Mutual Information-Driven Feature Reduction for Hyperspectral Image Classification
Abstract
1. Introduction
- We propose an MI-driven efficient FR approach for the effective classification of HSI.
- We introduce an NMI-based band grouping strategy for intrinsic FE by applying classical PCA transformation to each group of bands independently for effective FE from HSI.
- We propose an NMI-based mRMR FS method using the extracted features through our proposed transformation.
- We performed extensive experiments on two widely used benchmark HSI datasets captured by the AVIRIS and HYDICE sensors to validate the superiority of our proposed FR approach.
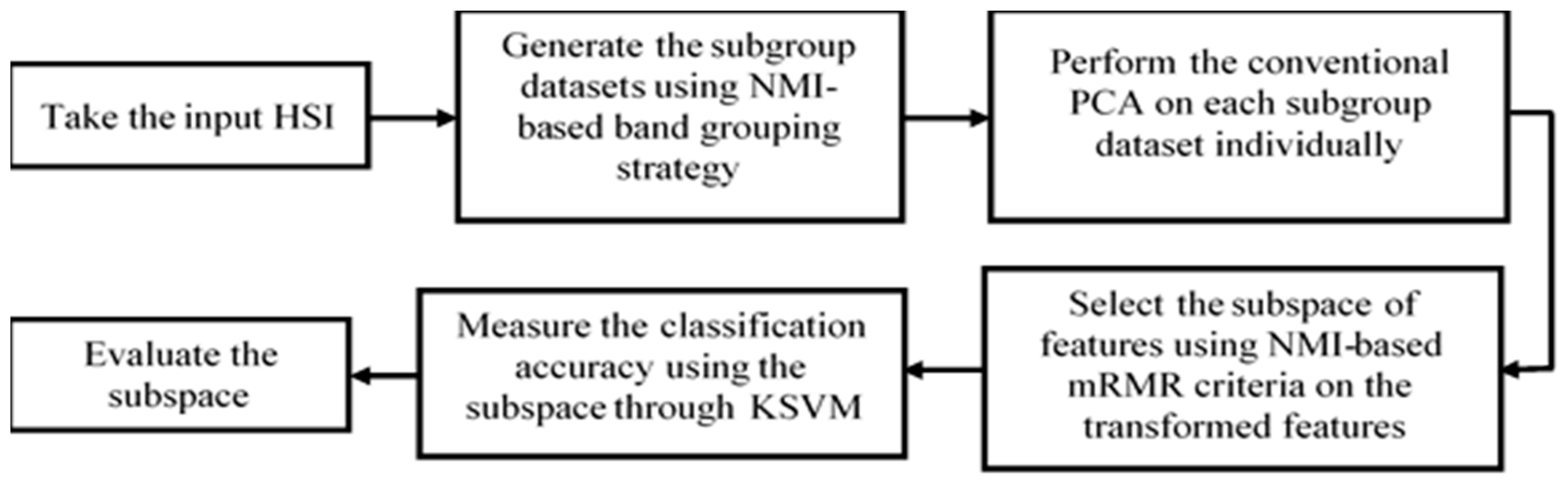
2. Methodology
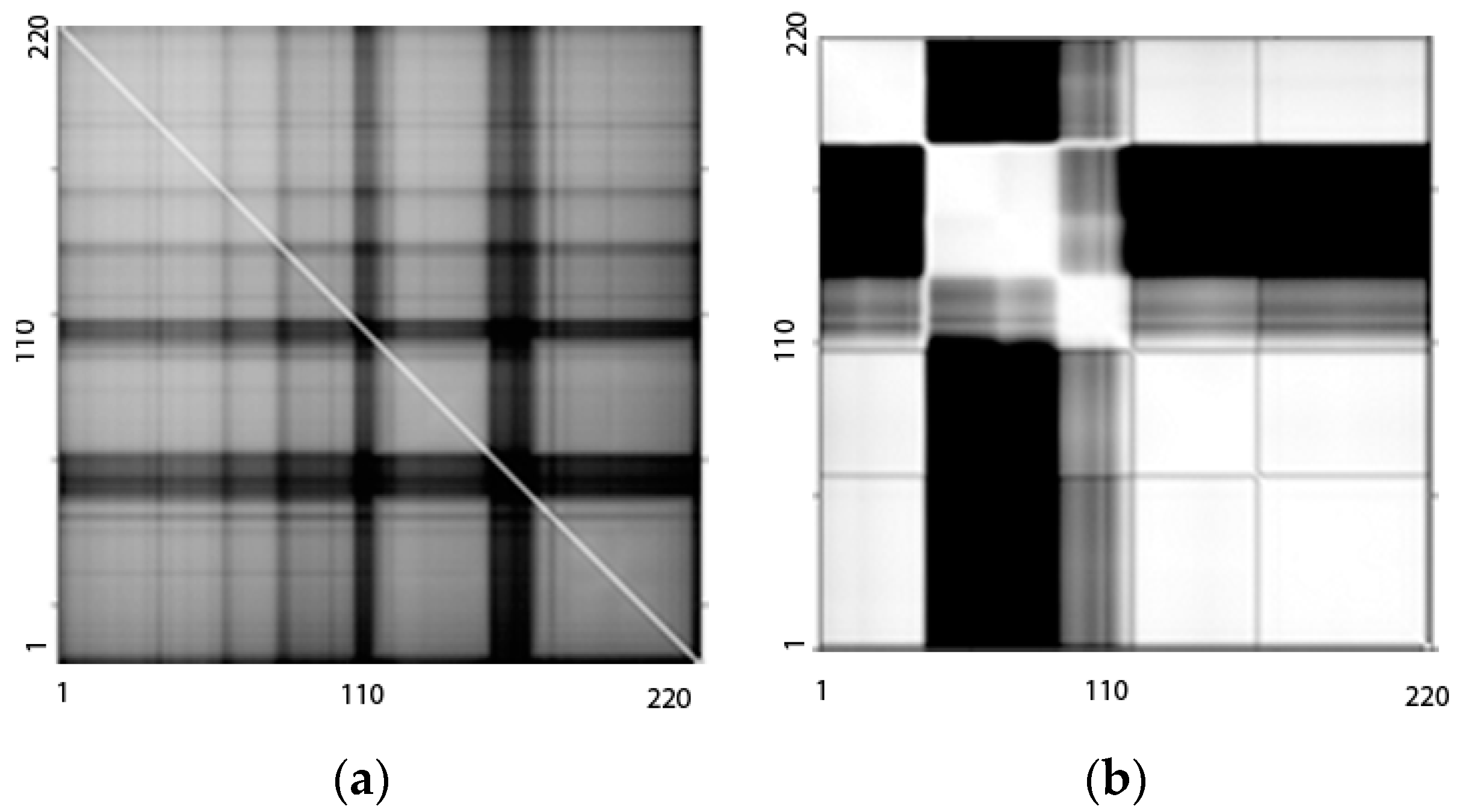
2.1. Proposed Band Grouping Strategy Based on NMI
2.2. PCA
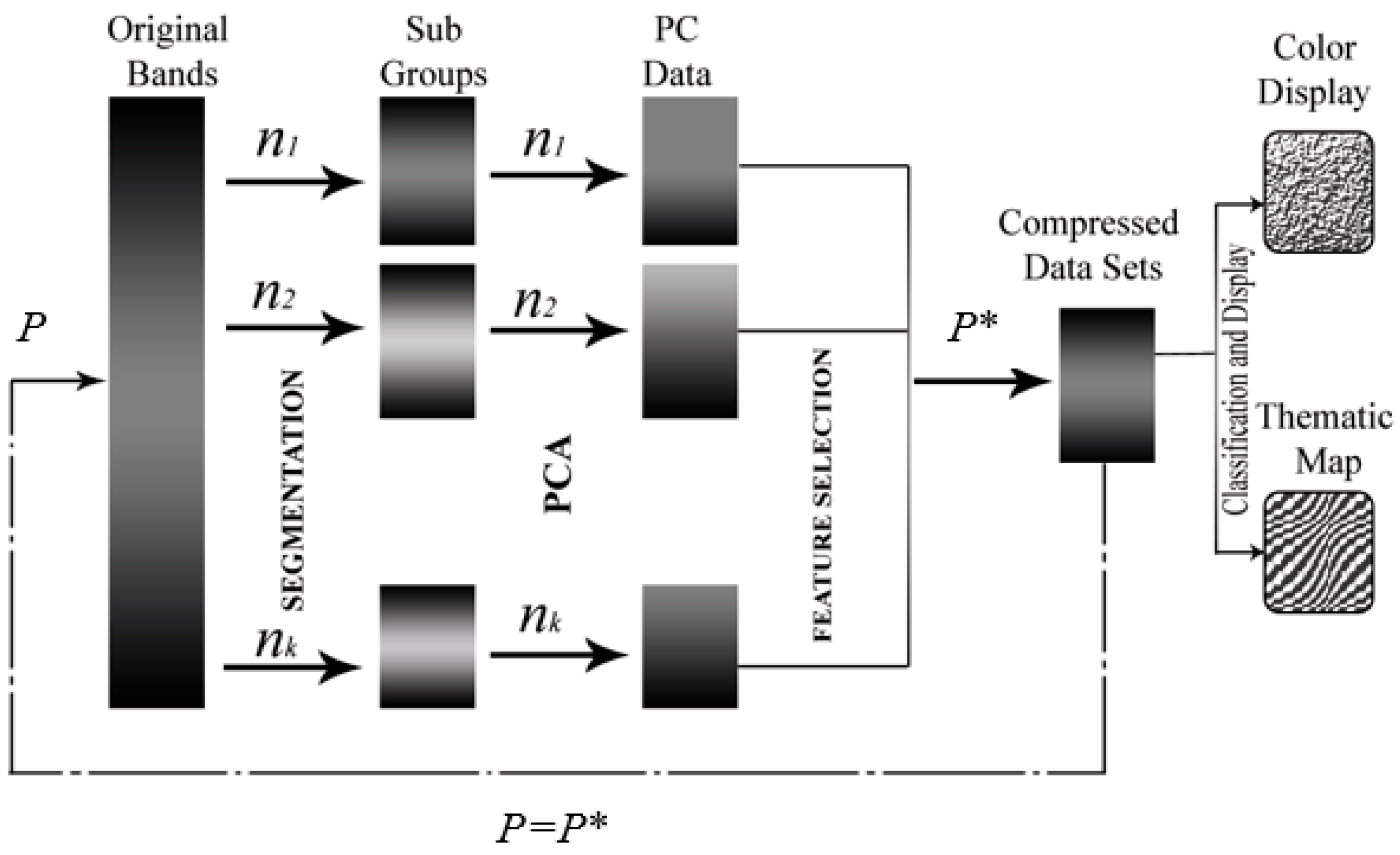
2.3. Proposed BgPCA
| Algorithm 1. BgPCA |
|
2.4. Proposed BgPCA-NMI
| Algorithm 2. BgPCA-NMI |
|
3. Experiment and Analysis of the Results
3.1. Description of the Dataset
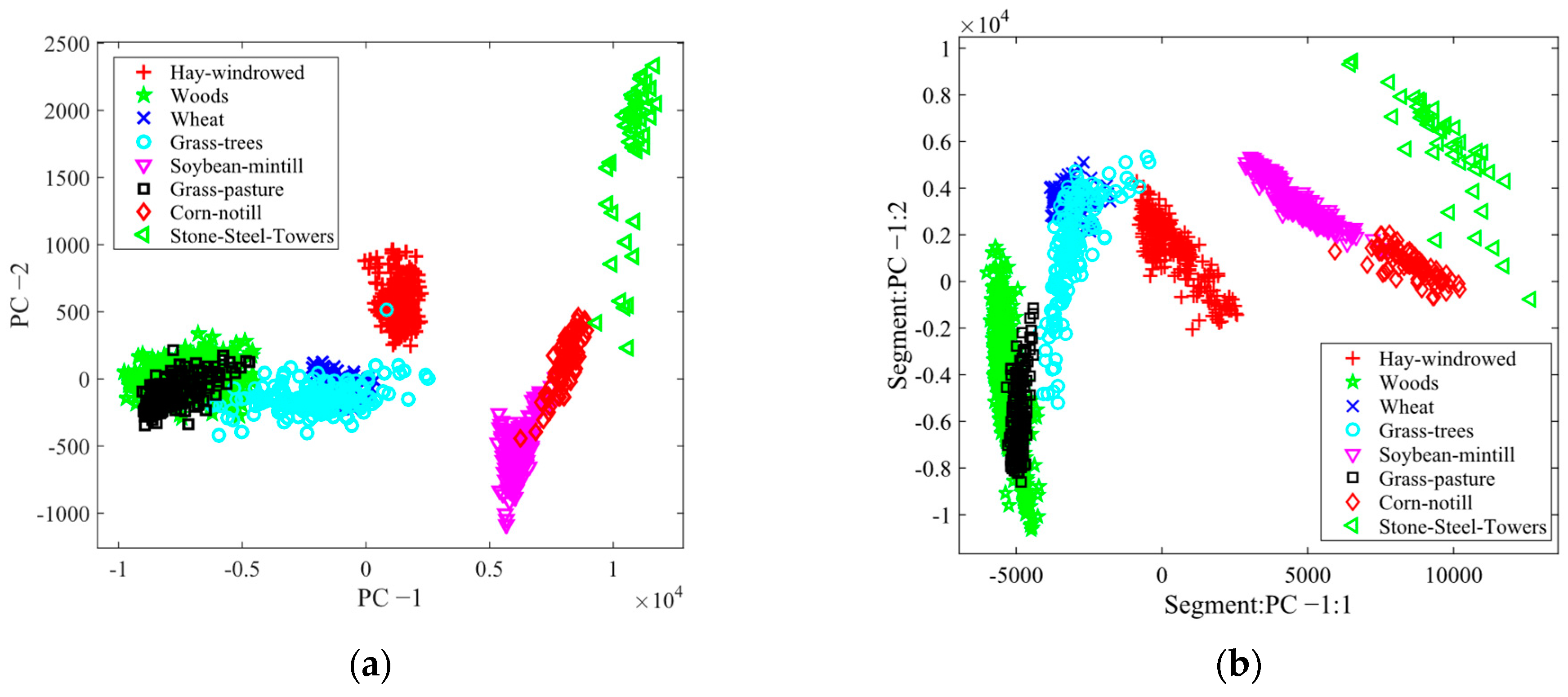
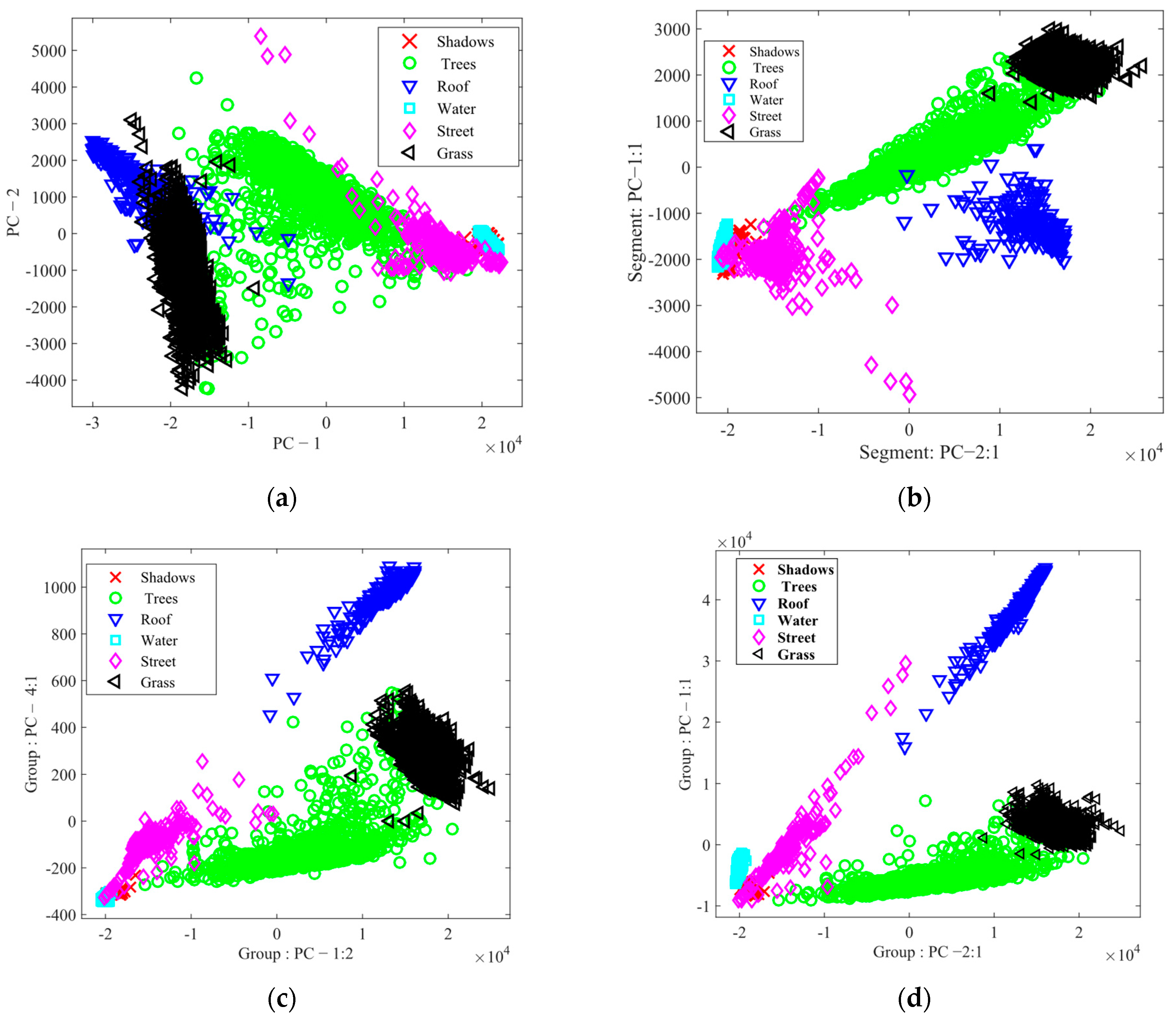
3.2. Results of FE and FS
3.3. Performance Evaluation Metrics
3.4. Classification Results and Evaluation
4. Conclusions and Future Work
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mohan, B.K.; Porwal, A. Hyperspectral image processing and analysis. Curr. Sci. 2015, 108, 833–841. [Google Scholar]
- Wang, N.; Zeng, X.; Duan, Y.; Deng, B.; Mo, Y.; Xie, Z.; Duan, P. Multi-Scale Superpixel-Guided Structural Profiles for Hyperspectral Image Classification. Sensors 2022, 22, 8502. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.A.; Jia, X. Remote Sensing Digital Image Analysis; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Tinega, H.; Chen, E.; Ma, L.; Mariita, R.M.; Nyasaka, D. Hyperspectral Image Classification Using Deep Genome Graph-Based Approach. Sensors 2021, 21, 6467. [Google Scholar] [CrossRef]
- Guo, B.; Gunn, S.R.; Damper, R.I.; Nelson, J.D.B. Band Selection for Hyperspectral Image Classification Using Mutual Information. IEEE Geosci. Remote Sens. Lett. 2006, 3, 522–526. [Google Scholar] [CrossRef]
- Manian, V.; Alfaro-Mejía, E.; Tokars, R.P. Hyperspectral Image Labeling and Classification Using an Ensemble Semi-Supervised Machine Learning Approach. Sensors 2022, 22, 1623. [Google Scholar] [CrossRef]
- Zabalza, J.; Ren, J.; Yang, M.; Zhang, Y.; Wang, J.; Marshall, S.; Han, J. Novel folded-PCA for improved feature extraction and data reduction with hyperspectral imaging and SAR in remote sensing. ELSEVIER ISPRS J. Photogramm. Remote Sens. 2014, 93, 112–122. [Google Scholar] [CrossRef]
- Rasti, B.; Scheunders, P.; Ghamisi, P.; Licciardi, G.; Chanussot, J. Noise Reduction in Hyperspectral Imagery: Overview and Application. Remote Sens. 2018, 10, 482. [Google Scholar] [CrossRef]
- Hossain, M.A.; Jia, X.; Pickering, M. Subspace Detection Using a Mutual Information Measure for Hyperspectral Image Classification. IEEE Geosci. Remote Sens. Lett. 2014, 11, 424–428. [Google Scholar] [CrossRef]
- Du, W.; Qiang, W.; Lv, M.; Hou, Q.; Zhen, L.; Jing, L. Semi-supervised dimension reduction based on hypergraph embedding for hyperspectral images. Int. J. Remote. Sens. 2018, 39, 1696–1712. [Google Scholar] [CrossRef]
- Hughes, G. On the Mean Accuracy of Statistical Pattern Recognizers. IEEE Trans. Inf. Theory 1968, IT-14, 55–63. [Google Scholar] [CrossRef]
- Hossain, M.A.; Jia, X.; Pickering, M. Subspace detection based on the combination of nonlinear feature extraction and feature selection. In Proceedings of the IEEE Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing, Gainesville, FL, USA, 26–28 June 2013. [Google Scholar]
- Islam, M.R.; Ahmed, B.; Hossain, M.A. Feature Reduction Based on Segmented Principal Component Analysis for Hyperspectral Images Classification. In Proceedings of the International Conference on Electrical, Computer and Communication Engineering (ECCE), Cox’s Bazar, Bangladesh, 7–9 February 2019. [Google Scholar]
- Haque, M.R.; Mishu, S.Z.; Uddin, M.P.; Mamun, M.A. A lightweight 3D-2D convolutional neural network for spectral-spatial classification of hyperspectral images. J. Intell. Fuzzy Syst. 2022, 43, 1241–1258. [Google Scholar] [CrossRef]
- Islam, M.R.; Hossain, M.A.; Ahmed, B. Improved Subspace Detection Based on Minimum Noise Fraction and Mutual Information for Hyperspectral Image Classification. In Proceedings of the International Joint Conference on Computational Intelligence, Dhaka, Bangladesh, 14–15 December 2018. [Google Scholar]
- Siddiqa, A.; Afzal, M.I.; Islam, M.R.; Nitu, A.M. Spectral Subset Detection for Hyperspectral Image Classification. In Proceedings of the International Conference on Innovation in Engineering and Technology (ICIET), Dhaka, Bangladesh, 26–27 November 2020. [Google Scholar]
- Ying, L.; Yanfeng, G.; Ye, Z. Hyperspectral feature extraction using selective PCA based on genetic algorithm with subgroups. In Proceedings of the International Conference on Innovative Computing, Information and Control, Beijing, China, 30 August–1 September 2006. [Google Scholar]
- Munishamaiaha, K.; Rajagopal, G.; Venkatesan, D.K.; Arif, M.; Vicoveanu, D.; Chiuchisan, I.; Izdrui, D.; Geman, O. Robust Spatial–Spectral Squeeze–Excitation AdaBound Dense Network (SE-AB-Densenet) for Hyperspectral Image Classification. Sensors 2022, 22, 3229. [Google Scholar] [CrossRef] [PubMed]
- Rodarmel, C.; Shan, J. Principal Component analysis for hyper-spectral image classification. ACM Surv. Land Inf. Syst. 2002, 62, 115–122. [Google Scholar]
- Arslan, O.; Akyürek, O.; Kaya, S.; Şeker, D.Z. Dimension reduction methods applied to coastline extraction on hyperspectral imagery. Geocarto Int. 2018, 35, 376–390. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, T.; Tang, X.; Hu, X.; Peng, Y. CAEVT: Convolutional Autoencoder Meets Lightweight Vision Transformer for Hyperspectral Image Classification. Sensors 2022, 22, 3902. [Google Scholar] [CrossRef]
- Jia, X.; Richards, J.A. Segmented Principal Components Transformation for Efficient Hyperspectral Remote-Sensing Image Display and Classification. IEEE Trans. Geosci. Remote Sens. 1999, 37, 538–542. [Google Scholar]
- Chen, G.; Qian, S.; Gleason, S. Denoising of hyperspectral imagery by combining PCA with block-matching 3-D filtering. Can. J. Remote Sens. 2012, 37, 590–595. [Google Scholar] [CrossRef]
- Ibarrola-Ulzurrun, E.; Marcello-Ruiz, J.; Gonzalo-Martín, C. Assessment of Component Selection Strategies in Hyperspectral Imagery. Entropy 2017, 19, 666. [Google Scholar] [CrossRef]
- Uddin, M.P.; Mamun, M.A.; Hossain, M.A. Feature Extraction for Hyperspectral Image Classification. In Proceedings of the IEEE 5th Region 10 Humanitarian Technology Conference (R10-HTC), Dhaka, Bangladesh, 21–23 December 2017. [Google Scholar]
- Uddin, M.P.; Mamun, M.A.; Hossain, M.A. Segmented FPCA for Hyperspectral Image Classification. In Proceedings of the IEEE 3rd International Conference on Electrical Information and Communication Technology (EICT), Khulna, Bangladesh, 7–9 December 2017. [Google Scholar]
- Uddin, M.P.; Mamun, M.A.; Hossain, M.A. Improved Feature Extraction Using Segmented FPCA for Hyperspectral Image Classification. In Proceedings of the International Conference on Electrical & Electronic Engineering, Rajshahi, Bangladesh, 27–29 December 2017. [Google Scholar]
- Uddin, M.P.; Mamun, M.A.; Hossain, M.A. Effective feature extraction through segmentation-based folded-PCA for hyperspectral image classification. Int. J. Remote Sens. 2019, 40, 7190–7220. [Google Scholar] [CrossRef]
- Uddin, M.P.; Mamun, M.A.; Hossain, M.A. PCA-based Feature Reduction for Efficient Hyperspectral Image Classification. IETE Tech. Rev. 2020, 38, 1740615. [Google Scholar]
- Wang, L.; Wang, X. Dual-Coupled CNN-GCN-Based Classification for Hyperspectral and LiDAR Data. Sensors 2022, 22, 5735. [Google Scholar] [CrossRef] [PubMed]
- Qasim, M.; Khan, S.D. Detection and Relative Quantification of Neodymium in Sillai Patti Carbonatite Using Decision Tree Classification of the Hyperspectral Data. Sensors 2022, 22, 7537. [Google Scholar] [CrossRef] [PubMed]
- Mukundan, A.; Huang, C.-C.; Men, T.-C.; Lin, F.-C.; Wang, H.-C. Air Pollution Detection Using a Novel Snap-Shot Hyperspectral Imaging Technique. Sensors 2022, 22, 6231. [Google Scholar] [CrossRef] [PubMed]
- Dionisio, A.; Menezes, R.; Mendes, D.A. Mutual information: A measure of dependency for nonlinear time series. Phys. A: Stat. Mech. Its Appl. 2004, 344, 326–329. [Google Scholar] [CrossRef]
- Lixin, G.; Weixin, X.; Jihong, P. Segmented minimum noise fraction transformation for efficient feature extraction of hyperspectral images. Pattern Recognit. 2015, 48, 3216–3226. [Google Scholar] [CrossRef]
- Kuo, B.; Chang, W.; Li, C.; Hung, C. Correlation matrix feature extraction based on spectral clustering for hyperspectral image segmentation. In Proceedings of the Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), Shanghai, China, 4–7 June 2012. [Google Scholar]
- Tarabalka, Y.; Benediktsson, J.A.; Chanussot, J. Spectral-spatial classification of hyperspectral imagery Based on partitional clustering techniques. IEEE Trans. Geosci. Remote Sens. 2009, 47, 2973–2987. [Google Scholar] [CrossRef]
- Tsai, F.; Lin, E.-K.; Yoshino, K. Spectrally segmented principal component analysis of hyperspectral imagery for mapping invasive plant species. Int. J. Remote Sens. 2007, 28, 1023–1039. [Google Scholar] [CrossRef]
- Uddin, M.P.; Mamun, M.A.; Afjal, M.I.; Hossain, M.A. Information-theoretic feature selection with segmentation-based folded principal component analysis (PCA) for hyperspectral image classification. Int. J. Remote Sens. 2020, 42, 286–321. [Google Scholar] [CrossRef]
- Baumgartner, M.F.; Labial, L.; Landgrebe, D.A. 220 Band AVIRIS Hyperspectral Image Data Set: June 12, 1992 Indian Pine Test Site 3; Purdue University Research Repository: West Lafayette, IN, USA, 2015. [Google Scholar]
- Landgrebe, D.A. Available online: https://engineering.purdue.edu/~biehl/MultiSpec/hyperspectral.html (accessed on 3 March 2022).
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 27:1–27:27. [Google Scholar] [CrossRef]




| SPCA (Baseline Approach) | BgPCA (Proposed Approach) | |||||
|---|---|---|---|---|---|---|
| Group | Range of Bands | # of Bands | Average Correlation | Range of Bands | # of Bands | Average NMI |
| 1 | 1–35 | 35 | 0.8770 | 1–102 | 102 | 0.3400 |
| 2 | 36–103 | 68 | 0.7171 | 103–143 | 41 | 0.7171 |
| 3 | 104–200 | 97 | 0.6950 | 144–200 | 67 | 0.6950 |
| SPCA (Baseline Approach) | BgPCA (Proposed Approach) | |||||
|---|---|---|---|---|---|---|
| Group | Range of Bands | # of Bands | Average Correlation | Range of Bands | # of Bands | Average NMI |
| 1 | 1–56 | 56 | 0.9443 | 1–58 | 58 | 0.4500 |
| 2 | 57–102 | 46 | 0.8842 | 59–108 | 50 | 0.5460 |
| 3 | 103–191 | 89 | 0.9813 | 109–159 | 51 | 0.5460 |
| 4 | - | - | - | 159–191 | 33 | 0.5600 |
| Name of the Dataset | Capturing Sensor | P | Wavelength Range (nm) | H | W | Ground Classes | Ground Sampling Distance (m) |
|---|---|---|---|---|---|---|---|
| Indian Pines | AVIRIS | 220 | 400–2500 | 145 | 145 | 16 | 20 |
| Washington DC Mall | HYDICE | 191 | 400–2400 | 1280 | 307 | 7 | 3 |
| Method Name | Best C | Best γ | Training Accuracy | |
|---|---|---|---|---|
| AVIRIS | PCA | 10 | 3 | 98.55 |
| SPCA | 3.5 | 2.8 | 94.65 | |
| SPCA-NMI | 5 | 2 | 98.50 | |
| BgPCA | 1.8 | 3.7 | 96.85 | |
| BgPCA-NMI | 7 | 1.2 | 98.88 | |
| HYDICE | PCA | 10 | 3 | 97.55 |
| SPCA | 4 | 2.1 | 95.83 | |
| SPCA-NMI | 2.5 | 3.9 | 97.68 | |
| BgPCA | 3 | 1.5 | 98.15 | |
| BgPCA-NMI | 7 | 1.2 | 98.95 |
| Class | PCA | SPCA | SPCA-NMI | BgPCA | BgPCA-NMI |
|---|---|---|---|---|---|
| Hay—windrowed | 97.66 | 90.60 | 90.60 | 94.41 | 96.43 |
| Soybean—no till | 80.41 | 76.67 | 94.81 | 84.54 | 84.69 |
| Woods | 94.96 | 92.27 | 91.27 | 96.19 | 96.71 |
| Wheat | 100.00 | 98.41 | 100 | 94.03 | 100.00 |
| Grass—trees | 100.00 | 100.00 | 100 | 100 | 100.00 |
| Soybean– min. till | 89.67 | 70.21 | 92.64 | 94.34 | 94.94 |
| Grass—pasture | 94.74 | 60.00 | 90.00 | 77.42 | 85.71 |
| Corn –no till | 96.67 | 95.24 | 95.12 | 100 | 100.00 |
| Corn | 88.10 | 92.68 | 76.47 | 88.00 | 97.78 |
| Corn—min. till | 100.00 | 100.00 | 68.75 | 87.50 | 100.00 |
| Stone, steel, towers | 100.00 | 100.00 | 77.27 | 100 | 100.00 |
| Alfalfa | 54.55 | 100.00 | 100 | 100 | 100.00 |
| Soybean—clean | 100.00 | 69.23 | 83.33 | 100 | 100.00 |
| Buildings, grass, trees, roads | 80.00 | 75.00 | 83.33 | 88.89 | 81.82 |
| AA | 91.20 | 87.17 | 88.83 | 93.24 | 95.58 |
| OA | 92.45 | 83.40 | 91.35 | 92.54 | 94.93 |
| Kappa | 91.24 | 80.77 | 90.00 | 91.41 | 94.16 |
| Precision | 80.23 | 70.45 | 82.22 | 82.23 | 91.87 |
| Recall | 91.20 | 87.17 | 88.83 | 93.24 | 95.58 |
| F1 score | 85.36 | 77.92 | 85.40 | 87.95 | 93.69 |
| Class | PCA | SPCA | SPCA-NMI | BgPCA | BgPCA-NMI |
|---|---|---|---|---|---|
| Shadow | 34.04 | 57.14 | 59.26 | 72.73 | 88.89 |
| Tree | 99.27 | 99.88 | 99 | 99.79 | 99.81 |
| Roof | 100 | 99.05 | 99.08 | 100 | 100 |
| Water | 100 | 100 | 100 | 100 | 100 |
| Street | 93.63 | 81.51 | 89.41 | 83.86 | 97.62 |
| Grass | 69.12 | 86.46 | 87.42 | 95.82 | 98.26 |
| AA | 95.05 | 87.34 | 89.03 | 92.03 | 97.43 |
| OA | 92.80 | 92.07 | 93.57 | 95.97 | 99.03 |
| Kappa | 90.22 | 89.32 | 91.30 | 94.54 | 98.67 |
| Precision | 93.62 | 95.49 | 96.79 | 97.77 | 99.51 |
| Recall | 95.05 | 87.34 | 89.02 | 92.03 | 97.43 |
| F1 score | 94.33 | 91.23 | 92.75 | 94.81 | 98.46 |
| Stage | AVIRIS | HYDICE | ||
|---|---|---|---|---|
| PCA | BgPCA-NMI | PCA | BgPCA-NMI | |
| FE | 0.098 s | 0.017 s | 0.120 s | 0.067 s |
| FS | 1.200 s | 0.980 s | 1.100 s | 0.670 s |
| Total cost | 1.298 s | 0.997 s | 1.220 s | 0.737 s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, M.R.; Ahmed, B.; Hossain, M.A.; Uddin, M.P. Mutual Information-Driven Feature Reduction for Hyperspectral Image Classification. Sensors 2023, 23, 657. https://doi.org/10.3390/s23020657
Islam MR, Ahmed B, Hossain MA, Uddin MP. Mutual Information-Driven Feature Reduction for Hyperspectral Image Classification. Sensors. 2023; 23(2):657. https://doi.org/10.3390/s23020657
Chicago/Turabian StyleIslam, Md Rashedul, Boshir Ahmed, Md Ali Hossain, and Md Palash Uddin. 2023. "Mutual Information-Driven Feature Reduction for Hyperspectral Image Classification" Sensors 23, no. 2: 657. https://doi.org/10.3390/s23020657
APA StyleIslam, M. R., Ahmed, B., Hossain, M. A., & Uddin, M. P. (2023). Mutual Information-Driven Feature Reduction for Hyperspectral Image Classification. Sensors, 23(2), 657. https://doi.org/10.3390/s23020657







