A Novel 3D Reversible Data Hiding Scheme Based on Integer–Reversible Krawtchouk Transform for IoMT
Abstract
:1. Introduction
- The original image cannot be perfectly reconstructed from the Krawtchouk coefficients, even if the Krawtchouk transform itself is orthogonal, due to rounding errors associated with the limited representation of significant digits. This can have an impact on the performance of lossless applications requiring exact calculations;
- Floating-point Krawtchouk coefficients require a more complex representation than integer image pixels (spatial domain), which means a higher cost in terms of memory and computing resources, especially when input images are large or voluminous (3D images). This can impact the performance of applications that require fast and efficient calculations;
- The Krawtchouk transform is unsuitable for applications where exact integer arithmetic is desired, such as lossless compression, RDH, digital communication systems, and embedded systems with limited precision.
- A precise, exact integer representation in the Krawtchouk domain is provided without rounding errors or accuracy limitations;
- IRKT guarantees that transformed coefficients remain integers (without the need for quantization), enabling lossless data reconstruction;
- The proposed IRKT can be easily generalized to accommodate 2D and 3D data representations;
- A novel 3D RDH algorithm suitable for 3D medical images is proposed, to the best of our knowledge, for the first time;
- Embedding data into 3D medical images does not increase their original size, thus optimizing infrastructure and maximizing resource utilization in the IoMT;
- Medical images are recovered without any loss or damage after extracting additional data;
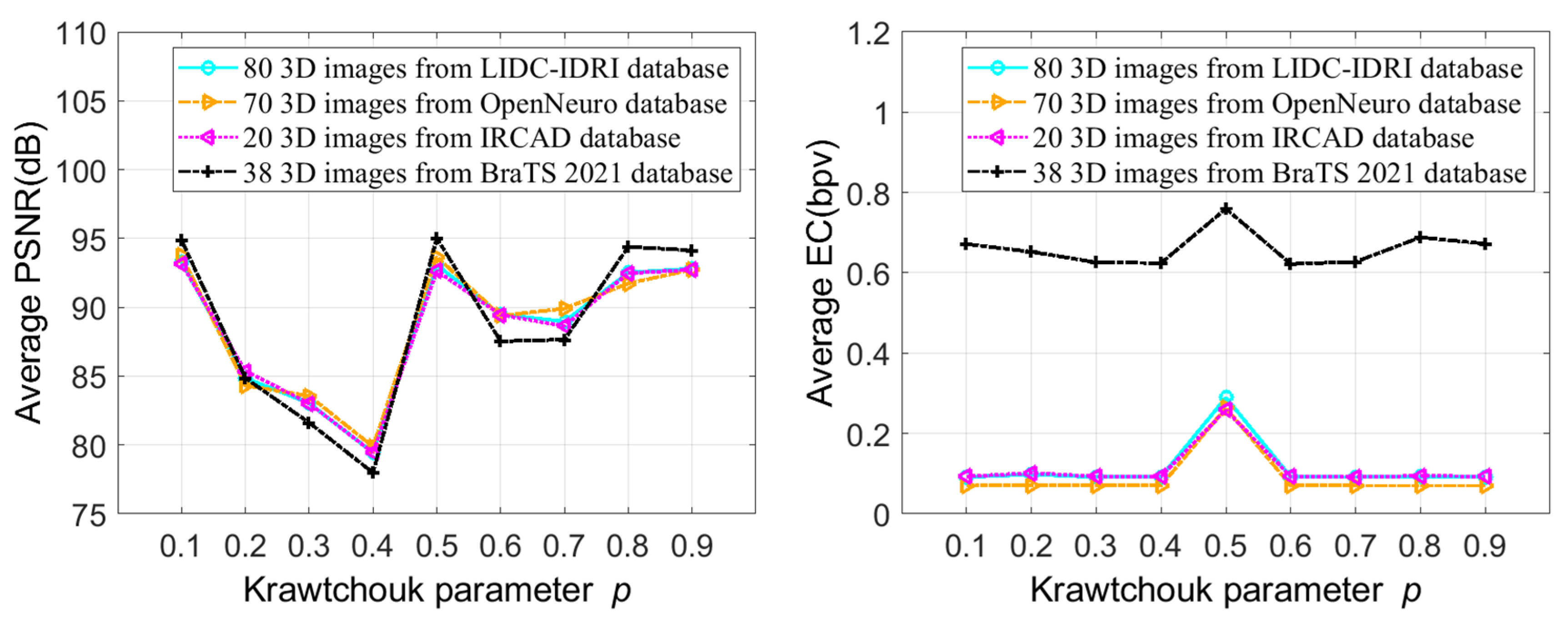
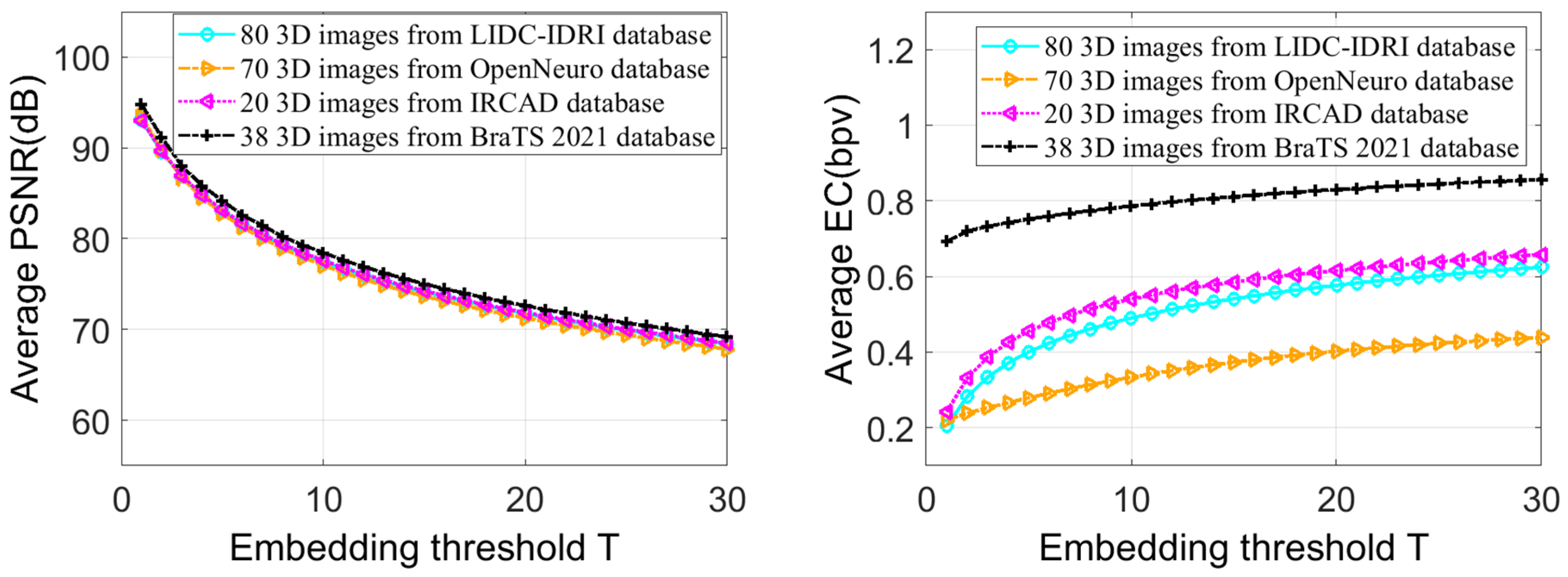
- The embedding capacity and quality of the 3D stego image (image after data embedding) can be adjusted using a threshold-based embedding technique.
2. Preliminaries
3. Construction of the Integer–Reversible Krawtchouk Transform
3.1. SERM Factorization of the Krawtchouk Polynomial Matrix
- Property 1: K is orthogonal, that is, KKT = KTK = I, KT is the transpose of K, and I is the identity matrix [13] of the appropriate size;
- Property 2: K is nonsingular, that is, KK−1 = K−1K = I;
- Property 3: The determinant of K is det(K) = 1;
- Property 4: The minors of the leading principal submatrices of K are all ones.
3.2. Integer–Reversible Krawtchouk Transform
| Algorithm 1 3D Integer–Reversible Krawtchouk Transform. |
| Input: C a volumetric cuboid of size N × N × N, P the permutation matrix of K, Sj(j = 0, 1,…, N) the SERMs of K. Output: Transformed cuboid Q of size N × N × N Initialize empty matrices Q, Q1, Q2 of size N × N × N. for i = 1 to N do A = P⌊SN⌊…⌊ S1⌊S0C(:,:,i) ⌋⌋…⌋;//C(:,:,i) is the i-th plane along the z-axis. Q1(:,:,i) = P⌊SN⌊…⌊ S1⌊S0AT⌋⌋…⌋;//AT is the transpose of A. end //Transpose the 3D matrix Q1 into another 3D matrix Q2. for i = 1 to N do for j = 1 to N do Q2(j,:,i) = Q1(i,:,j); end end //Generate the IRKT matrix Q for i = 1 to N do A = P⌊SN⌊…⌊S1⌊S0Q2(:,:,i) ⌋⌋…⌋; Q(:,:,i) = P⌊SN⌊…⌊S1⌊S0AT⌋…⌋; end Return the IRKT matrix Q. |
| Algorithm 2 Inverse 3D Integer–Reversible Krawtchouk Transform. |
| Input: IRKT matrixQ of size N × N × N,P the permutation matrix of K, Sj(j = 0, 1,…, N) the SERMs of K. Output: Reconstructed cuboid R of size N × N×N. Initialize empty matrices R, R1, R2 of size N × N × N. Compute the inverse of Sj. for i = 1 to N do A= ⌊⌊…⌊PTQ(:,:,i)⌋…⌋⌋;//PT is the transpose of P. R1(:,:,i) = ⌊⌊…⌊PTAT⌋…⌋⌋; end for i = 1 to N do for j = 1 to N do R2(j,:,i) = R1(i,:,j); end end for i = 1 to N do A= ⌊⌊…⌊PTR2 (:,:,i)⌋…⌋⌋; R(:,:,i) = ⌊…⌊PTAT⌋…⌋⌋; end Return the Reconstructed cuboid R. |
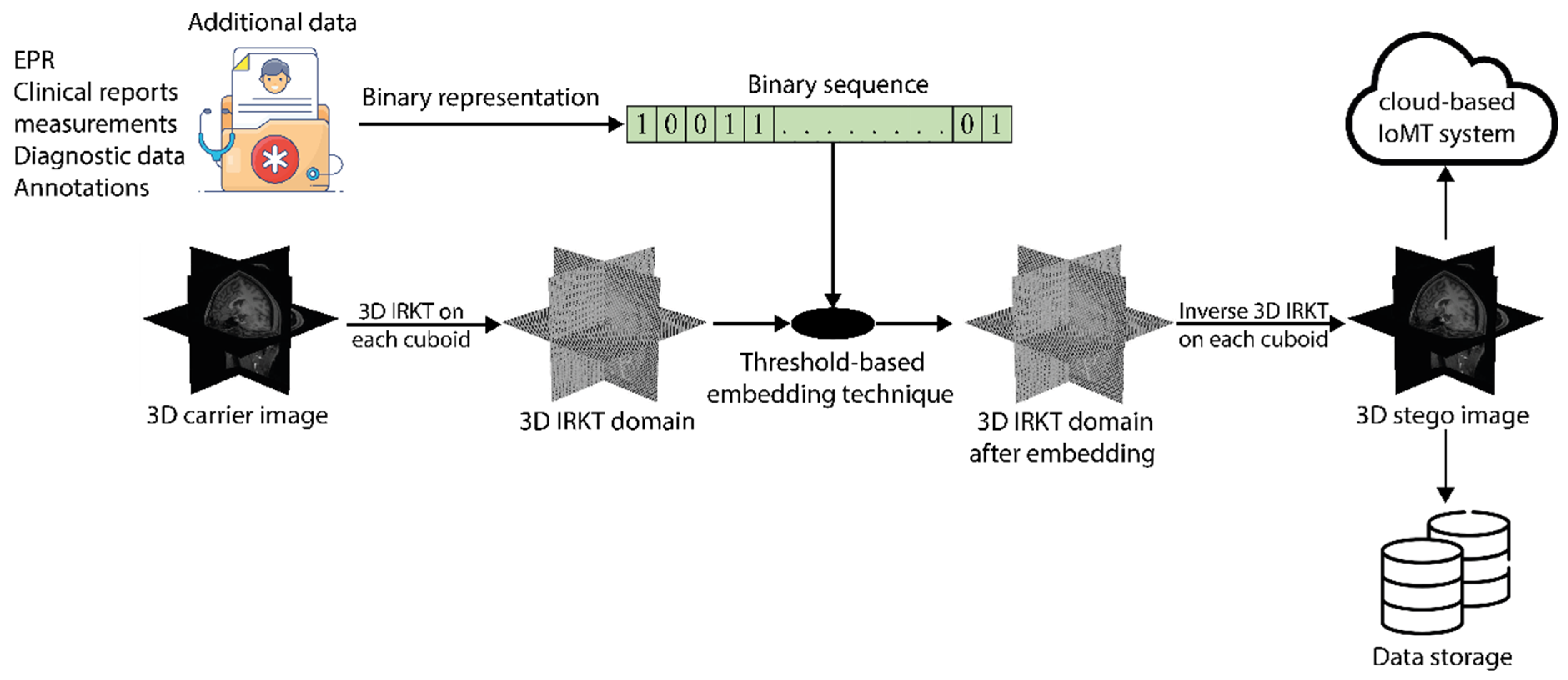
4. Application in RDH for 3D Medical Images
4.1. The Usefulness of 3D RDH in IoMT
- Due to the substantial volume of data generated by IoMT, storage and transmission may encounter limitations. By storing and transmitting a significant amount of medical data within the same carrier image, efficient resource utilization and management can be achieved within IoMT applications;
- Protecting patient privacy is crucial. The 3D RDH can selectively embed confidential patient identifiers or sensitive information within 3D medical images, ensuring that only authorized personnel can access these details;
- By embedding additional diagnostic information into 3D medical images, the accuracy of diagnoses can be enhanced, thereby granting healthcare professionals extended access to relevant data during remote consultations;
- In telemedicine scenarios, real-time interaction is limited. The 3D RDH can hide additional explanations, annotations, or visual cues within 3D medical images, offering remote healthcare professionals a more detailed understanding of the patient’s condition;
- In the context of IoMT, medical data are frequently employed for research and analysis purposes. The adoption of 3D RDH alongside 3D medical images enables the integration of research data, metadata, or annotations into the images. This facilitates comprehensive data analysis, data exploration, and collaboration among researchers, thereby enabling them to gain deeper insights into medical conditions and treatment outcomes.
4.2. Embedding Procedure
4.3. Additional Data Extraction and Carrier Image Restoration
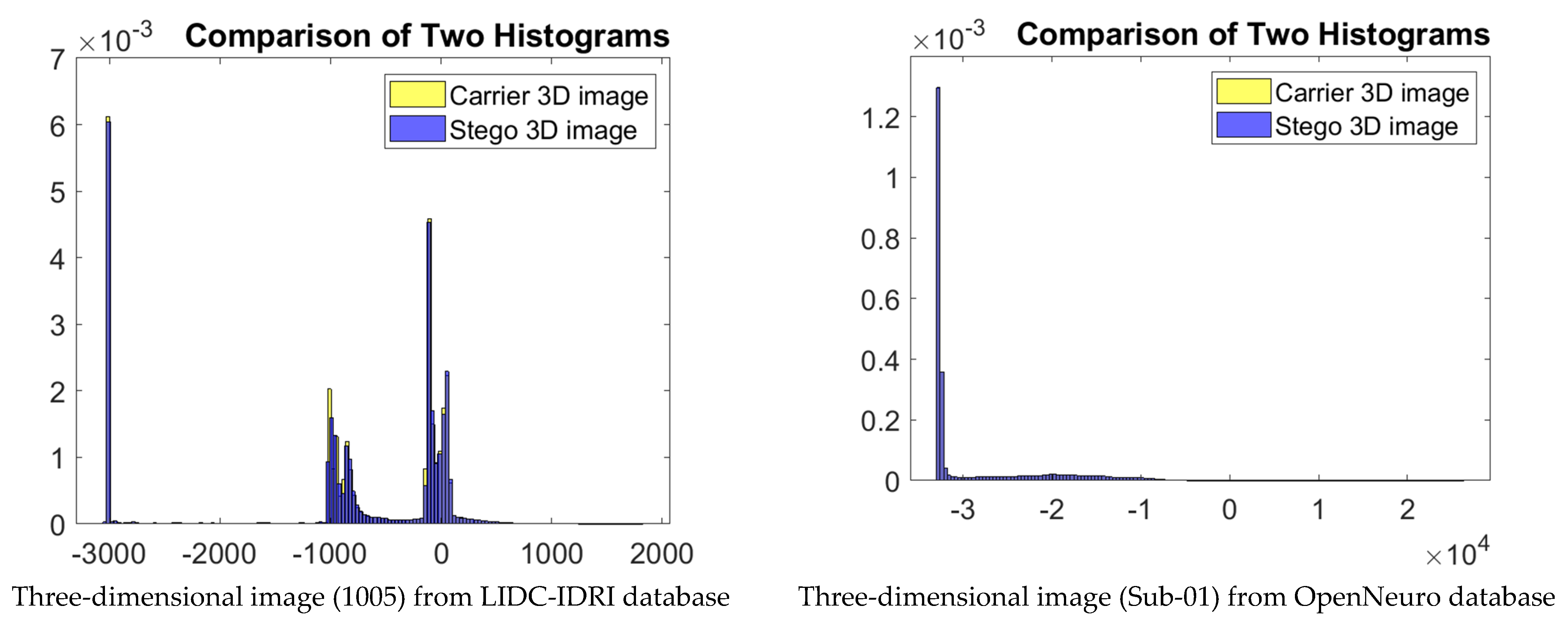
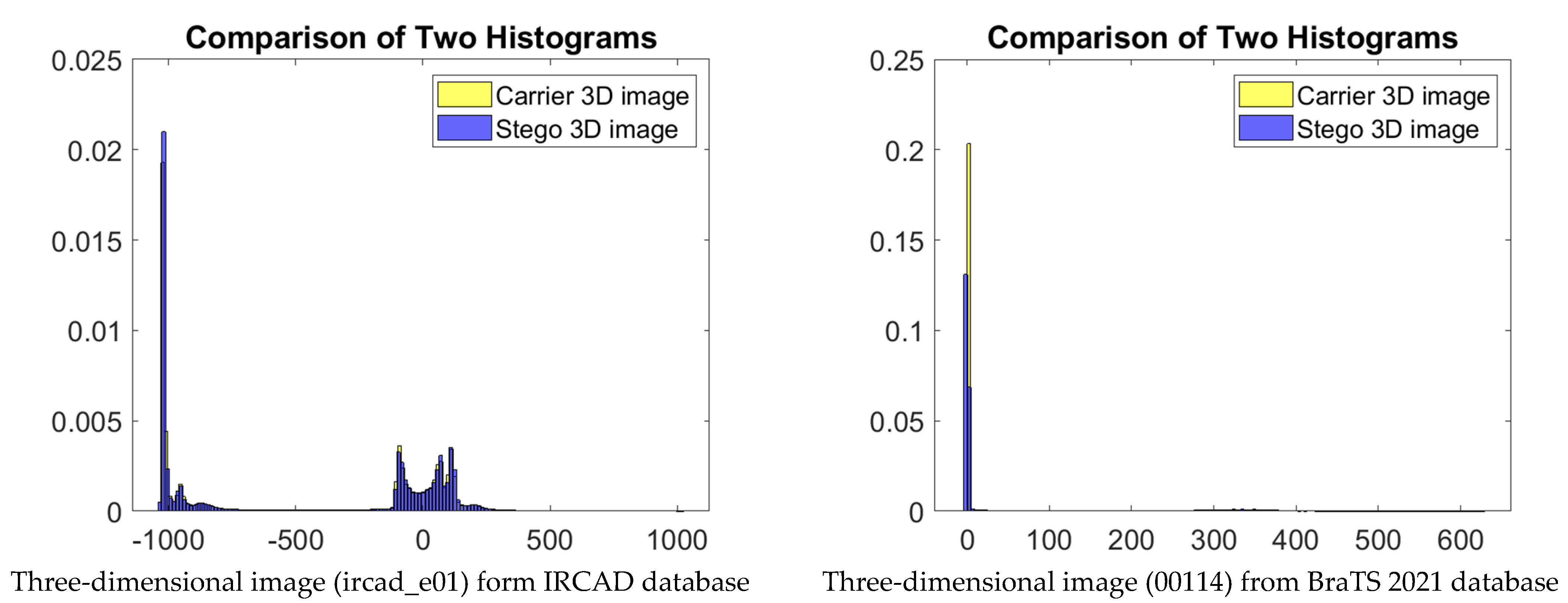
5. Experimental Results
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Calderbank, A.R.; Daubechies, I.; Sweldens, W.; Yeo, B.-L. Wavelet transforms that map integers to integers. Appl. Comput. Harmon. Anal. 1998, 5, 332–369. [Google Scholar] [CrossRef]
- Chen, Y.; Hao, P. Integer reversible transformation to make JPEG lossless. In Proceedings of the 7th International Conference on Signal Processing, 2004, Beijing, China, 31 August–4 September 2004; Volume 1, pp. 835–838. [Google Scholar]
- Gormish, M.J.; Schwartz, E.L.; Keith, A.F.; Boliek, M.P.; Zandi, A. Lossless and nearly lossless compression for high-quality images. In Proceedings of the Very High Resolution and Quality Imaging II, San Jose, CA, USA, 8–14 February 1997; Volume 3025, pp. 62–70. [Google Scholar]
- Said, A.; Pearlman, W.A. An image multiresolution representation for lossless and lossy compression. IEEE Trans. Image Process. 1996, 5, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Zandi, A.; Allen, J.D.; Schwartz, E.L.; Boliek, M. CREW: Compression with reversible embedded wavelets. In Proceedings of the DCC’95 Data Compression Conference, Snowbird, UT, USA, 28–30 March 1995; pp. 212–221. [Google Scholar]
- Xiao, B.; Lu, G.; Zhang, Y.; Li, W.; Wang, G. Lossless image compression based on integer Discrete Tchebichef Transform. Neurocomputing 2016, 214, 587–593. [Google Scholar] [CrossRef]
- Xuan, G.; Zhu, J.; Chen, J.; Shi, Y.Q.; Ni, Z.; Su, W. Distortionless data hiding based on integer wavelet transform. Electron. Lett. 2002, 38, 1646–1648. [Google Scholar] [CrossRef]
- Xuan, G.; Yang, C.; Zhen, Y.; Shi, Y.Q.; Ni, Z. Reversible data hiding using integer wavelet transform and companding technique. In IWDW 2004: Digital Watermarking: Proceedings of the 3rd International Workshop, Seoul, Republic of Korea, 30 October–1 November 2004; Revised Selected Papers 3; Springer: Berlin/Heidelberg, Germany, 2005; pp. 115–124. [Google Scholar]
- Shaik, A.; Thanikaiselvan, V. Comparative analysis of integer wavelet transforms in reversible data hiding using threshold based histogram modification. J. King Saud Univ.-Comput. Inf. Sci. 2021, 33, 878–889. [Google Scholar] [CrossRef]
- Lin, Y.-K. High capacity reversible data hiding scheme based upon discrete cosine transformation. J. Syst. Softw. 2012, 85, 2395–2404. [Google Scholar] [CrossRef]
- Pei, S.-C.; Ding, J.-J. Integer discrete Fourier transform and its extension to integer trigonometric transforms. In Proceedings of the 2000 IEEE International Symposium on Circuits and Systems (ISCAS), Geneva, Switzerland, 28–31 May 2000; Volume 5, pp. 513–516. [Google Scholar]
- Oraintara, S.; Chen, Y.-J.; Nguyen, T.Q. Integer fast Fourier transform. IEEE Trans. Signal Process. 2002, 50, 607–618. [Google Scholar] [CrossRef]
- Yap, P.-T.; Paramesran, R.; Ong, S.-H. Image analysis by Krawtchouk moments. IEEE Trans. Image Process. 2003, 12, 1367–1377. [Google Scholar]
- Yap, P.T.; Raveendran, P.; Ong, S.H. Krawtchouk moments as a new set of discrete orthogonal moments for image reconstruction. In Proceedings of the 2002 International Joint Conference on Neural Networks, IJCNN’02 (Cat. No.02CH37290), Honolulu, HI, USA, 12–17 May 2002; Volume 1, pp. 908–912. [Google Scholar]
- Venkataramana, A.; Raj, P.A. Image Watermarking Using Krawtchouk Moments. In Proceedings of the 2007 International Conference on Computing: Theory and Applications (ICCTA’07), Kolkata, India, 5–7 March 2007; pp. 676–680. [Google Scholar]
- Papakostas, G.A.; Tsougenis, E.D.; Koulouriotis, D.E. Near optimum local image watermarking using Krawtchouk moments. In Proceedings of the 2010 IEEE International Conference on Imaging Systems and Techniques, Thessaloniki, Greece, 1–2 July 2010; pp. 464–467. [Google Scholar]
- Papakostas, G.A.; Tsougenis, E.D.; Koulouriotis, D.E. Moment-based local image watermarking via genetic optimization. Appl. Math. Comput. 2014, 227, 222–236. [Google Scholar] [CrossRef]
- Yamni, M.; Daoui, A.; Karmouni, H.; Elmalih, S.; Ben-fares, A.; Sayyouri, M.; Qjidaa, H.; Maaroufi, M.; Alami, B.; Jamil, M.O. Copyright protection of multiple CT images using Octonion Krawtchouk moments and grey Wolf optimizer. J. Frankl. Inst. 2023, 360, 4719–4752. [Google Scholar] [CrossRef]
- Yamni, M.; Karmouni, H.; Daoui, A.; Sayyouri, M.; Qjidaa, H. Blind image zero-watermarking algorithm based on radial krawtchouk moments and chaotic system. In Proceedings of the 2020 International Conference on Intelligent Systems and Computer Vision (ISCV), Fez, Morocco, 9–11 June 2020; pp. 1–7. [Google Scholar]
- Atakishiyev, N.M.; Wolf, K.B. Fractional Fourier–Kravchuk transform. JOSA A 1997, 14, 1467–1477. [Google Scholar] [CrossRef]
- Hao, P.; Shi, Q. Matrix factorizations for reversible integer mapping. IEEE Trans. Signal Process. 2001, 49, 2314–2324. [Google Scholar]
- Huang, L.-C.; Tseng, L.-Y.; Hwang, M.-S. A reversible data hiding method by histogram shifting in high quality medical images. J. Syst. Softw. 2013, 86, 716–727. [Google Scholar] [CrossRef]
- Kumar, C.V.; Natarajan, V.; Bhogadi, D. High capacity reversible data hiding based on histogram shifting for medical images. In Proceedings of the 2013 International Conference on Communication and Signal Processing, Melmaruvathur, India, 3–5 April 2013; pp. 730–733. [Google Scholar]
- Girdhar, A.; Kumar, V. Comprehensive survey of 3D image steganography techniques. IET Image Process. 2018, 12, 1–10. [Google Scholar] [CrossRef]
- Li, M.-T.; Huang, N.-C.; Wang, C.-M. A novel high capacity 3D steganographic algorithm. Int. J. Innov. Comput. Inf. Control 2011, 7, 1055–1074. [Google Scholar]
- Chao, M.-W.; Lin, C.; Yu, C.-W.; Lee, T.-Y. A high capacity 3D steganography algorithm. IEEE Trans. Vis. Comput. Graph. 2008, 15, 274–284. [Google Scholar] [CrossRef]
- Tsai, Y.-Y. An adaptive steganographic algorithm for 3D polygonal models using vertex decimation. Multimed. Tools Appl. 2014, 69, 859–876. [Google Scholar] [CrossRef]
- Anish, K.; Arpita, N.; Nikhil, H.; Sumant, K.; Bhagya, S.; Desai, S.D. Intelligence system security based on 3D image. In Proceedings of the 5th International Conference on Frontiers in Intelligent Computing: Theory and Applications, Bhubaneswar, India, 16–17 September 2016; Advances in Intelligent Systems and Computing; 2016. Springer: Singapore, 2017; Volume 1, pp. 159–167. [Google Scholar]
- Chuang, C.-H.; Cheng, C.-W.; Yen, Z.-Y. Reversible Data Hiding with Affine Invariance for 3D Models. In Proceedings of the IET International Conference on Frontier Computing. Theory, Technologies and Applications, Taichung, Taiwan, 4–6 August 2010. [Google Scholar]
- Jhou, C.-Y.; Pan, J.-S.; Chou, D. Reversible data hiding base on histogram shift for 3D vertex. In Proceedings of the Third International Conference on Intelligent Information Hiding and Multimedia Signal Processing (IIH-MSP 2007), Kaohsiung, Taiwan, 26–28 November 2007; Volume 1, pp. 365–370. [Google Scholar]
- Huang, Y.-H.; Tsai, Y.-Y. A reversible data hiding scheme for 3D polygonal models based on histogram shifting with high embedding capacity. 3D Res. 2015, 6, 20. [Google Scholar] [CrossRef]
- Ni, Z.; Shi, Y.-Q.; Ansari, N.; Su, W. Reversible data hiding. IEEE Trans. Circuits Syst. Video Technol. 2006, 16, 354–362. [Google Scholar]
- Mademlis, A.; Axenopoulos, A.; Daras, P.; Tzovaras, D.; Strintzis, M.G. 3D content-based search based on 3D Krawtchouk moments. In Proceedings of the Third International Symposium on 3D Data Processing, Visualization, and Transmission (3DPVT’06), Chapel Hill, NC, USA, 14–16 June 2006; pp. 743–749. [Google Scholar]
- Kumar, N.; Kumar, R.; Caldelli, R. Local moment driven PVO based reversible data hiding. IEEE Signal Process. Lett. 2021, 28, 1335–1339. [Google Scholar] [CrossRef]
- Wang, X.; Wang, X.; Ma, B.; Li, Q.; Wang, C.; Shi, Y. High-performance reversible data hiding based on ridge regression prediction algorithm. Signal Process. 2023, 204, 108818. [Google Scholar] [CrossRef]
- Singh, Y.V.; Khan, S.; Shukla, S.K.; Jung, K.-H. Local-Moment-Driven Robust Reversible Data Hiding. Appl. Sci. 2022, 12, 11826. [Google Scholar] [CrossRef]
- Shi, Y.-Q.; Li, X.; Zhang, X.; Wu, H.-T.; Ma, B. Reversible data hiding: Advances in the past two decades. IEEE Access 2016, 4, 3210–3237. [Google Scholar] [CrossRef]
- Mandal, P.C.; Mukherjee, I.; Chatterji, B.N. High capacity reversible and secured data hiding in images using interpolation and difference expansion technique. Multimed. Tools Appl. 2021, 80, 3623–3644. [Google Scholar] [CrossRef]
- Lin, J.-Y.; Horng, J.-H.; Chang, C.-C.; Li, Y.-H. Asymmetric Orientation Combination for Reversible and Authenticable Data Hiding of Dual Stego-images. Symmetry 2022, 14, 819. [Google Scholar] [CrossRef]
- Mehbodniya, A.; Douraki, B.K.; Webber, J.L.; Alkhazaleh, H.A.; Elbasi, E.; Dameshghi, M.; Abu Zitar, R.; Abualigah, L. Multilayer reversible data hiding based on the difference expansion method using multilevel thresholding of host images based on the slime mould algorithm. Processes 2022, 10, 858. [Google Scholar] [CrossRef]
- Ghebleh, M.; Kanso, A. A robust chaotic algorithm for digital image steganography. Commun. Nonlinear Sci. Numer. Simul. 2014, 19, 1898–1907. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, X.; Guo, Y.; Qin, C.; Liu, F. Zernike moment-based spatial image steganography resisting scaling attack and statistic detection. IEEE Access 2019, 7, 24282–24289. [Google Scholar] [CrossRef]
- Li, Q.; Wang, X.; Ma, B.; Wang, X.; Wang, C.; Xia, Z.; Shi, Y. Image steganography based on style transfer and quaternion exponent moments. Appl. Soft Comput. 2021, 110, 107618. [Google Scholar] [CrossRef]
- Soria-Lorente, A.; Berres, S.; Díaz-Nuñez, Y.; Avila-Domenech, E. Hiding data inside images using orthogonal moments. J. Inf. Secur. Appl. 2022, 67, 103192. [Google Scholar] [CrossRef]
- Radeaf, H.S.; Mahmmod, B.M.; Abdulhussain, S.H.; Al-Jumaeily, D. A steganography based on orthogonal moments. In Proceedings of the International Conference on Information and Communication Technology, Baghdad, Iraq, 15–16 April 2019; pp. 147–153. [Google Scholar]
- Tahiri, M.A.; Bencherqui, A.; Karmouni, H.; Amakdouf, H.; Mirjalili, S.; Motahhir, S.; Abouhawwash, M.; Askar, S.S.; Sayyouri, M.; Qjidaa, H. Implementation of a Steganography System Based on Hybrid Square Quaternion Moment Compression in IoMT. J. King Saud Univ.-Comput. Inf. Sci. 2023, 35, 101604. [Google Scholar] [CrossRef]
- Arsalan, M.; Malik, S.A.; Khan, A. Intelligent reversible watermarking in integer wavelet domain for medical images. J. Syst. Softw. 2012, 85, 883–894. [Google Scholar] [CrossRef]
- Jinna, S.K.; Ganesan, L. Reversible image data hiding using lifting wavelet transform and histogram shifting. arXiv 2010, arXiv:1004.1791. [Google Scholar]
- Muhammad, N.; Bibi, N.; Mahmood, Z.; Akram, T.; Naqvi, S.R. Reversible integer wavelet transform for blind image hiding method. PLoS ONE 2017, 12, e0176979. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-C.; Fang, W.-C. Integrity preservation and privacy protection for medical images with histogram-based reversible data hiding. In Proceedings of the 2011 IEEE/NIH Life Science Systems and Applications Workshop (LiSSA), Bethesda, MD, USA, 7–8 April 2011; pp. 108–111. [Google Scholar]
- Daoui, A.; Yamni, M.; Karmouni, H.; Sayyouri, M.; Qjidaa, H.; Motahhir, S.; Jamil, O.; El-Shafai, W.; Algarni, A.D.; Soliman, N.F. Efficient Biomedical Signal Security Algorithm for Smart Internet of Medical Things (IoMTs) Applications. Electronics 2022, 11, 3867. [Google Scholar] [CrossRef]
- Mirsky, Y.; Mahler, T.; Shelef, I.; Elovici, Y. CT-GAN: Malicious Tampering of 3D Medical Imagery using Deep Learning. In Proceedings of the USENIX Security Symposium, Santa Clara, CA, USA, 14–16 August 2019; Volume 2019. [Google Scholar]
- Grössinger, D.; Fischmeister, F.P.S.; Witte, M.; Koschutnig, K.; Ninaus, M.; Neuper, C.; Kober, S.E.; Wood, G. The role of superstition of cognitive control during neurofeedback training. bioRxiv 2021. [Google Scholar] [CrossRef]
- Soler, L.; Hostettler, A.; Agnus, V.; Charnoz, A.; Fasquel, J.; Moreau, J.; Osswald, A.; Bouhadjar, M.; Marescaux, J. 3D image reconstruction for comparison of algorithm database: A patient specific anatomical and medical image database. IRCAD Strasbg. Fr. Tech Rep 2010, 1. [Google Scholar]
- Baid, U.; Ghodasara, S.; Mohan, S.; Bilello, M.; Calabrese, E.; Colak, E.; Farahani, K.; Kalpathy-Cramer, J.; Kitamura, F.C.; Pati, S. The RSNA-ASNR-MICCAI BraTS 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv 2021, arXiv:2107.02314. [Google Scholar]
- Kuo, W.-C.; Jiang, D.-J.; Huang, Y.-C. A reversible data hiding scheme based on block division. In Proceedings of the 2008 Congress on Image and Signal Processing, Sanya, China, 27–30 May 2008; Volume 1, pp. 365–369. [Google Scholar]
- Chang, C.-C.; Lin, C.-C.; Tseng, C.-S.; Tai, W.-L. Reversible hiding in DCT-based compressed images. Inf. Sci. 2007, 177, 2768–2786. [Google Scholar] [CrossRef]
- Huang, H.-Y.; Chang, S.-H. A 9/7 wavelet-based lossless data hiding. In Proceedings of the 2011 IEEE Symposium On Computational Intelligence For Multimedia, Signal And Vision Processing, Paris, France, 11–15 April 2011; pp. 1–6. [Google Scholar]
- Kailath, T. The divergence and Bhattacharyya distance measures in signal selection. IEEE Trans. Commun. Technol. 1967, 15, 52–60. [Google Scholar] [CrossRef]
- Daoui, A.; Yamni, M.; Chelloug, S.A.; Wani, M.A.; El-Latif, A.A.A. Efficient Image Encryption Scheme Using Novel 1D Multiparametric Dynamical Tent Map and Parallel Computing. Mathematics 2023, 11, 1589. [Google Scholar] [CrossRef]




| = [ | 0.63299 | −1.00113 | 1.00113 | 0 | ] |
| = [ | 0 | −0.25951 | 0.25951 | −0.61237 | ] |
| = [ | 0.12976 | 0 | 0.22474 | 0.43301 | ] |
| = [ | 0.68330 | −0.81650 | 0 | 0.35355 | ] |
| = [ | 0.93265 | −0.57735 | −1.09638 | 0 | ] |
| = [ | 0.912366 | −1.293027 | −1.738221 | −1.857055 | 1.923238 | 4.164748 | 4.787527 | 0 | ] |
| = [ | 0 | −0.478498 | −1.251264 | −1.236242 | 1.270850 | 2.520126 | 2.305815 | −0.522913 | ] |
| = [ | 0.035496 | 0 | 0.836863 | 0.453748 | −1.166437 | −1.687982 | −1.561738 | 0.423608 | ] |
| = [ | 0.025099 | −0.129757 | 0 | 0.320848 | −0.824795 | −0.785335 | −1.811422 | 0.299536 | ] |
| = [ | 0.022913 | 0.527046 | 0.205163 | 0 | −0.045825 | −1.629780 | −1.653595 | 0.273437 | ] |
| = [ | −0.021909 | 0.150701 | 0.181791 | 0.449090 | 0 | 1.558368 | 1.581139 | −0.261456 | ] |
| = [ | 0.750597 | −0.709915 | 0.848661 | 0.782428 | −0.522623 | 0 | 2.956796 | −0.286411 | ] |
| = [ | 0.612860 | 0.142044 | 0.442929 | 0.079832 | 0.234718 | −0.239146 | 0 | −0.233854 | ] |
| = [ | −1.775545 | −2.571369 | −0.004222 | 0.503776 | −2.003696 | −0.941328 | 6.166002 | 0 | ] |
| Database | Number of 3D Images | Body Part Examined | Modality | Format and Size | Class and Bit Depth |
|---|---|---|---|---|---|
| LIDC-IDRI [52] | 80 | Human lungs | CT | DICOM | Signed |
| 160 × 160 × 160 | 16 bit | ||||
| OpenNeuro [53] | 70 | Head | MRI | NIfTI | Signed |
| 192 × 192 × 192 | 16 bit | ||||
| IRCAD [54] | 20 | Liver | CT | NIfTI | Signed |
| 128 × 128 × 128 | 16 bit | ||||
| BraTS 2021 [55] | 38 | Brain | MRI | DICOM | Unsigned |
| 96 × 96 × 96 | 16 bit |
| 3D Medical Images Database | Metric | Classical Krawtchouk Transform for p = 0.5 | |
|---|---|---|---|
| 80 images from LIDC-IDRI [52] | PSNR (dB) | ∞ | 341.5722 |
| MSE | 0 | 2.9904 × 10−25 | |
| 70 images from OpenNeuro [53] | PSNR (dB) | ∞ | 312.5929 |
| MSE | 0 | 2.3640 × 10−22 | |
| 20 images from IRCAD [54] | PSNR (dB) | ∞ | 343.7595 |
| MSE | 0 | 1.8072 × 10−25 | |
| 38 images from BraTS 2021 [55] | PSNR | ∞ | 360.6442 |
| MSE | 0 | 3.7028 × 10−27 |
| Image Name from the Database | Average EC (bits) | Average EC (bpv) | Average PSNR (dB) |
|---|---|---|---|
| 1003 | 2,156,809 | 0.5266 | 75.7744 |
| 1005 | 2,125,935 | 0.5190 | 75.7660 |
| 1009 | 2,207,080 | 0.5388 | 75.8119 |
| 1045 | 2,191,740 | 0.5351 | 75.7590 |
| 1067 | 2,190,836 | 0.5349 | 75.7844 |
| 1201 | 2,037,064 | 0.4973 | 75.7113 |
| 1219 | 2,115,337 | 0.5164 | 75.7513 |
| Image Name from the Database | Average EC (bits) | Average EC (bpv) | Average PSNR (dB) |
|---|---|---|---|
| sub-01 | 2,434,181 | 0.3439 | 75.3041 |
| sub-02 | 2,271,334 | 0.3209 | 75.2472 |
| sub-08 | 2,944,946 | 0.4161 | 75.4962 |
| sub-09 | 2,649,708 | 0.3744 | 75.3851 |
| sub-10 | 2,538,429 | 0.3586 | 75.3416 |
| sub-11 | 2,641,653 | 0.3732 | 75.3831 |
| sub-12 | 2,664,451 | 0.3764 | 75.3923 |
| Image Name from the Database | Average EC (bits) | Average EC (bpv) | Average PSNR (dB) |
|---|---|---|---|
| ircad_e01 | 1,127,532 | 0.5376 | 75.8147 |
| ircad_e02 | 1,216,511 | 0.5801 | 75.9162 |
| ircad_e03 | 1,084,896 | 0.5173 | 75.7673 |
| ircad_e04 | 1,222,973 | 0.5832 | 75.8694 |
| ircad_e05 | 1,149,071 | 0.5479 | 75.8471 |
| ircad_e06 | 1,138,679 | 0.5430 | 75.8361 |
| ircad_e07 | 1,217,345 | 0.5805 | 75.9177 |
| Image Name from the Database | Average EC (bits) | Average EC (bpv) | Average PSNR (dB) |
|---|---|---|---|
| 00114 | 759,584 | 0.8585 | 76.9145 |
| 00119 | 707,022 | 0.7991 | 76.6859 |
| 00125 | 786,924 | 0.8894 | 77.0169 |
| 00153 | 784,908 | 0.8872 | 77.0111 |
| 00161 | 716,177 | 0.8095 | 76.7315 |
| 00174 | 704,289 | 0.7960 | 76.6777 |
| 00181 | 736,130 | 0.8320 | 76.8149 |
| IRKT-Based 3D RDH Method with | 80 Images from LIDC-IDRI | 70 Images from OpenNeuro | 20 Images from IRCAD | 38 Images from BraTS 2021 |
|---|---|---|---|---|
| For T = 1 | 1.4416 × 10−7 | 7.8335 × 10−7 | 2.8873 × 10−5 | 2.2724 × 10−6 |
| For all thresholds | 3.49 × 10−2 | 1.2647 × 10−4 | 8.87 × 10−2 | 4.9249 × 10−4 |
| 3D Medical Image Database | MSE | PSNR (dB) |
|---|---|---|
| 80 3D images from LIDC-IDRI database | 0 | ∞ |
| 70 3D images from OpenNeuro database | 0 | ∞ |
| 20 3D images from IRCAD database | 0 | ∞ |
| 38 3D images from BraTS 2021 database | 0 | ∞ |
| Algorithm | Embedding Domain | Type of Carrier Data | Suitable for Medical Image | Average EC |
|---|---|---|---|---|
| [31] | Spatial | 3D image models | No | 0.2460 bits/vertex |
| [30] | Spatial | 3D image models | No | 0.1725 bits/vertex |
| [29] | Spatial | 3D image models | No | 0.2118 bits/vertex |
| Our | IRKT | 3D grayscale medical image | Yes | 0.4228 bits/voxel |
| Method | Embedding Domain | Unsign 16-bit Grayscale Medical Image | Sign 16-bit Grayscale Medical Image | ||
|---|---|---|---|---|---|
| Average EC | Average PSNR | Average EC | Average PSNR | ||
| [22] | Spatial | 0.248 bits/pixel | 74.66 | 0.124 bits/pixel | 74.77 |
| Proposed | IRKM | 0.803 bits/voxel | 76.70 | 0.422 bits/voxel | 75.51 |
| Method | Embedding Domain | Lena | Mandrill | Airplane | Boat | Average | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| EC (bits) | PSNR | EC (bits) | PSNR | EC (bits) | PSNR | EC (bits) | PSNR | EC (bits) | PSNR | ||
| [32] | Spatial | 5460 | 48.20 | 5421 | 48.20 | 16,171 | 48.3 | 7301 | 48.20 | 8588.3 | 48.2250 |
| [56] | Spatial | 8835 | 48.2 | 5423 | 48.20 | 23,199 | 48.3 | 10,217 | 48.30 | 11,919 | 48.25 |
| [8] | CDF transform | 52,429 | 46.23 | 26,214 | 43.45 | ----- | ----- | ----- | ----- | 39,322 | 44.8400 |
| [9] | Integer Haar | 40,108 | 48.39 | 13,107 | 48.21 | 57,147 | 48.53 | 27,787 | 48.33 | 34,537 | 48.3650 |
| [9] | Integer 2/6 transform | 44,827 | 48.25 | 14,680 | 48.03 | 61,604 | 48.36 | 31,195 | 48.22 | 38,077 | 48.2150 |
| [9] | Integer 9/7-F | 50,070 | 45.25 | 17,564 | 45.46 | 67,633 | 45.35 | 35,914 | 45.26 | 42,795 | 45.3300 |
| [10] | Integer DCT | 64,345 | 41.787 | 23,904 | 41.505 | 93,951 | 42.039 | 50,848 | 41.697 | 58,262 | 41.7570 |
| [57] | Standard DCT | 36,857 | 30.34 | 35,389 | 26.46 | 36,805 | 29.98 | 36,700 | 29.75 | 36,438 | 29.1325 |
| [58] | Standard DWT | 39,872 | 30.08 | ----- | ----- | 39,453 | 27.84 | 39,138 | 31.41 | 39,488 | 29.7767 |
| Proposed | IRKT | 80,569 | 45.978 | 19,982 | 46.030 | 63,374 | 45.986 | 70,263 | 46.016 | 58,547 | 46.0031 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamni, M.; Daoui, A.; Pławiak, P.; Mao, H.; Alfarraj, O.; El-Latif, A.A.A. A Novel 3D Reversible Data Hiding Scheme Based on Integer–Reversible Krawtchouk Transform for IoMT. Sensors 2023, 23, 7914. https://doi.org/10.3390/s23187914
Yamni M, Daoui A, Pławiak P, Mao H, Alfarraj O, El-Latif AAA. A Novel 3D Reversible Data Hiding Scheme Based on Integer–Reversible Krawtchouk Transform for IoMT. Sensors. 2023; 23(18):7914. https://doi.org/10.3390/s23187914
Chicago/Turabian StyleYamni, Mohamed, Achraf Daoui, Paweł Pławiak, Haokun Mao, Osama Alfarraj, and Ahmed A. Abd El-Latif. 2023. "A Novel 3D Reversible Data Hiding Scheme Based on Integer–Reversible Krawtchouk Transform for IoMT" Sensors 23, no. 18: 7914. https://doi.org/10.3390/s23187914
APA StyleYamni, M., Daoui, A., Pławiak, P., Mao, H., Alfarraj, O., & El-Latif, A. A. A. (2023). A Novel 3D Reversible Data Hiding Scheme Based on Integer–Reversible Krawtchouk Transform for IoMT. Sensors, 23(18), 7914. https://doi.org/10.3390/s23187914








