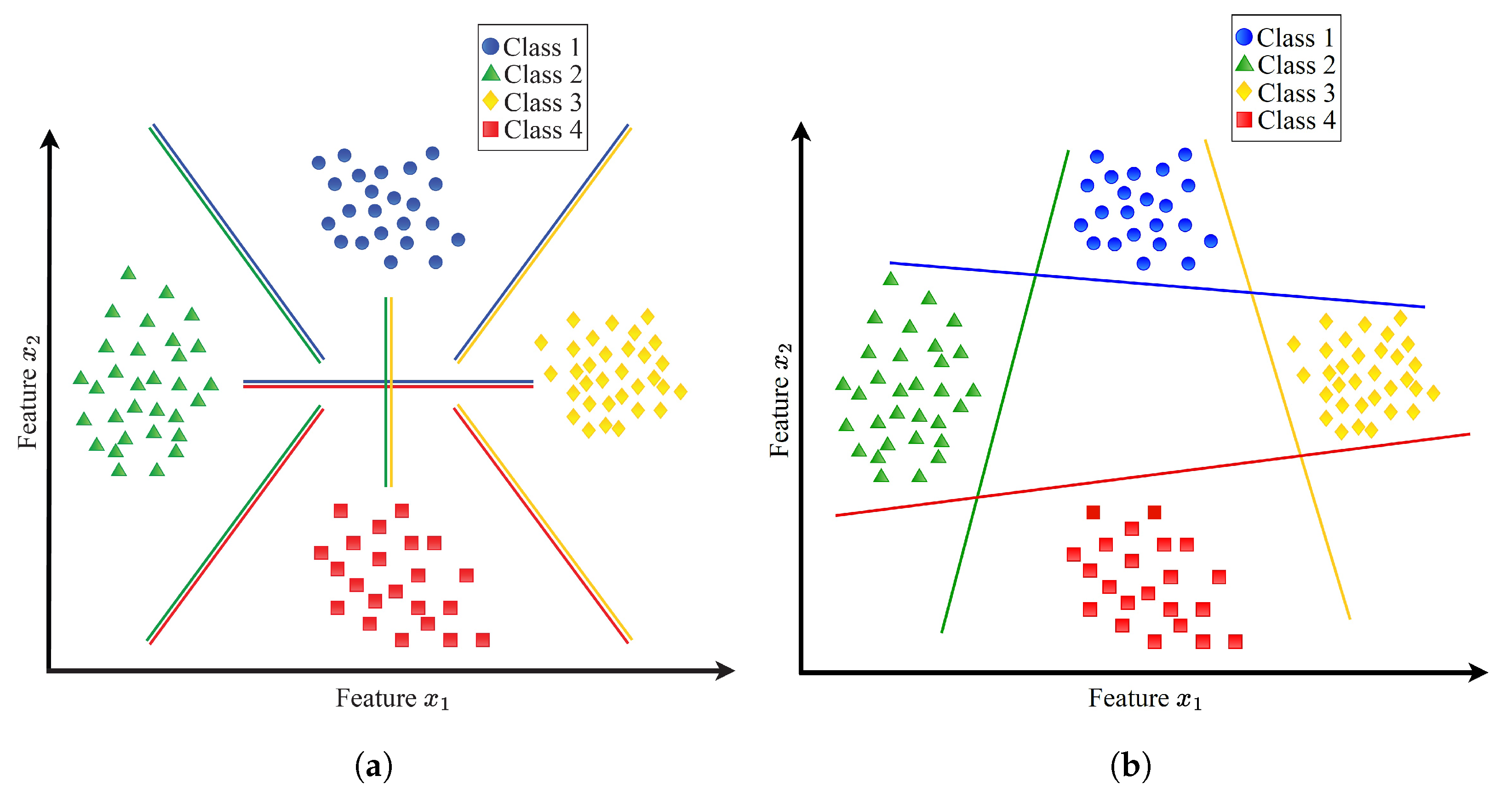
Constructing an MCSVM classifier typically involves creating multiple two-class classifiers.
Figure 3 illustrates two conventional techniques for building multi-class classifiers using SVM: the one-versus-all (1VA) and one-versus-one (1V1) approaches. Four classes are used in the figure.
In the 1V1 approach, a hyperplane is computed between every pair of classes, resulting in a
k-class classifier. Consequently,
is the total number of hyperplanes. In the example shown in
Figure 3, the 1V1 approach establishes six binary classifiers. On the other hand, the 1VA approach involves computing a hyperplane between every class and the remaining classes. In this case, only
k hyperplanes are needed. In the example shown in
Figure 3, four binary classifiers are established by the 1VA approach. Obviously, compared to the 1V1 approach, the 1VA approach creates fewer classifiers.
Due to its efficiency, we opt for the 1VA approach. To implement this method, we train
SVM models. During the training phase, each classifier uses class
j as the +
ve class and the remaining samples as the −
ve class. The decision function of the MCSVM classifier during the testing phase, denoted as
, where
is a given normalized test sample, can be calculated as follows:
where
represents the decision function of the
jth classifier and can be computed as follows:
3.3. Function-Hiding Inner-Product Encryption Cryptosystem
The research in [
15] introduced a functional encryption method enabling the computation of the inner product of two vectors using their ciphertexts without revealing the vectors themselves. In this scheme, the inner product
can be calculated, where
and
are the ciphertexts of vectors. This inner product encryption system is classified as function-hiding since it discloses no additional information about
and
other than the result of the inner product.
Choose as generators for the multiplicative group , and as generators for the multiplicative group . Let the two groups be of prime order q. Define the bilinear pairing map , which maps elements from and to elements in the target group of prime order q. Consider a polynomial-sized subset S of . Define , where . Here, represents the general linear group of matrices over .
The public key and the master secret key are denoted as and , respectively, where , and and . The encryption scheme involves three phases as follows:
: Given and a vector , the algorithm selects a uniformly random element and produces the secret key pair , where and are the secret key and n elements vector that has values from , respectively.
: Given and a vector , the algorithm selects a uniformly random element and produces , where and are the ciphertext pair and n element vector that has values from , respectively.
: Given
,
, and
, the algorithm computes the inner product
. The decryption algorithm produces
and
, where
. The algorithm then checks whether there exists
z such that
by computing a discrete logarithm in
using methods such as the baby-step giant-step algorithm [
35]. If a valid
z is found, it is output; otherwise, ⊥ is output to indicate that no valid
z exists.
This encryption scheme is originally designed for a single encryptor, which is MO in our scheme, and a single decryptor, which is MU in our scheme. The encryptor must share a unique key pair and with every decryptor. If this method is employed in our research, the model parameters would need to be encrypted n times, where n represents the number of MOs. This results in significant computation and communication overhead, which is not feasible. In this research paper, we have adapted this scheme to accommodate a scenario with a single model owner and multiple model users. In this setup, each model user and the model owner utilize unique keys for their operations. In this modified scheme, even though the model parameters are encrypted using the model owner’s specific key, the inner product computation can be performed when the medical data are encrypted using any of the model users’ keys. This approach significantly reduces the computation and communication overhead, as the model parameters need to be encrypted only once, regardless of the number of model users.
4. Proposed Scheme
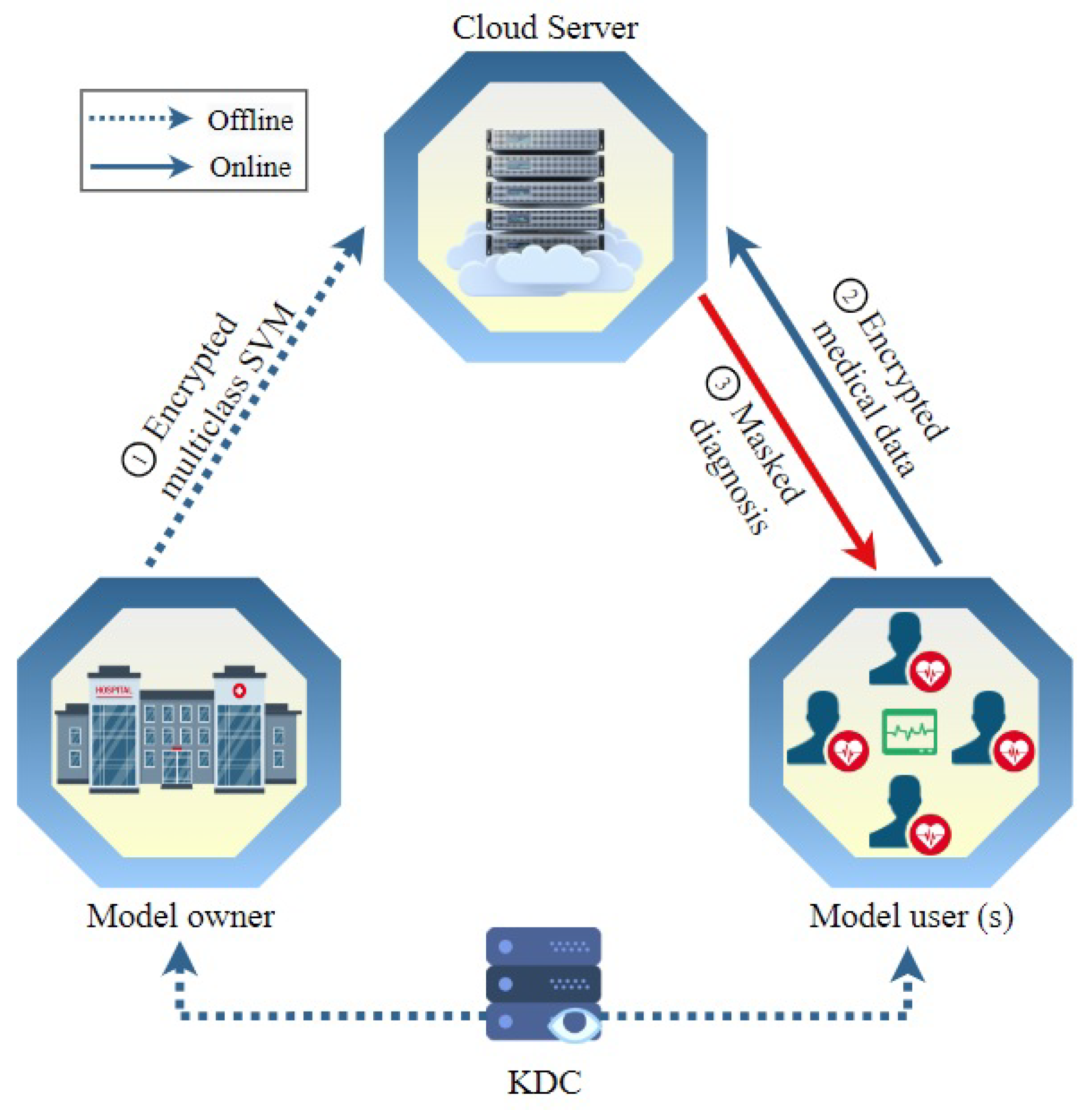
In this section, the term “hospital”, denoted as , signifies the entity owning the model, while “patient”, represented by , refers to the user of the model. The proposed methodology unfolds through four distinct phases. During the system initialization stage, the key distribution center (KDC) undertakes the computation and distribution of secret keys, disseminating them to both and . Moving on to the model encryption phase, each support vector machine (SVM) model’s parameter vector is encrypted by . Subsequently, all these encrypted parameters, along with random numbers employed for masking classification results, are outsourced to the cloud server (CS). Transitioning to the medical data encryption step, encrypts their medical data vector, including symptoms and vital data. The encrypted data are then sent to the server to input to the diagnosis model and compute the masked classification (or diagnosis) score in the medical diagnosis step. These masked classifications are then transmitted back to for the process of unmasking and subsequent understanding.
4.1. Design Objectives
We aim to accomplish specific design objectives in our approach:
Preserving Privacy. In our proposed scheme, it is imperative to protect the confidentiality of the model user’s medical data. The outsourced medical data of the MU must remain confidential, ensuring that neither the cloud server (CS) nor the model owner (MO) can access any information about it. Additionally, the classification results should remain concealed from the CS, with only the MU having access to this information.
Protecting Intellectual Property. Our scheme is designed to preserve the confidentiality of the diagnosis model’s parameters from potential threats posed by the CS, MU, and external eavesdroppers.
High Diagnosis Accuracy and Low Communication/Computation Cost. The proposed scheme aims to deliver precise medical diagnoses for MUs while minimizing computational and communication burdens. Considering the limited computational and communication resources typically available to MUs, it is crucial to optimize their involvement in the online diagnosis process. The encryption methods utilized for user requests should be lightweight, allowing data users to remain offline during the online diagnosis process until they receive the classification results. Moreover, heavy computation and communication tasks should be offloaded to the CS, which possesses ample communication and computation resources. Notably, the cloud server should be able to compute the medical diagnosis without the need for the participation of the model owner because requiring the model owner to be online and interactive all the time to help the server in the diagnosis computations diminishes the benefits of outsourcing the medical diagnosis to a third party.
4.2. System Initialization
The following algorithms are used by the key distribution center (KDC) to generate the secret keys of the patients and hospitals.
. This algorithm takes as the security parameter input, where represents the security parameter, and produces and , which represent the public parameters and the master secret key, respectively.
The key distribution center (KDC) generates the master secret key randomly from the set of invertible matrices , where are matrices of dimensions . Here, m represents the size of the patient’s medical data, and denotes the general linear group of matrices over the field . Subsequently, the output of this process yields the public parameters .
. For hospital
, the algorithm yields secret key
, crucial for encrypting the parameters of each SVM binary classifier. The computation is outlined as follows:
Subsequently, KDC sends to the .
. The secret key
is computed according to the following scheme for every patient
:
where
represents
matrices of randomly generated values, satisfying
. Subsequently, the patient
obtains
from the
KDC. This key is utilized by the patient to encrypt their medical data, which are then transmitted to the cloud for medical diagnosis.
4.3. Model Encryption
To securely transmit the encrypted SVM models’ parameters to the cloud server, performs the following procedure.
. This operation uses the public parameters , the hospital secret key , and the model’s parameter vector as input and produces the encrypted SVM model parameters .
The parameters of each model
j are denoted as
, where
, and
k represents the total number of the models.
then constructs the
vector with
-element where the first
elements will be filled the model parameters and places one at the following
element. In order to mask the classification, the
element will be multiplied by the corresponding element in the patient’s data vector, which stores the masking number. To encrypt
, the hospital selects a uniformly random element
and generates,
Here, and are vectors of size elements. then transfers the encrypted model’s parameters to the cloud server.
4.4. Encryption of Medical Data
During this stage, the subsequent algorithm is utilized to encrypt the medical data by and transmits it to the cloud server for classification.
. This algorithm takes the public parameters , the patient’s secret keys , and the medical data vector as inputs, producing the encrypted medical data . The ’s medical data for the patient is denoted as . The patient then constructs with -elements, encompassing the medical data in the first elements, using the element for storing the randomly generated masking number .
After that,
selects a uniformly random element
to encrypt
as follows
It is noteworthy that and are arrays of size elements. Ultimately, the patient sends to for the diagnosis.
4.5. Classification
Using the ciphertext of the model parameters and the ciphertext of the medical data of users, in this phase, the server computes the inner product result of the the model parameters and medical data which results in masked classification score (i.e., the medical diagnosis).
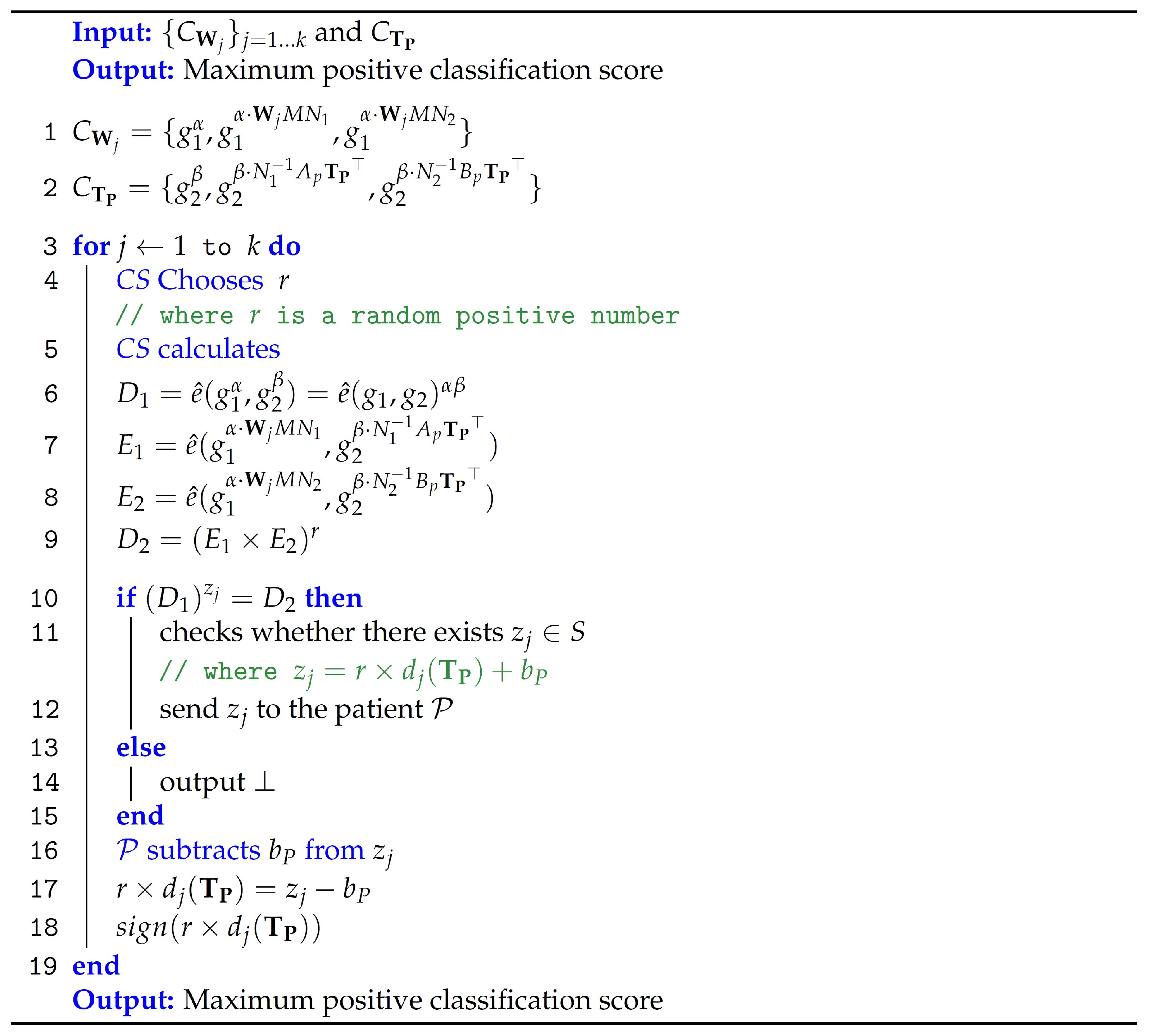
Figure 4 outlines the computations performed by
CS to classify the medical diagnosis. The specifics are as follows.
: The encrypted model’s parameters and the encrypted patient’s medical data are the algorithm’s input, which then produces the masked classification .
A random positive number
r is used by the cloud server for the model’s intellectual property protection. This protection can be accomplished by multiplying
r with the first
elements of
.
and
are used to compute the inner product of
and
for every model
j, where
and
Subsequently, the algorithm checks for the existence of its output
satisfying
, where
S is a polynomial-sized subset of
,
and
. If no such
exists, the algorithm outputs ⊥. Note that this algorithm demonstrates efficiency as
.
Figure 5 illustrates the structure of vectors used in computing the classification result.
Theorem 1. For each SVM model, CS uses the encrypted patients’ data to calculate the masked classification result.
Proof. Therefore, if , such that , the secure classification algorithm outputs the masked classification result . □
Finally, the CS transmits all the multiclass SVM model’s masked results to . To obtain the unmasked scores, the patient can subtract from . Then, the final classification result is the highest positive unmasked score.
5. Security Evaluations
This section uses a preposition/proof format to present the main security/privacy features provided by our proposal.
Proposition 1. Our proposal can resist attacks launched by external eavesdroppers.
Proof. As discussed earlier in the threat model section, attackers can be external eavesdroppers who have the capability to capture all messages exchanged in our scheme and they analyze these messages looking for vulnerabilities to infer sensitive information or compute the parameters of the diagnosis model. As explained earlier, the messages exchanged in our system are as follows: (1) the ciphertext of the diagnosis model sent from the model owner to the server; (2) the ciphertexts of the medical data sent by users to the server for classifications; and (3) the masked diagnosis returned by the server to the users. The first two types of messages are encrypted using private keys and without learning the keys, the external attackers cannot infer the messages. This is proved in the original paper of the inner product encryption we modified. For the diagnosis returned by the server to the users, as explained earlier, random numbers that are known only to the users are used to mask the diagnosis, and thus, because the attackers do not know these numbers, they cannot unmask the diagnosis to learn the diagnosis. □
Proposition 2. Our proposal is robust against the known-ciphertext model attacks when the master secret key is unknown to the attackers.
Proof. As explained earlier, the matrices and are used to compute the secret key of the model users (), and this key is used to compute the ciphertexts of the medical data. Additionally, the matrices and are used to create the secret key of the model owner (), which is used to compute the ciphertext of the parameters of the diagnosis model. Our proposal is secure under the known-ciphertext model if the attacker does not know the master secret key , which includes the matrices , , and M. The attackers can capture the ciphertexts of the medical data and the ciphertext of the model parameters, and without knowing the matrices , , and M, it cannot infer the medical data or the model parameters. □
Proposition 3. Our proposal is secure under the known-background model, i.e., external attackers and the cloud server cannot compute the classification result (diagnosis).
Proof. Under the known-background model, attackers are equipped with some background information, such as the distribution of diseases in a certain area to infer sensitive information. For instance, the work in [
22] is not secure under the known-background model because the server can link the ciphertexts of the same diagnosis, so by using the distribution of a disease and the frequency of a ciphertext, the server can figure out the diagnosis of the users. In order to secure our proposal under the known-background model, we mask the classification result of the diagnosis model using random numbers that frequently change. These random numbers are known only by the users so they are the only ones who can unmask the results of the model and learn the diagnosis. In summary, external eavesdroppers and the server cannot learn the diagnosis because they are masked by a secret random number and cannot use background information to deduce the diagnosis because the random number frequently changes. □
Proposition 4. Our proposal can resist the known-plaintext model attacks when the attackers do not know the users’ random numbers utilized to mask the diagnosis model’s output.
Proof. In the Known-plaintext model, attackers can gather pairs of plaintext and corresponding ciphertext messages exchanged in the system and then analyze these pairs to infer any information that can be used to figure out new patients’ medical data. Because our proposal uses inner product operations to evaluate the diagnosis model, one way to launch the Known-plaintext model attack is by creating equations and solving them. Specifically, if each vector (either the medical data or the model) has elements, the attacker needs to create equations to solve them and obtain the unknowns. To do that, the attacker needs to learn medical data vectors and the result of the classification to make equations and solve them to compute the model’s parameters. The attack can also be launched with the objective of computing the medical data of the users. This attack can be launched by external eavesdroppers and/or the cloud server. In order to secure our proposal against this attack, we allocate the th element in the medical data vector for a random number that should change every time the user sends data for classification. By doing that, we introduce a new variable each time the inner product is computed, i.e., the result of the inner product of the medical data and the diagnosis model is masked by the random number and is not known to the attackers. In summary, our proposal is secure against Known-plaintext model attacks because the attackers (either the cloud server or the data users) know only masked classifications (the result of the inner product of two vectors), and by changing the random number in each medical data, attackers cannot make equations and solve them to infer sensitive data because this random number adds a new variable in each equation. □
Proposition 5. To preserve privacy, the same medical data (or same classification) of a patient sent at different diagnosis occasions are not linkable in our proposed approach.
Proof. Even if attackers are not able to decrypt the ciphertexts in our system, sensitive information can be inferred if ciphertexts of the same data are linkable. For instance, if a user sends the same medical data on different occasions and attackers can link the ciphertexts, side information can be inferred such as no change in the users’ symptoms and vital data and thus no change in the health condition. Similarly, even if the attacker cannot unmask the masked diagnosis score that the server returns, linking the masked classifications of the same diagnosis reveals side information, such as no change in the health status of the users. To secure our proposed approach against this attack, in the encryption of the medical data, a random number () is used so that the ciphertexts of the same data look different when it is encrypted multiple times on different occasions. Additionally, by masking the output scores of the diagnosis model by different random numbers, the masked diagnosis scores of the same classifications look different. □
Proposition 6. Our proposed approach can protect the confidentiality of the diagnosis model’s parameters.
Prepositions 4 explains how the cloud server cannot compute the diagnosis model’s parameters leading to the protection of the intellectual property. Moreover, a random number (r) is used by the cloud server to prevent the users from computing the parameters of the model. Without using this random number, the users can compute the model’s parameters because they can use the medical data vectors and the diagnosis scores returned by the server to create equations and solve them to obtain the parameters of the model. Specifically, if the size of the vectors is elements, then attackers need equations by requesting diagnosis. Note that the diagnosis score is the inner product result of the vectors of the medical data and the model parameters. To protect our proposed scheme against this attack, a random number r is used by the cloud server to add a masking level to the output score resulting from the evaluation of the model. In this way, the user does not know the exact result of the product of the two vectors of the medical data and the model’s parameters and for each diagnosis operation, the random number r adds a new variable in the equations the user tries to solve. The server needs to change the random number each time it performs a diagnosis process. Note that the classification is determined by the sign of the classification score after unmasking the classification received by the users. This necessitates that the random number (r) is positive to avoid changing the sign of the classification value.
7. Literature Review
Our objective is to protect the confidentiality of the users’ data, health status, and model’s parameters and enhance the computational and communication efficiency. Additionally, we aim to create a diagnosis system that needs a single cloud server and does not need to involve the model owner in the computation of the classification result. To the best of our knowledge, our scheme stands out as the only one capable of achieving all of them. We refer to
Table 2 for a concise comparison between our approach and the most relevant state-of-art works. Furthermore, this section delves into more detailed explanations of other schemes and compares them to our proposal.
Due to its potential, machine learning has been widely used in different medical diagnoses [
37,
38,
39]. Artificial neural networks, SVMs, and regression trees are used in [
40] for diagnosing Parkinson’s disease in early stages, while in [
41], linear SVM is used to diagnose Alzheimer’s disease. In another example, random forest, Naive Bayes (NB), and SVM are used in [
42] to diagnose COVID-19. There is no doubt that machine learning will play a major role in the realization of smart healthcare systems. Nevertheless, in most of the existing schemes, including the aforementioned works, machine learning models are evaluated using plaintext data and the outsourcing of these models to a third party that provides cloud diagnosis service endangers the privacy of the users and the model’s parameters. In comparison to these schemes, our proposal in this paper can achieve the objective of protecting the confidentiality of the model’s parameters and the users’ data and health status, which cannot be accomplished by the aforementioned schemes.
The privacy problem in machine learning-based medical diagnosis can be divided into two parts, including training and testing. While our paper aims to preserve privacy during the testing phase, several works in the literature have studied privacy in the training phase such as [
11,
16,
17,
18]. These works aim to train SVM models without exposing the sensitive information of the dataset of the training. The approach proposed in [
43] trains the model using an encrypted dataset instead of using plaintext data to avoid revealing sensitive information. In [
44], different entities have datasets for drug formulas and the work investigates an approach to enable a central unit to compute a model trained on all the datasets without sharing the data or exposing sensitive information. The work in [
45,
46] has investigated the use of blockchain technology to create a privacy-preserving approach for training SVM models, where the data are owned by different parties that do not want to reveal them. To do that, each entity stores the ciphertexts of their data on the blockchain and then these ciphertexts can be used to train the SVM model without being able to compute the plaintext data. The work in [
28] investigates the use of mimic learning techniques to train a machine learning model without revealing the datasets that are owned by different parties. Each data owner trains a model, called teacher, using its local dataset, and then uses it to label insensitive unlabeled data used to train the usable model, called student. This process actually transfers the learning from the teacher model to the student without exposing the sensitive data to preserve privacy. In contrast to these works, which primarily focus on training machine learning models without exposing sensitive information, our emphasis in this paper lies in executing a medical diagnosis model while protecting the confidentiality of the users and the model’s parameters and building an efficient scheme by using lightweight cryptosystems and ensuring that it does not need involving the model owner in computing the diagnosis (or classification) scores.
Few works have investigated the evaluation of medical diagnosis models with privacy preservation. In [
47], homomorphic encryption cryptosystem [
48] is used to develop a medical diagnosing approach that can be run by a distrusted third-party server that provides cloud service. In [
49], homomorphic encryption is used to evaluate a single decision tree medical diagnosis without revealing sensitive information on the patient’s data. The work in [
9] has investigated the use of homomorphic encryption to evaluate a multiclass SVM medical diagnosis system hosted by a cloud server. However, as explained in [
10], this work suffers from several limitations, where the most prominent one is exposing sensitive information by the server. Compared to these schemes, our approach can protect the diagnosis model’s confidentiality and preserve the privacy of the users’ data and health status, and it does not need the participation of the MO to compute the diagnosis scores while maintaining low resources in term of communication and computation.
The approach of [
12] is close to our proposal. It uses the Okamoto–Uchiyama (OU) cryptosystem [
50] to build an online medical diagnosis system based on multi-class SVM. It is worth noting that the homomorphic operations are used in the OU cryptosystem to evaluate the model over encrypted data. Nevertheless, there are several shortcomings associated with the approach proposed in [
12], which can be summarized as follows. First of all, under the known-background model, the proposed approach reveals the classification (diagnosis) results to the cloud server, which creates a serious privacy issue. Specifically, because the server can link encrypted classifications that are for the same diagnosis, the server can easily determine these diagnoses by using some contextual data, like the prevalence of a particular disease in a certain area. The second shortcoming is that the MOs participate in the proposed computation of each diagnosis score in [
12]. By doing this, the advantage of outsourcing the diagnosis model to a third party that provides a cloud service is diminished because it has to be always online and allocate computation and communication resources for the medical diagnosis process. The third shortcoming is that the MO has to send some model parameters to all users, if it needs to update the diagnosis model run by the cloude server, i.e., to add more diseases or increase the accuracy. The reason for this is that the proposed approach requires that the users execute a part of the model. In another notable shortcoming, because the users need to compute a part of the model, as just explained, the proposed approach imposes high computation overhead on the users. This requirement may be difficult to achieve practically because some users may use resource-limited devices like tablets and cellphones. Compared to the proposed approach in [
12], our approach can hide diagnosis results without the need for online and interactive model owners to compute the diagnosis scores, in addition to requiring low resources in terms of communication and computation.
In [
13], the authors attempted to address the limitations of [
12] discussed above. The work in [
13] proposes a multi-class SVM-based diagnosis system using two cloud servers that are assumed non-colluding. Although this system was able to address several limitations such as protecting the confidentiality of the diagnosis model’s parameters and the classification results, and computing the classification without the involvement of the model owner, it suffers from several limitations. Much more communication and computation resources are needed especially as more diseases (or classes) are diagnosed by the model. Additionally, two servers are needed to execute the proposed approach in [
13], and the servers should not be colluding to ensure the security of the approach. Such a requirement is costly compared to our approach which needs only one server and the requirement that the two servers are not colluding is hard to achieve. Finally, the proposed approach uses a non-linear SVM model, and evaluating this model in the ciphertext domain needs extensive computation and communication resources because it is not easy to compute non-linear operations using encrypted data. In some applications, employing non-linear SVMs might seem appealing due to making a more sophisticated decision boundary compared to linear SVM. However, this choice often adds complexity to the scheme’s implementation in the ciphertext domain to preserve privacy. Our experiments in this paper showed that linear SVM gives good accuracy and, at the same time, it is easier to implement in the ciphertext domain. Compared to the approach proposed in [
13], our proposal in this paper develops a single-cloud system and offers more efficient computation and communication overheads.