A Deep Reinforcement Learning Approach to Droplet Routing for Erroneous Digital Microfluidic Biochips †
Abstract
:1. Introduction
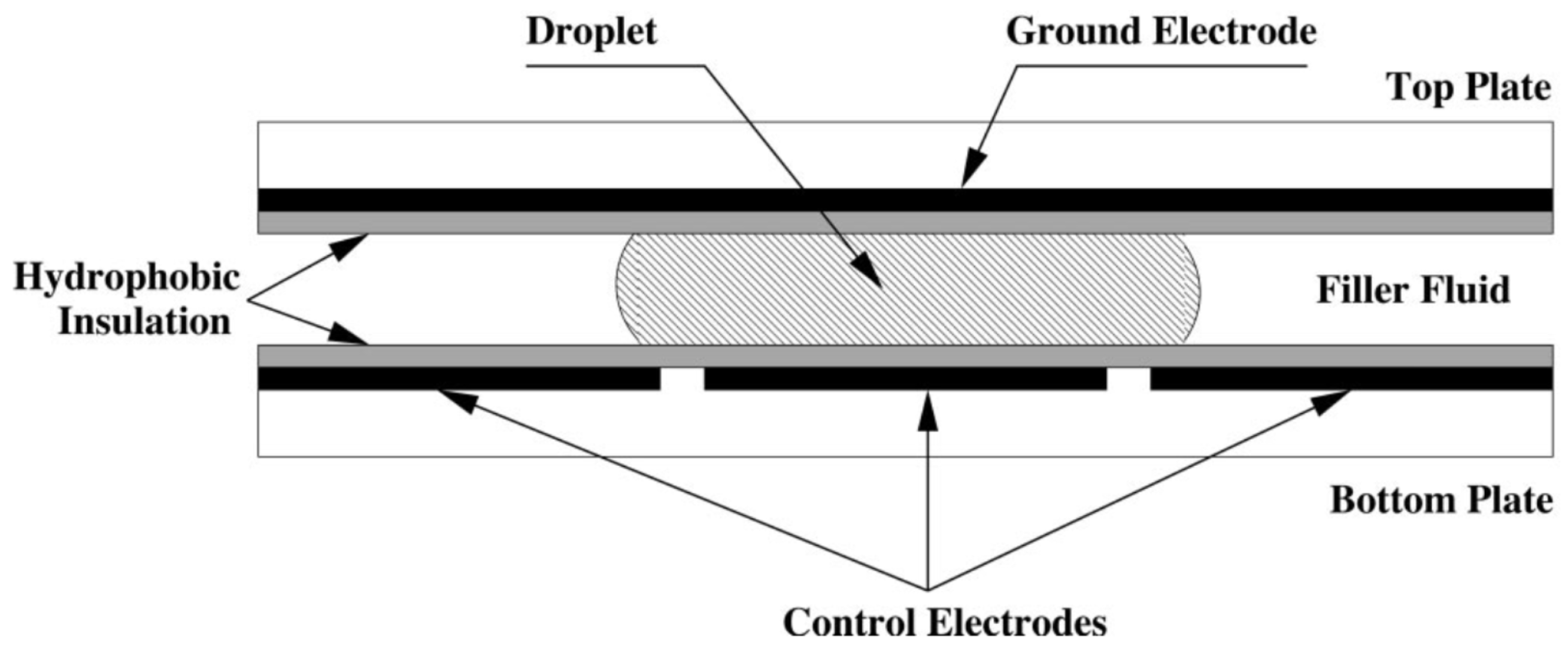
1.1. Digital Microfluidic Biochips (DMFBs)
1.2. Deep Reinforcement Learning (DRL)
1.3. Related Works
1.4. Paper Contributions
- This paper presents a new deep reinforcement learning-based routing algorithm for digital microfluidic biochips (DMFBs);
- It contributes to the field by addressing the crucial issue of error management in DMFBs, specifically both known and unknown errors. It proposes and tests an algorithm that can effectively handle different types of errors, potentially boosting the reliability and efficiency of biochips;
- In addition to proposing a new algorithm, this paper conducted extensive experiments to compare the performance of this algorithm against existing ones. The comprehensive results demonstrated the superior performance of the proposed algorithm in terms of accuracy, optimality of the routing path, and error detection capability.
2. Proposed DRL-Based Routing Algorithm
2.1. Framework Description
2.2. Environment
2.3. Agent
3. Simulation Experiments
3.1. Simulation Setup
- GPU: GeForce RTX 3060 LHR
- CPU: Core i7-12700F
- RAM: 80 GB
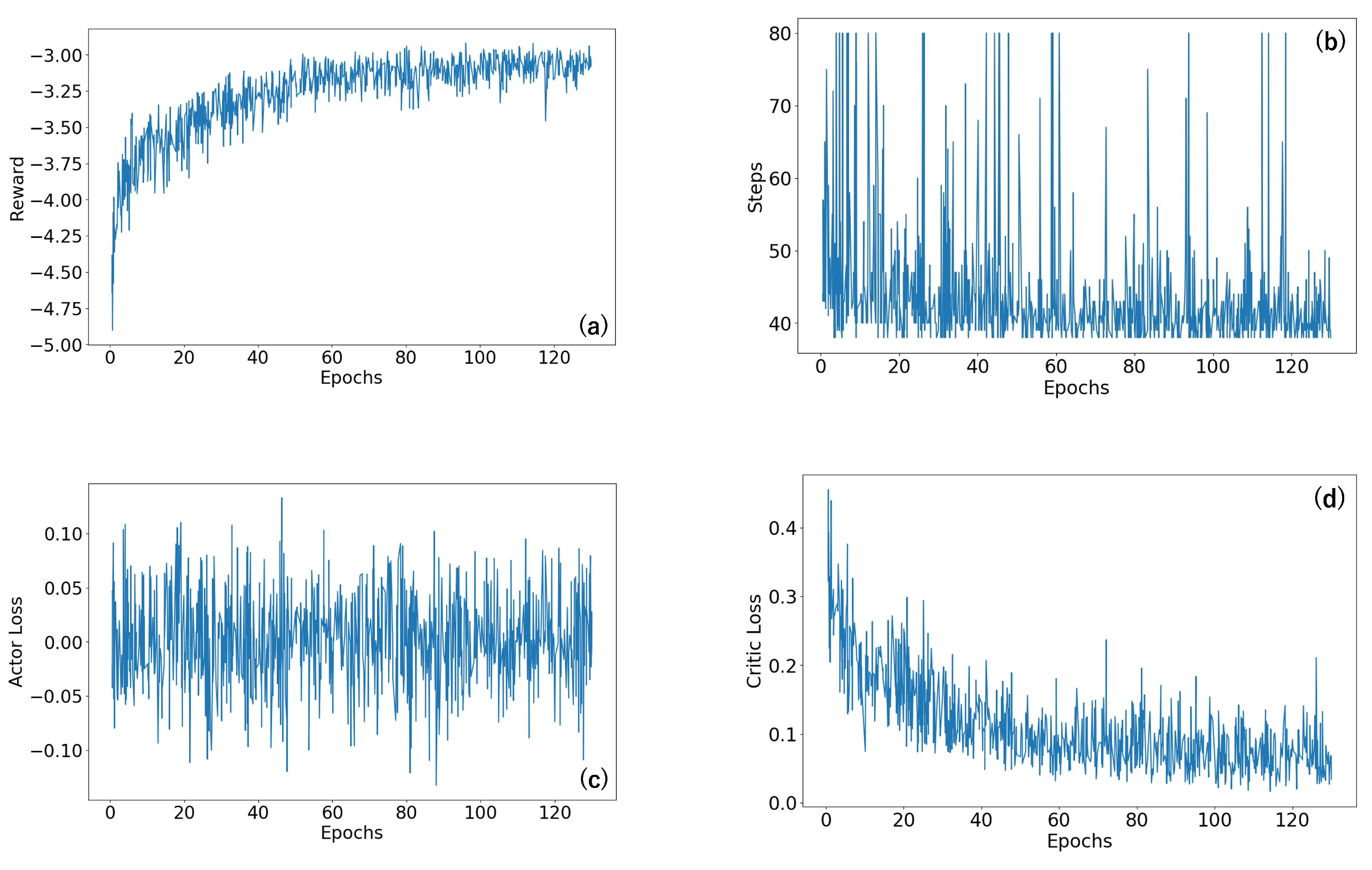
3.2. Agent Training
3.3. Simulation Results
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Azizipour, N.; Avazpour, R.; Rosenzweig, D.H.; Sawan, M.; Ajji, A. Evolution of biochip technology: A review from lab-on-a-chip to organ-on-a-chip. Micromachines 2020, 11, 599. [Google Scholar] [CrossRef]
- Su, F.; Chakrabarty, K. High-level synthesis of digital microfluidic biochips. ACM J. Emerg. Technol. Comput. Syst. (JETC) 2008, 3, 1–32. [Google Scholar] [CrossRef]
- Sista, R.S.; Ng, R.; Nuffer, M.; Basmajian, M.; Coyne, J.; Elderbroom, J.; Hull, D.; Kay, K.; Krishnamurthy, M.; Roberts, C.; et al. Digital microfluidic platform to maximize diagnostic tests with low sample volumes from newborns and pediatric patients. Diagnostics 2020, 10, 21. [Google Scholar] [CrossRef]
- Huang, S.; Connolly, J.; Khlystov, A.; Fair, R.B. Digital microfluidics for the detection of selected inorganic ions in aerosols. Sensors 2020, 20, 1281. [Google Scholar] [CrossRef]
- Ganguli, A.; Mostafa, A.; Berger, J.; Aydin, M.Y.; Sun, F.; Ramirez, S.A.S.d.; Valera, E.; Cunningham, B.T.; King, W.P.; Bashir, R. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 22727–22735. [Google Scholar] [CrossRef]
- Yang, C.; Gan, X.; Zeng, Y.; Xu, Z.; Xu, L.; Hu, C.; Ma, H.; Chai, B.; Hu, S.; Chai, Y. Advanced design and applications of digital microfluidics in biomedical fields: An update of recent progress. Biosens. Bioelectron. 2023, 242, 115723. [Google Scholar] [CrossRef]
- Schachter, S.C.; Dunlap, D.R.; Lam, W.A.; Manabe, Y.C.; Martin, G.S.; McFall, S.M. Future potential of Rapid Acceleration of Diagnostics (RADx Tech) in molecular diagnostics. Expert Rev. Mol. Diagn. 2021, 21, 251–253. [Google Scholar] [CrossRef]
- Dkhar, D.S.; Kumari, R.; Malode, S.J.; Shetti, N.P.; Chandra, P. Integrated lab-on-a-chip devices: Fabrication methodologies, transduction system for sensing purposes. Pharm. Biomed. Anal. 2023, 223, 115120. [Google Scholar] [CrossRef]
- Chaudhary, V.; Khanna, V.; Awan, H.T.A.; Singh, K.; Khalid, M.; Mishra, Y.K.; Bhansali, S.; Li, C.Z.; Kaushik, A. Towards hospital-on-chip supported by 2D MXenes-based 5th generation intelligent biosensors. Biosens. Bioelectron. 2023, 220, 114847. [Google Scholar] [CrossRef]
- Thorsen, T.; Maerkl, S.J.; Quake, S.R. Microfluidic large-scale integration. Science 2002, 298, 580–584. [Google Scholar] [CrossRef]
- Verpoorte, E.; De Rooij, N.F. Microfluidics meets MEMS. Proc. IEEE 2003, 91, 930–953. [Google Scholar] [CrossRef]
- Pollack, M.G. Electrowetting-Based Microactuation of Droplets for Digital Microfluidics; The Duke University: Durham, NC, USA, 2001. [Google Scholar]
- Cho, S.K.; Moon, H.; Kim, C.J. Creating, transporting, cutting, and merging liquid droplets by electrowetting-based actuation for digital microfluidic circuits. J. Microelectromech. Syst. 2003, 12, 70–80. [Google Scholar]
- Pollack, M.G.; Fair, R.B.; Shenderov, A.D. Electrowetting-based actuation of liquid droplets for microfluidic applications. Appl. Phys. Lett. 2000, 77, 1725–1726. [Google Scholar] [CrossRef]
- Verheijen, H.; Prins, M. Reversible electrowetting and trapping of charge: Model and experiments. Langmuir 1999, 15, 6616–6620. [Google Scholar] [CrossRef]
- Welch, E.R.F.; Lin, Y.Y.; Madison, A.; Fair, R.B. Picoliter DNA sequencing chemistry on an electrowetting-based digital microfluidic platform. Biotechnol. J. 2011, 6, 165–176. [Google Scholar] [CrossRef]
- Su, F.; Chakrabarty, K.; Fair, R.B. Microfluidics-based biochips: Technology issues, implementation platforms, and design-automation challenges. IEEE Trans. Comput.-Aided Des. Integr. Circuits Syst. 2006, 25, 211–223. [Google Scholar] [CrossRef]
- Zhao, Y.; Chakrabarty, K. Cross-contamination avoidance for droplet routing in digital microfluidic biochips. IEEE Trans. Comput.-Aided Des. Integr. Circuits Syst. 2012, 31, 817–830. [Google Scholar] [CrossRef]
- Liang, T.C.; Zhong, Z. Adaptive droplet routing in digital microfluidic biochips using deep reinforcement learning. In Proceedings of the 37th International Conference on Machine Learning, ICML 2020, Online, 13–18 July 2020. [Google Scholar]
- Li, J.; Kim, C.-J. Current commercialization status of electrowetting-on-dielectric (EWOD) digital microfluidics. Lab Chip 2020, 20, 1705–1712. [Google Scholar] [CrossRef]
- Sutton, R.S. Dyna, an integrated architecture for learning, planning, and reacting. ACM Sigart Bull. 1991, 2, 160–163. [Google Scholar] [CrossRef]
- Mnih, V.; Kavukcuoglu, K.; Silver, D.; Graves, A.; Antonoglou, I.; Wierstra, D.; Riedmiller, M. Playing atari with deep reinforcement learning. arXiv 2013, arXiv:1312.5602. [Google Scholar]
- Mnih, V.; Kavukcuoglu, K.; Silver, D.; Rusu, A.A.; Veness, J.; Bellemare, M.G.; Graves, A.; Riedmiller, M.; Fidjeland, A.K.; Ostrovski, G.; et al. Human-level control through deep reinforcement learning. Nature 2015, 518, 529–533. [Google Scholar] [CrossRef]
- Silver, D.; Huang, A.; Maddison, C.J.; Guez, A.; Sifre, L.; Van Den Driessche, G.; Schrittwieser, J.; Antonoglou, I.; Panneershelvam, V.; Lanctot, M.; et al. Mastering the game of Go with deep neural networks and tree search. Nature 2016, 529, 484–489. [Google Scholar] [CrossRef]
- Vinyals, O.; Babuschkin, I.; Czarnecki, W.M.; Mathieu, M.; Dudzik, A.; Chung, J.; Choi, D.H.; Powell, R.; Ewalds, T.; Georgiev, P.; et al. Grandmaster level in StarCraft II using multi-agent reinforcement learning. Nature 2019, 575, 350–354. [Google Scholar] [CrossRef]
- Ouyang, L.; Wu, J.; Jiang, X.; Almeida, D.; Wainwright, C.; Mishkin, P.; Zhang, C.; Agarwal, S.; Slama, K.; Ray, A.; et al. Training language models to follow instructions with human feedback. Adv. Neural Inf. Process. Syst. 2022, 35, 27730–27744. [Google Scholar]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Su, F.; Chakrabarty, K. Architectural-level synthesis of digital microfluidics-based biochips. In Proceedings of the IEEE/ACM International Conference on Computer Aided Design, 2004. ICCAD-2004, San Jose, CA, USA, 7–11 November 2004; pp. 223–228. [Google Scholar]
- Chakrabarty, K.; Fair, R.B.; Zeng, J. Design tools for digital microfluidic biochips: Toward functional diversification and more than moore. IEEE Trans. Comput.-Aided Des. Integr. Circuits Syst. 2010, 29, 1001–1017. [Google Scholar] [CrossRef]
- Huang, T.W.; Ho, T.Y. A fast routability-and performance-driven droplet routing algorithm for digital microfluidic biochips. In Proceedings of the 2009 IEEE International Conference on Computer Design, Lake Tahoe, CA, USA, 4–7 October 2009; pp. 445–450. [Google Scholar]
- Keszocze, O.; Wille, R.; Drechsler, R. Exact routing for digital microfluidic biochips with temporary blockages. In Proceedings of the 2014 IEEE/ACM International Conference on Computer-Aided Design (ICCAD), San Jose, CA, USA, 2–6 November 2014; pp. 405–410. [Google Scholar]
- Pan, I.; Samanta, T. Weighted optimization of various parameters for droplet routing in digital microfluidic biochips. In Recent Advances in Intelligent Informatics, Proceedings of the Second International Symposium on Intelligent Informatics (ISI’13), Mysore, India, 23–24 August 2013; Springer: Cham, Switzerland, 2013; pp. 131–139. [Google Scholar]
- Su, F.; Chakrabarty, K. Yield enhancement of reconfigurable microfluidics-based biochips using interstitial redundancy. ACM J. Emerg. Technol. Comput. Syst. (JETC) 2006, 2, 104–128. [Google Scholar] [CrossRef]
- Xu, T.; Chakrabarty, K. Integrated droplet routing in the synthesis of microfluidic biochips. In Proceedings of the DAC07: The 44th Annual Design Automation Conference 2007, San Diego, CA, USA, 4–8 June 2007; pp. 948–953. [Google Scholar]
- Zhao, Y.; Chakrabarty, K. Simultaneous optimization of droplet routing and control-pin mapping to electrodes in digital microfluidic biochips. IEEE Trans. Comput.-Aided Des. Integr. Circuits Syst. 2012, 31, 242–254. [Google Scholar] [CrossRef]
- Luo, Y.; Chakrabarty, K.; Ho, T.Y. Error recovery in cyberphysical digital microfluidic biochips. IEEE Trans. Comput.-Aided Des. Integr. Circuits Syst. 2012, 32, 59–72. [Google Scholar] [CrossRef]
- Willsey, M.; Stephenson, A.P.; Takahashi, C.; Vaid, P.; Nguyen, B.H.; Piszczek, M.; Betts, C.; Newman, S.; Joshi, S.; Strauss, K.; et al. Puddle: A dynamic, error-correcting, full-stack microfluidics platform. In Proceedings of the ASPLOS’19: Architectural Support for Programming Languages and Operating Systems, Providence, RI, USA, 13–17 April 2019; pp. 183–197. [Google Scholar]
- Su, F.; Chakrabarty, K. Design of fault-tolerant and dynamically-reconfigurable microfluidic biochips. In Proceedings of the Design, Automation and Test in Europe, Munich, Germany, 7–11 March 2005; pp. 1202–1207. [Google Scholar]
- Kawakami, T.; Shiro, C.; Nishikawa, H.; Kong, X.; Tomiyama, H.; Yamashita, S. A Deep Reinforcement Learning-based Routing Algorithm for Unknown Erroneous Cells in DMFBs. In Proceedings of the 2023 21st IEEE Interregional NEWCAS Conference (NEWCAS), Edinburgh, UK, 26–28 June 2023; pp. 1–5. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25. [Google Scholar] [CrossRef]
- Schulman, J.; Wolski, F.; Dhariwal, P.; Radford, A.; Klimov, O. Proximal policy optimization algorithms. arXiv 2017, arXiv:1707.06347. [Google Scholar]
- Mnih, V.; Badia, A.P.; Mirza, M.; Graves, A.; Lillicrap, T.; Harley, T.; Silver, D.; Kavukcuoglu, K. Asynchronous methods for deep reinforcement learning. In Proceedings of the 33rd International Conference on Machine Learning, New York, NY, USA, 20–22 June 2016; pp. 1928–1937. [Google Scholar]
- Schulman, J.; Levine, S.; Abbeel, P.; Jordan, M.; Moritz, P. Trust region policy optimization. In Proceedings of the 32nd International Conference on Machine Learning, Lille, France, 7–9 July 2015; pp. 1889–1897. [Google Scholar]




| State | Reward |
|---|---|
| Reach the goal state | 0 |
| Reach the maximum step number | −1.0 |
| Any other state | −0.1 |
| Type | Depth | Activation | Kernel | Padding |
|---|---|---|---|---|
| Convolution | 32 | ReLU | 3 | 1 |
| Convolution | 64 | ReLU | 3 | 1 |
| Convolution | 64 | ReLU | 3 | 0 |
| Linear | 256 | ReLU | N/A | N/A |
| Linear | 4 (1) a | Softmax | N/A | N/A |
| Chip Size | Error Rate | Existing Method | Proposed Method | ||
|---|---|---|---|---|---|
| Routing Success | Routing Success | Optimal Path Rate | Unknown Error Detection | ||
| (0, 5) | 62 | 100 | 100 | 0.34 | |
| (5, 5) | 58 | 100 | 99 | 0.42 | |
| (0, 10) | 58 | 100 | 99 | 0.74 | |
| (0, 5) | 74 | 100 | 100 | 0.35 | |
| (5, 5) | 69 | 100 | 100 | 0.30 | |
| (0, 10) | 32 | 100 | 99 | 0.82 | |
| (0, 5) | 52 | 100 | 100 | 0.54 | |
| (5, 5) | 55 | 100 | 93 | 0.51 | |
| (0, 10) | 2 | 100 | 94 | 1.25 | |
| (0, 5) | 53 | 100 | 100 | 0.49 | |
| (5, 5) | 53 | 100 | 98 | 0.60 | |
| (0, 10) | 22 | 100 | 94 | 1.27 | |
| (0, 5) | 62 | 100 | 99 | 0.79 | |
| (5, 5) | 42 | 100 | 97 | 0.76 | |
| (0, 10) | 16 | 100 | 95 | 1.36 | |
| (0, 5) | 46 | 100 | 100 | 0.74 | |
| (5, 5) | 43 | 100 | 93 | 0.87 | |
| (0, 10) | 9 | 100 | 95 | 1.36 | |
| (0, 5) | 38 | 100 | 98 | 0.98 | |
| (5, 5) | 30 | 100 | 91 | 0.91 | |
| (0, 10) | 17 | 100 | 89 | 0.89 | |
| (0, 5) | 31 | 100 | 98 | 1.18 | |
| (5, 5) | 34 | 100 | 91 | 1.01 | |
| (0, 10) | 6 | 100 | 92 | 2.06 | |
| (0, 5) | 27 | 100 | 93 | 0.93 | |
| (5, 5) | 33 | 100 | 90 | 1.30 | |
| (0, 10) | 8 | 100 | 84 | 2.55 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawakami, T.; Shiro, C.; Nishikawa, H.; Kong, X.; Tomiyama, H.; Yamashita, S. A Deep Reinforcement Learning Approach to Droplet Routing for Erroneous Digital Microfluidic Biochips. Sensors 2023, 23, 8924. https://doi.org/10.3390/s23218924
Kawakami T, Shiro C, Nishikawa H, Kong X, Tomiyama H, Yamashita S. A Deep Reinforcement Learning Approach to Droplet Routing for Erroneous Digital Microfluidic Biochips. Sensors. 2023; 23(21):8924. https://doi.org/10.3390/s23218924
Chicago/Turabian StyleKawakami, Tomohisa, Chiharu Shiro, Hiroki Nishikawa, Xiangbo Kong, Hiroyuki Tomiyama, and Shigeru Yamashita. 2023. "A Deep Reinforcement Learning Approach to Droplet Routing for Erroneous Digital Microfluidic Biochips" Sensors 23, no. 21: 8924. https://doi.org/10.3390/s23218924
APA StyleKawakami, T., Shiro, C., Nishikawa, H., Kong, X., Tomiyama, H., & Yamashita, S. (2023). A Deep Reinforcement Learning Approach to Droplet Routing for Erroneous Digital Microfluidic Biochips. Sensors, 23(21), 8924. https://doi.org/10.3390/s23218924







