1. Introduction
Humans spend almost one-third of their lifetime sleeping. Healthy sleep is vital for human physical and mental health. Sleep disorders, such as insomnia, apnea, and parasomnias, affect the quality of sleep, causing medical conditions, such as depression, difficulty concentrating, and weight gain [
1]. Delays in the treatment of some sleep disorders can lead to more serious diseases, such as heart disease, diabetes, and memory loss [
2,
3,
4]. Polysomnography, or a sleep study, is commonly used to diagnose sleep disorders. A polysomnography includes various biosignals measured during eight hours of overnight sleep [
5]. Electroencephalogram (EEG), which measures the brain’s electrical activity, is the main biosignal measure used in sleep staging, the time-intensive step of identifying the five different stages in the EEG signal. Sleep stage scoring is typically performed by dividing the EEG signal into epochs and assigning each epoch to one of the five sleep stages defined by the American Academy of Sleep Medicine (AASM) [
6], namely wake stage (W), rapid eye movement stage (REM), and non-random eye movement stages (N1, N2, and N3). N3 is what is known as the deep sleep stage. The amount of sleep spent in the deep sleep stage is a key indicator of healthy sleep.
Manual sleep stage scoring is costly and a resource- and time-consuming process. Therefore, there is an increased interest in automatic sleep stage scoring techniques to support experts in their diagnosis. Several approaches have been proposed to automatically classify each epoch to one of the five sleep stages, including shallow classifiers, such as support vector machine [
7], random forest [
8], decision tree [
9], as well as deep learning techniques [
10], such as convolutional neural network (CNN) [
11] and long short-term memory (LSTM) [
12,
13]. Most of the recent automatic sleep stage scoring methods rely on a signal from a single EEG channel [
12,
13,
14,
15,
16,
17,
18,
19]. A typical EEG-based sleep stage scoring method consists of pre-processing and feature extraction modules followed by the classifier module. However, recent end-to-end deep learning methods use the raw EEG signal as an input to a trainable front-end feature extraction network. The most commonly used architecture for the feature extraction network is the multi-resolution convolutional neural networks (CNNs) [
18,
20].
In polysomnography, multiple EEG channels are collected using a cap placed on the head, which patients find uncomfortable and disruptive. Collecting data from just a single EEG channel would be more comfortable for the patient and enable home monitoring. It would also make the collection of large datasets needed to train data-hungry algorithms for sleep stage scoring, such as deep learning methods, more feasible. Knowing which channel is the most effective is, thus, crucial. Due to the limitation of available sleep datasets in terms of the number of channels and the number and varieties of participants, most of the current work has been applied to a limited number of channels and focused only on adult sleep signals.
Children’s sleep is known to have significant variations across different age groups and within the same age group [
21]. However, due to the unavailability of adequate child sleep data, very little work has been carried out on automatically analyzing child sleep [
13,
16,
22].
Therefore, in this paper, the following questions were investigated:
How does the type of EEG channel used impact the accuracy of automatic sleep staging?
How does the accuracy of automatic sleep staging vary with the subject’s age?
How effective is a model trained on adult EEG data for sleep staging on pediatric EEG signals?
To answer these questions, the performance of two state-of-the-art (SOTA) automatic sleep staging algorithms, DeepSleepNet [
20] and AttnSleep [
18], was compared on seven different EEG channels and over 19 age ranges between the ages of 6 days to ~58 years.
The remainder of the paper is organized as follows.
Section 2 highlights recent work on automatic sleep staging. The two adopted sleep staging algorithms are summarized in
Section 3 along with the utilized dataset. The experimental setup is detailed in
Section 4. The results are presented in
Section 5 and discussed in
Section 6. Finally, the conclusion is drawn in
Section 7.
2. Related Work
Existing sleep staging techniques can be divided into two main categories: classical (shallow) machine learning and deep learning-based. Although classical machine learning techniques are computationally inexpensive, their accuracy is highly dependent on the selection of hand-crafted features.
Deep learning techniques have recently made significant advancements in their classification capabilities. These techniques can extract features of EEG signals directly and capture the transition relationship between sleep stages. However, a considerable amount of training data is needed to train a reliable deep learning sleep staging model.
In [
16], a bi-stream adversarial learning network was proposed for classifying sleep stages of pediatric data. The proposed system consists of two stream networks (i.e., Student and Teacher) in addition to a similarity function to reduce the difference between the output of the aforementioned networks. Furthermore, two datasets were used to evaluate the proposed system. The first one named NPH contains O1-M2 EEG data of 15 pediatric participants, while the second dataset is a subset of the Sleep-EDF dataset that contains Fpz-Cz EEG data of eight adult participants. The proposed systems achieved remarkable results (i.e., 80% and 91% over NPH and a subset of the Sleep-EDF, respectively) compared with other systems, although they struggled with the N1 sleep stage.
In [
22], a hybrid multi-domain neural network was proposed. The proposed system used CNN and bidirectional LSTM (BLSTM) models for classifying sleep stages of pediatric EEG data. Two layers of CNN were used to model the frequency and time domain information followed by the LSTM layer to model the dependency of the data within every epoch. The proposed system was tested using a dataset of 115 female and 103 male pediatric participants with 32 EEG channels. In addition, the effect of epoch length, the number of channels to be used, and the use of frequency and time domain information was investigated. The proposed system achieved 92% accuracy but over three sleeping stages only (i.e., W, N1, and N2).
A model that utilized the CNN and recurrent neural network (RNN) was introduced in [
15]. This model is composed of three layers of convolutional neural networks and two layers of recurrent neural networks. The proposed model was trained using a dataset of ECG signals of two groups of participants (i.e., controlled and uncontrolled) with a total of 71 male participants and 41 female participants. All participants were above 48 years old. The model accuracy was high in the case of three-stage classification (i.e., 86.4%) and low with five-stage classification (i.e., 74.2%). One of the drawbacks of this model is its higher computational load compared to other models.
Enhancing feature extraction from EEG signals was performed in [
17]. The authors added an attention component to the CNN to learn local correlations of EEG signals. The CNN layers were followed by two layers of BLSTM to extract global correlations of sequential epochs. The sleep-EDF dataset with its two versions, 2013 and 2018, was used to evaluate the proposed model. It achieved 84.14% and 82.58% accuracy over the Fpz-Cz and Pz-Oz channels, respectively, on the 2013 version. However, less accuracy was achieved with the 2018 version.
The authors of [
14] used classical machine learning algorithms to classify sleep stages. In the feature extraction phase, they extracted a set of frequency and time domain features from EEG signals. In addition, they used assigned weights for each feature to determine its importance. These weights were used to decrease the number of features from 59 to only 11. In the classification phase, decision trees, support vector machine, random forest, and backpropagation neural network were tested. The random forest classifier achieved the best accuracy over the Fpz-Cz channel from the expanded Sleep-EDF dataset. It achieved 83.56% and 82.53% using 59 and 11 features, respectively.
Similarly, time and frequency domain features were extracted from EEG signals in [
23]. The authors extracted a set of 136 features and proposed a technique based on ant colony optimization (ACO) to reduce the features to only 40 features. In addition, they utilized a random forest classifier for sleep stage classification. Furthermore, they improved the classification accuracy using the hidden Markov model (HMM), which takes into account knowledge about the temporal pattern of sleep stage transitions. The proposed model was faster than RNN- and CNN-based models and less susceptible to modifications of hyperparameters, but its feature extraction process was time-consuming.
The authors of [
24] used the C4-M1 channel to develop clinical decision support systems (CDSSs). The authors proposed a deep learning model that utilized CNN and transformers to classify three sleep stages. Five layers of CNN were used to extract features from EEG data, then two transformers were used to encode the current and previous epochs. In addition, a large dataset with 2274 participants was used to train and evaluate the proposed model that achieved high accuracy. The disadvantage of this model is that the training of the second transformer requires a successive number of epochs (i.e., seven epochs).
Using the EEG signal in addition to time-frequency images was proposed in [
25]. The authors proposed a sequence-to-sequence model able to learn from both EEG signals and images of time-frequency. This model utilized a CNN to extract features from raw signals and a bidirectional RNN to extract time–frequency features. In addition, they designed this model to take into consideration the complementary effect of both network streams (i.e., CNN and RNN) and the avoidance of overfitting. Although they achieved a high accuracy of sleep staging classification over five datasets, using two-stream networks consumes more time and compactional resources.
Ref. [
26] proposed using a multi-level fusion technique over EEG and electromyography (EMG) data. The authors tried one level of fusion (i.e., data, features, and decision fusion) and found that multi-level fusion achieved better accuracy. They suggested a fusion technique constructed with the CNN and suited to the properties of various EEG and EMG inputs. In addition, they tested the proposed method over the Sleep-EDF dataset using three layers of CNN to extract features from Fpz-Cz, Pz-Oz, and EMG data. Their methods achieved high accuracy (i.e., 87.3%) and improved the N1 stage accuracy.
Very limited methods have been applied to children’s EEG signals. In [
27], a deep neural network model was proposed for classifying children’s sleep stages. The proposed model utilized a modularized architecture that enables the neural network to have many layers without being constrained by the increasing number of hyperparameters. In addition, a dataset containing a sleep study of 344 patients was developed to train and evaluate the proposed system. The age of patients was between 2 and 18 years. Furthermore, the system achieved an accuracy of 83.36% over five stages of classification. The limitations of this system are its low accuracy for N1 and the lack of results from children under 2 years old.
A stacked 1D CNN- and LSTM-based method has been introduced in [
13] and applied to a small private dataset of 26 children ranging from 2–12 years old. The method employed data from a single EEG channel and trained using the edge AI paradigm. Six EEG channels were explored, namely F3-M2, F4-M1, C3-M2, C4-M1, O1-M2, and O2-M1. The best F1 score of 76.5 was achieved using the F4-M1 channel.
It has been noted that most of the literature work used a small publicly available dataset, such as sleep-EDF [
28], or a large private dataset. In addition, most of the experiments have been conducted using an adult dataset and applied to limited EEG channels.
In this study, a large-scale dataset is used with a wide age range of participants, and seven different EEG channels are explored.
4. Experimental Setup
As previously mentioned, the main objective of this paper is to study the performance of the single EEG channel-based sleep staging algorithms over different types of channels and different subjects’ age ranges. Several experiments were conducted to better understand the impact of the type of EEG channel and the age of the participant on automatic sleep staging. All experiments have been performed using the two sleep staging algorithms detailed above, DeepSleepNet and AttnSleep.
The dataset is first divided into 19 subsets representing different age groups with a one-year step. That is, group 0 contains data from subjects aged from 6 days to 52 weeks, while group 19 contains data from subjects aged 18 years and over. Subjects of each age group are further split into training, validation, and testing sets. Of the subjects, 80% were used for training, while the other 20% was split equally between the validation and testing subsets. All three subsets had the same male/female distribution of subjects.
Figure 3 shows the number of training epochs in each age range.
Seven EEG channels were employed in this study, namely C3-M2, O1-M2, O2-M1, CZ-O1, C4-M1, F4-M1, and F3-M2, as they are available in the recordings of ~99% of the participants [
29]. For each EEG channel, recordings of participants from each age group were used to train two automatic sleep staging models, one for each automatic sleep staging algorithm. Given the seven channels and the 19 age groups, a total number of 126 models for each algorithm were trained. All models were trained to classify each epoch of 30 s duration as one of the five sleep stages, W, N1, N2, N3, and REM.
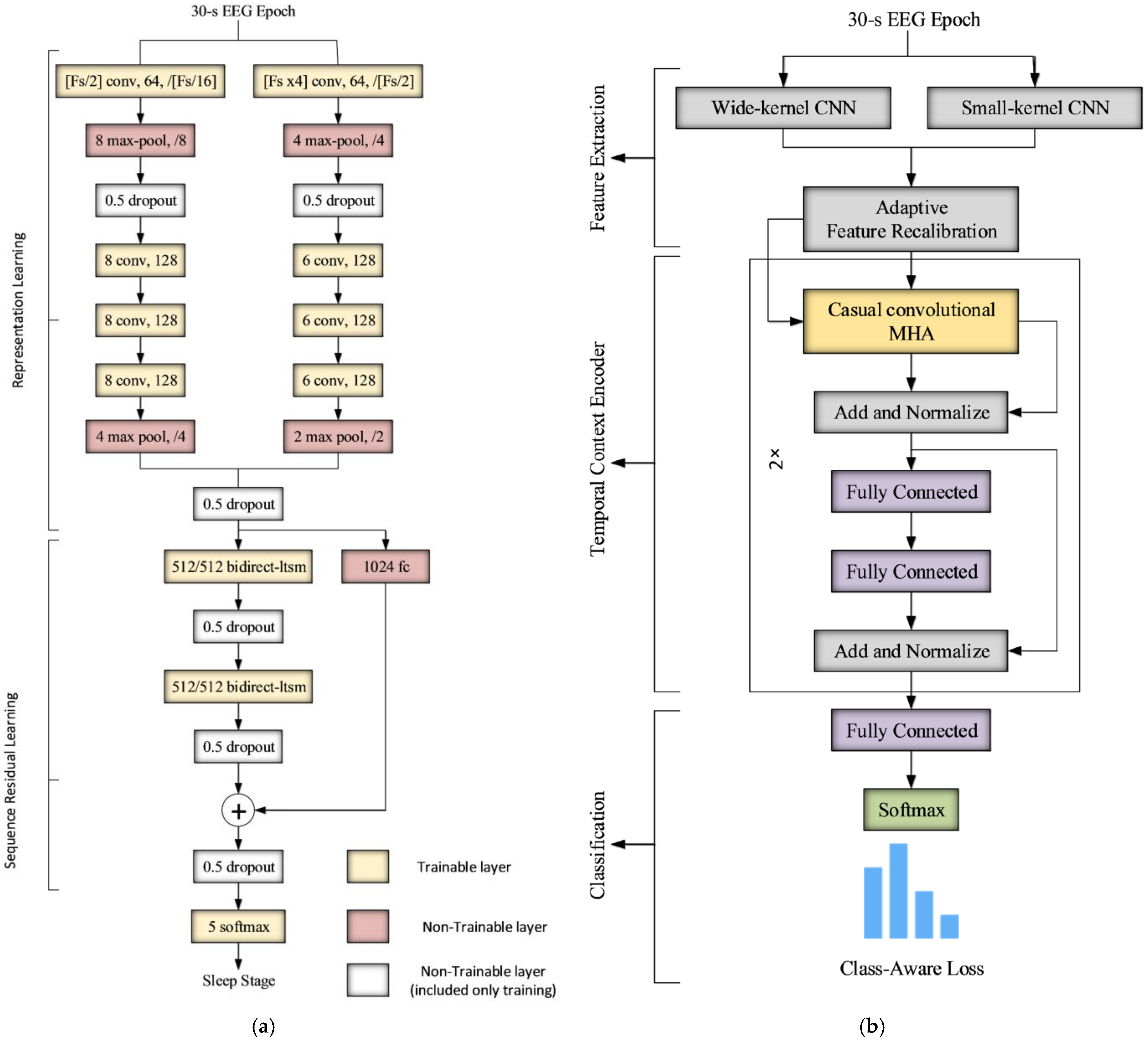
The parameters of the CNN feature extraction layers are the same in both algorithms, as they are a function of the sampling rate (see
Figure 2). The parameters of the classification network and the training parameters are empirically determined, and the best values are listed in
Table 1.
The sampling rate of the recordings was fixed to 256 Hz, and recordings with higher sampling rates were downsampled to 256 Hz. Both algorithms receive the raw EEG signal of a single epoch of 30 s as an input at each time step.
The F1 score was used to evaluate the classification performance of each stage
as follows:
The overall performance was reported as a weighted average of the
scores of all stages as follows:
where
is the number of test samples of the stage
, and
is the total number of samples in the test set.
5. Results
5.1. Channel Analysis
As discussed, to investigate the impact of EEG channels on the accuracy of automatic sleep staging, seven DeepSleepNet models and seven AttnSleep models were trained and tested using data from one of the EEG channels.
Figure 4a,b show the average
of each channel over all age groups for both DeepSleepNet and AttnSleep, respectively. Overall, AttnSleep achieved a higher average
WF1 compared with DeepSleepNet overall EEG channels, which is in line with the performance reported in [
18] on other datasets. For AttnSleep, the highest average
WF1 of ~77.8% was achieved on C3-M2 and F3-M2, followed by F4-M1, with an average
WF1 of ~77.3%. On the other hand, with DeepSleepNet, the highest average
of ~78% was obtained on CZ-O1, followed by F3-M2 and C3-M2, with average
scores of ~75% and ~74.2%, respectively. For both algorithms, O1-M2 and O2-M1 channels obtained the lowest average
scores of ~75% and ~72.7% for AttnSleep and ~70% and ~66% for DeepSleepNet, respectively.
As seen in
Figure 4a,b, DeepSleepNet suffers from a high variation in performance over different age groups throughout all channels, with standard deviations ranging from ±5% on CZ-M1 to ±10% on O2-M1. On the contrary, AttnSleep was shown to have a more consistent performance over age groups, with standard deviations ranging from ±4% achieved on C3-M2 to ±6.6% achieved on O2-M1.
Figure 5a,b show a heatmap of the WF1 scores of each channel over different age groups for the two adopted algorithms. AttnSleep performance on F3-M2 and F4-M1 was superior in subjects aged 5 years and older, while at younger ages, it outperformed on C3-M2 compared with other channels. With DeepSleepNet, the best performance was achieved on CZ-O1 over other channels in most age ranges.
We further broke down the performance of the DeepSleepNet and AttnSleep algorithms into the five sleep stages, as depicted in
Figure 6a–e. The performance is measured by the F1-score computed using Equation (1). N3 is the most accurate stage, with an average F1 score ranging from 85.6% ± 4% achieved on C3-M2 to 82% ± 4.3% achieved on the O2-M1 channel for the AttnSleep algorithm. For DeepSleepNet, the best average N3 detection F1 score of 84% ± 6.5% was obtained on CZ-O1, while the lowest average F1 score of 77% ± 13.3% was attained on O2-M1.
It is obvious from the figures that N1 is the most challenging stage, with an F1 score below 35% for both AttnSleep and DeepSleepNet models over all ages and channels. It is also noticeable that N1 has the highest performance variations among age groups in both algorithms compared with other stages.
5.2. Age Analysis
To explore the performance of automatic sleep staging at different age ranges, the NCH dataset was split into 19 groups on the basis of the age of the subjects from 0 to 18. The 0 group contains subjects aged up to 52 weeks, while the 18 group includes subjects of 18 years and older. Each model was trained and tested using data from the same age group.
In the previous section, CZ-O1 performed the best with DeepSleepNet and C3-M2 channel with AttnSleep. Therefore, in the following experiment, the CZ-O1 channel is used with the DeepSleepNet, and the C3-M2 channel is used with the AttnSleep.
Figure 7a,b show the F1 scores of the five sleep stages (dotted lines) over all age groups along with the WF1 score (solid line) for DeepSleepNet and AttnSleep algorithms, respectively. Each model was trained and tested using data from the same age group. As shown in the two figures, infant data (group 0) has the lowest F1 score over all stages and for both DeepSleepNet and AttnSleep algorithms compared with other age groups. The highest degradation in the performance of the infant data occurred in N1 and N2 stages, with F1 scores in the N1 stage close to 0% for both algorithms.
For DeepSleepNet, the F1 score gradually increases with the age and is relatively saturated after group 5 (>5 years). On the other hand, N3 and N2 stages in AttnSleep were shown to be less affected by age, with the exception of infant (group 0), while REM and Wake suffered from inconsistency performance, with a high degradation occurring at ages 3, 9, and 14.
As most of the available EEG datasets were collected from adult participants, such as the PhysioBank [
28], Sleep-EDF, and UCD datasets, and SHHS [
36], we tested the effectiveness of a model trained on adult data (>18) in detecting sleep stages of younger age groups. The results of the sleep staging of the two algorithms using the seven EEG channels over different age groups are depicted in
Figure 8. The results clearly show that the performance of automatic sleep staging using the adult model improves gradually with age and starts to saturate after 11 years old and 14 years old for AttnSleep and DeepSleepNet, respectively. This is an important finding, demonstrating the development of the EEG signals with age and the cut-off age where these signals become more adult-like. In other words, there is a high degree of discrepancy between adult and child EEG signal at younger ages, which decreases gradually as the child grows up. Unlike the within-age group scenario where CZ-O1 and C3-M2 outperform other channels in DeepSleepNet and AttnSleep algorithms, respectively, here F4-M1 channel is superior to other channels in both algorithms.
Figure 9a,b show the breakdown of the performance over different sleep stages using the F4-M1 channel for AttnSleep and DeepSleepNet, respectively. In AttnSleep, the N3 stage was shown to be the stage least affected by the subject age, with an average F1 score of 82.7% ± 5.7%, while the REM stage has a significantly lower performance at younger ages compared with older ages, with an average F1 score of 54% ± 17%.
Similarly, in DeepSleepNet, N3 has low variation in the performance between younger and older age groups compared with other stages, with an average F1 score of 80% ± 7%, while the N2 stage has a lower average F1 score of 69% and significantly higher standard deviation of ±17%.
6. Discussions
This paper provided, for the first time, an extensive comparison of the performance of automatic sleep staging of adults and children from a single EEG channel covering seven different EEG channels and a wide age range. The NCH recently released dataset was utilised for the study. The NCH dataset contains 40,884 h of sleep recordings collected from 3984 patients during a single night’s sleep. The participants were split into 19 groups on the basis of their age, from 0 (participants < 52 weeks) to 18 (participants >= 18 years old).
All experiments were conducted using two well-known deep learning-based automatic sleep staging algorithms, namely DeepSleepNet and AttnSleep.
A comparison of the two algorithms showed that AttnSleep performed better than the DeepSleepNet, specifically at younger ages (<2 years old) and also in the detection of the N1 stage throughout all ages (see
Figure 6). Moreover, the performance of AttnSleep was more consistent over different ages compared with the DeepSleepNet, which suffers from high variations in performance among age groups. As explained in
Section 3, both DeepSleepNet and AttnSleep adopt the same multi-resolution CNN front end module; however, AttnSleep architecture has an additional recalibration network AFR that works as an adaptive feature selection layer. As reported in the AttnSleep paper [
18], this module is the most effective part of the network, as it has the ability to learn the inter-dependencies among features and adaptively select the most discriminative ones.
The channel analysis revealed that the models have the lowest average F1 score on O1-M2 and O2-M1 over almost all ages, indicating that these channels are not effective in training single-EEG channel automatic sleep staging models. The most effective three channels were C3-M2, F3-M2, and F4-M1 for AttnSleep and CZ-O1, F3-M2, and F4-M1 for DeepSleepNet, which have F3-M2 and F4-M1 in common (see
Figure 4). Furthermore, F4-M1 was shown to be less affected by the age mismatch when used to train an adult model and tested with children’s data from different age groups (see
Figure 8).
To demonstrate how significant the superiority of the model trained on a signal from the central and front EEG channels (C3-M2, CZ-O1, F3-M2, and F4-M1) was over the back channels (O1-M2 and O2-M1), we employed the almost stochastic order (ASO) significance test [
37,
38], as implemented by [
39]. The method compares scores from two deep learning models
and
by computing the significance score
. If
then
is better than
based on a pre-defined significance level
. The lower
is, the higher the confidence that
is better than
.
Table 2 shows the significance score
when applying the ASO test, with significance level
, to pairs of models trained on different EEG channels using both DeepSleepNet and AttnSleep algorithms. As shown in
Table 2,
for all channel pairs, which indicates that the models trained on central and front EEG channels data are significantly better than the models trained on back channels.
When the sleep staging models were trained and tested using data from the same age group, significant degradation in the performance of the detection of all stages for infant subjects (<52 weeks) was observed in both algorithms (see
Figure 7).
When a model trained on adult data was used in performing sleep staging for child data, the performance was significantly lower in infant data and started to improve gradually to saturate after 13 years old (see
Figure 8). Therefore, to achieve an accurate sleep staging for young subjects (<12 years old), the model should be trained on data from the age group, while an adult model can be reliable from 13 years old and older.
Overall, the N1 stage achieved the lowest classification accuracy compared with other stages in all experiments. This is a well-known problem that has been reported in several EEG-based sleep staging algorithms [
18,
20,
30,
40]. The possible reasons for this low classification accuracy are: Firstly, N1 is the shortest stage in the sleep cycle (~5%), which means that it has the lowest number of samples in the training set (see
Figure 3), and therefore, even with the oversampling of the N1 class samples, the variations in the training data are not adequate to accurately model the N1 stage. Secondly, N1 is the transition stage between wake and deep sleep, and according to the AASM sleep-scoring guidelines [
41], the characteristics of the EEG signal during the N1 stage is similar to those of the W stage, and therefore, high confusion between N1 and W stages occurs. Finally, Lee et. al. show in a recent study [
42] that N1 has a very low interrater reliability of 0.24, while W, N2, N3, and R have interrater reliability of 0.7, 0.57, 0.57, and 0.69, respectively. The low interrater reliability affects the quality of the labels of the training data and, in turn, affects the accuracy of the trained model.
Compared with existing literature, to the best of our knowledge, no other work investigates the effect of age on the sleep staging classification performance. Moreover, the performance of the automatic sleep staging algorithm on pediatric sleep data has been briefly investigated. Jeon et al. [
22] applied several deep and shallow machine learning methods to private pediatric data of children aged 10–15 years old and used multiple EEG channels. When using signals from 19 EEG channels, the average F1 score was 0.9 and dropped to 0.86 when the number of channels reached 6. In our study, the average F1 score of children of the same age range was ~0.8 when only one EEG channel was utilized. Although they used several EEG channels, they did not investigate which channel is the most effective but focused on the optimum number of channels to be used.
In [
27] a CNN-based sleep staging model has been applied to private data collected from 344 children aged 2–18 years old. Although the data they used has a wide age range, the model was trained and tested using data from all age ranges, and the reported performance was averaged over all participants due to the lack of sufficient participants at each age group to train age-specific models.
Table 3 compares the results of the NCH dataset to the work performed on the Sleep-EDF and SHHS datasets using DeepSleepNet and AttnSleep algorithms. For a fair comparison, the results of NCH were obtained from the adult model (>18), as both Sleep-EDF and SHHS datasets were collected from adult participants. The results of the Sleep-EDF and SHHS datasets are obtained from the AttnSleep paper [
18].
The NCH is shown to be more challenging than the other datasets; one possible reason is that most NCH participants have some type of sleep disorder, while other datasets are collected from participants with healthy sleep.
7. Conclusions
Intensive experiments were conducted using a recently released large-scale pediatric sleep dataset for the automatic sleep-scoring problem to investigate the impact of the type of the EEG channel and the patient’s age on the accuracy of the single EEG channel sleep scoring system.
Two recent and widely accepted algorithms of automatic sleep scoring were utilized in this work. Approximately 126 models were trained and evaluated covering 19 age groups and seven different EEG channels. The experimental results provide the following answers to the underlying research questions:
The experiments clearly showed that the performance of the automatic sleep scoring was significantly affected by the electrode position of the channel. The EEG signal obtained from the back electrodes (O1 and O2) achieved consistently lower accuracy compared with the central (Cz) and front (F3, F4) electrodes. A similar finding has been reported in [
13], where six EEG channels were compared and O1-M2 and O2-M1 obtained the lowest F1 score of ~0.67 and ~0.65, respectively, while F4-M1 achieved the highest F1 score of 0.76.
This finding suggests that when training a single EEG-based automatic sleep-scoring system, O1 and O2 should be avoided, and Cz, F3, and F4 are more likely to achieve higher classification accuracy.
A noticeable degradation in the accuracy of the infant participants using a model trained and tested on infant EEG signals occurred in both utilized algorithms. Elder ages achieved consistent performance when tested using models trained on a dataset of the same age group.
This finding suggests that, with the exception of infants, age-specific automatic sleep-scoring models, i.e., model trained and tested on a dataset from the same age group, are effective and achieved reasonable average classification accuracy over all age groups.
Experimental results showed that the classification accuracy of the sleep-scoring model trained on adult participants (>18 years) was maintained when tested on participants of 13 years and older. The accuracy of the adult model degraded gradually for ages younger than 13 years old, with the lowest accuracy obtained in participants in age groups 0 and 1 (<2 years).
This finding suggests that the model trained on adult EEG signals is not reliable for use in sleep scoring of younger ages. Training data from matching age groups is needed to achieve reasonable accuracy.
This work is considered the first work to study the influence of EEG channels and participants’ age using a large-scale dataset that includes participants as young as a few days old. However, the study is limited to one dataset and needs to be extended to other datasets to validate the generalization of the findings. Moreover, this study focuses on the performance of the single EEG channel sleep-scoring algorithms. However, using a multichannel (e.g., multiple EEG channels and EEG+EOG) model may improve the performance, specifically in younger ages.
Our future work includes investigating several domain adaptation techniques to alleviate the impact of age variations on the performance of the sleep-scoring algorithms.