Development of a Deep Learning-Based Epiglottis Obstruction Ratio Calculation System
Abstract
:1. Introduction
2. Methods
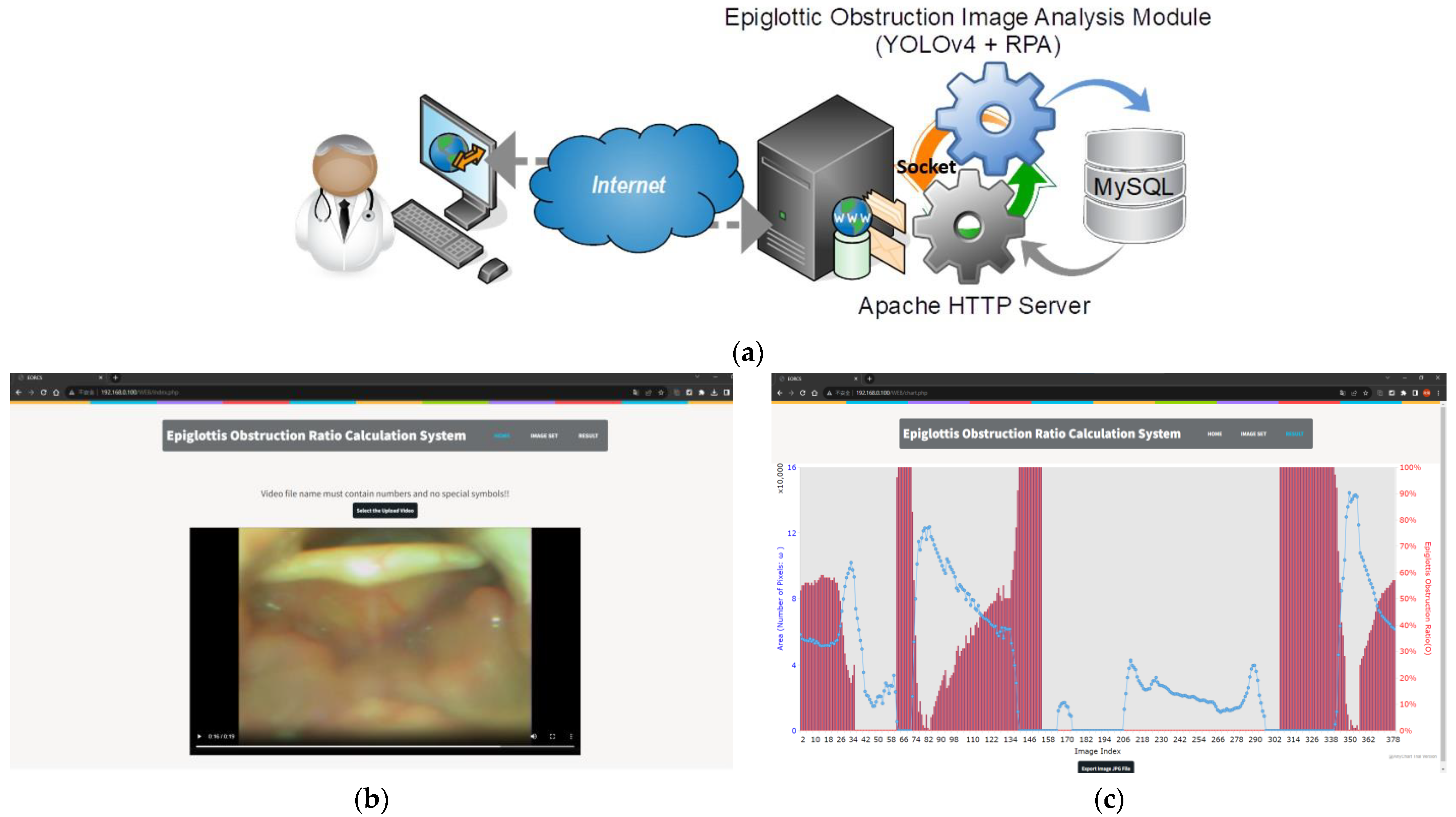
2.1. System Architecture and Processing Procedures
2.2. FOV Detection of Epiglottis Airway
2.3. You Only Look Once Version 4 Model
2.4. Region Puzzle Algorithm
2.4.1. EP-Region Merging
2.4.2. AE-Region Merging
- Step 1.
- Find the region covered by the range at the location as the initial region and fill any holes within .
- Step 2.
- In the th merging procedure (), check whether the region surrounding and region meets two conditions: (1) is surrounded by a single region , and (2) the range of does not exceed . If the two conditions are satisfied, will be directly merged with and the AE-region is updated to be . Fill in the holes in , and proceed to Step 3.
- Step 3.
- Perform the edge operation processing on to obtain the edge line and calculate the puzzle function .
- Step 4.
- Search for puzzle pieces adjacent to , and calculate the edge lines of .
- Step 5.
- Perform an XOR operation on each edge of with and calculate . If , merge with , and vice versa. After completing all the surrounding analysis of , fill in the holes of .
- Step 6.
- Compare the values of and . If , stop the algorithm. is the best result. If , update the AE-region result to be , and repeat steps 2 through 6.
3. Results
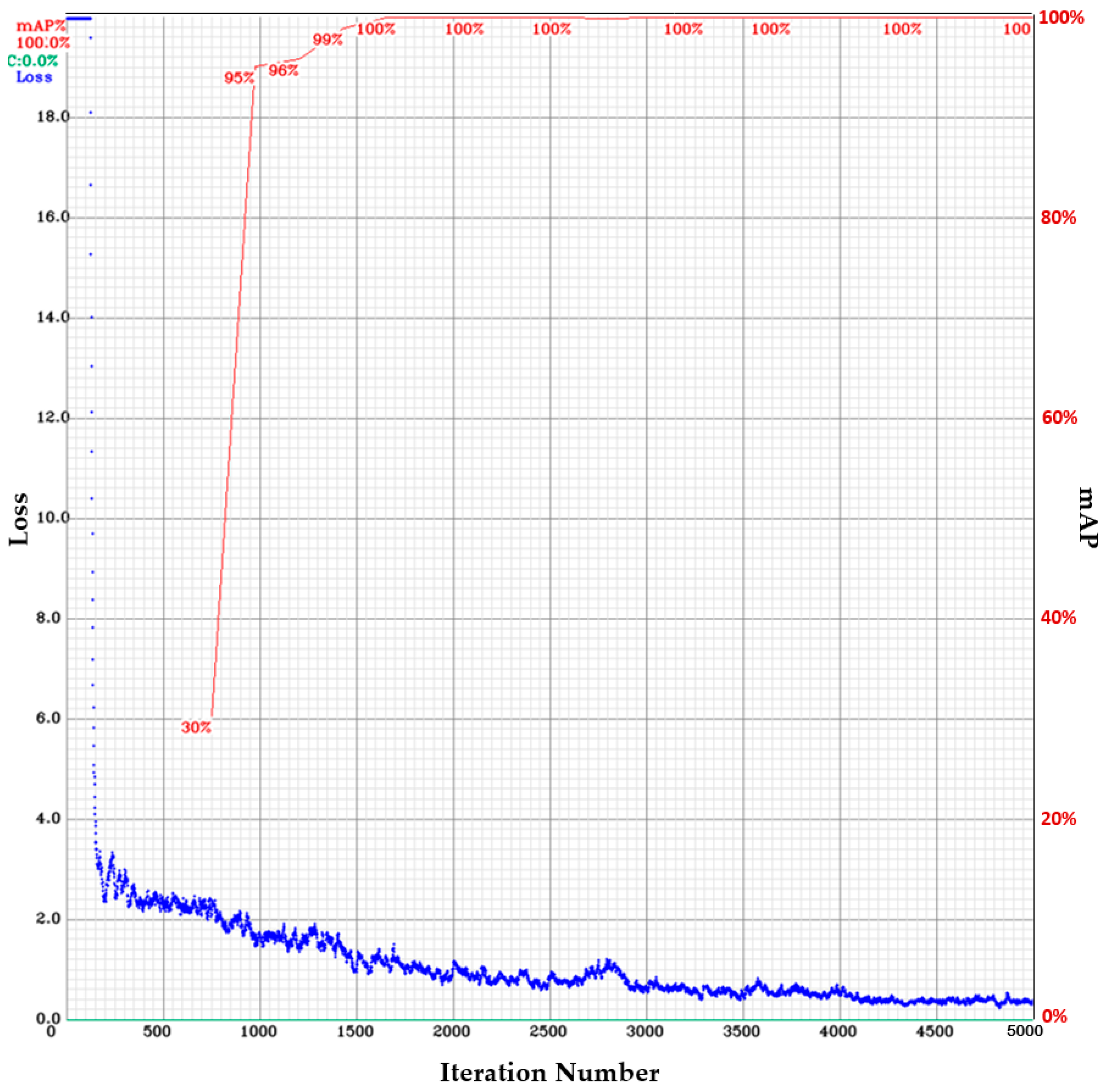
3.1. Epiglottis Image Positioning by the YOLOv4
- The batch size and the mini-batch size are 32 and 8, respectively.
- The image input to the network is 24-bit color images with 416 416 pixels.
- The activation function is Mish.
- The regional comparison rate is 0.9 (Momentum = 0.949).
- The weight reduction ratio is 0.0005 (Decay = 0.0005).
- The sample image diversification parameters are set to Angle = 0, Saturation = 1.5, Exposure = 1.5, and Hue = 0.1.
- The network learning rate is 0.001 (Learning Rate = 0.001).
- The maximum number of iterations is 5000 (Max Batches = 5000).
- The learning policy is “Step”.
- The number of feature generation filters is 32 (Filter = 32).
- The number of object classes is 1 (epiglottis) (Classes = 1).
3.2. EP-Region Merging
3.3. AE-Region Merging
3.4. The Effect of Color Quantification on the AE-Region
3.5. Continuous Image AE-Region Change Analysis
3.6. Comparing the Proposed Method with the Clinical Judgment Results
- In t1–t36, the epiglottis shrinks at least 50%;
- In t37–t62, there is tonsil interference (not included in the calculation of obstruction ratio);
- In t63–t73, there is no posterior pharyngeal wall in the image, but the epiglottic cartilage is sucked to the posterior pharyngeal wall, which is clinically judged to be 76–100% obstructed;
- In t74–t137, the epiglottis shrinks less than 50%;
- In t138–t155, there is no posterior pharyngeal wall in the image, but the epiglottic cartilage is sucked to the posterior pharyngeal wall, which is clinically judged to be 76–100% obstructed;
- In t156–t306, there is no posterior pharyngeal wall in the image, but there is tonsil interference (not included in the calculation of obstruction ratio);
- In t307–t343, the epiglottis cartilage is sucked to the direction of the posterior pharyngeal wall, which is clinically judged to be 76–100% obstructed;
- In t345–t380, the epiglottis shrinks less than 50%.
- (t1–t36): is 48.8% (clinical judgment CI = 0–49%).
- (t63–t73): is 98.1% (clinical judgment CI = 76–100%);
- (t74–t137): is 31.3% (clinical judgment CI = 0–49%);
- (t138–t155): is 96.4% (clinical judgment CI = 76–100%);
- (t307–t343): is 99.7% (clinical judgment CI = 76–100%);
- (t345–t380): is 34.5% (clinical judgment CI = 0–49%).
4. Discussion
4.1. Method Limitations
4.2. Work Limitations
- The study specifically calculates the precise extent of airway obstruction in the epiglottic region only. The method designed for the study cannot calculate the obstruction ratio for other obstruction sites.
- After calculating the AE-Region, a surgeon needs to determine the period affected by the tonsils, as this method cannot identify the tonsils.
- This research did not conduct a control group analysis, meaning the study did not analyze the images of the epiglottic region in healthy individuals who do not have OSA. However, theoretically, the research method is equally applicable.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, Y.-T.; Chang, R.; Chiu, L.-T.; Hung, Y.-M.; Wei, C.-C. Obstructive sleep apnea and influenza infection: A nationwide population-based cohort study. Sleep Med. 2021, 81, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Evans, E.C.; Sulyman, O.; Froymovich, O. The goals of treating obstructive sleep apnea. Otolaryngol. Clin. N. Am. 2020, 53, 319–328. [Google Scholar] [CrossRef]
- Lin, C.-H.; Chin, W.-C.; Huang, Y.-S.; Wang, P.-F.; Li, K.K.; Pirelli, P.; Chen, Y.-H.; Guilleminault, C. Objective and subjective long term outcome of maxillomandibular advancement in obstructive sleep apnea. Sleep Med. 2020, 74, 289–296. [Google Scholar] [CrossRef]
- Vanderveken, O.M.; Maurer, J.T.; Hohenhorst, W.; Hamans, E.; Lin, H.-S.; Vroegop, A.V.; Anders, C.; Nico, V.; Heyning, P.H.V. Evaluation of drug-induced sleep endoscopy as a patient selection tool for implanted upper airway stimulation for obstructive sleep apnea. J. Clin. Sleep Med. 2013, 9, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-B.; Lan, M.-Y.; Huang, Y.-C.; Huang, T.-T.; Lan, M.-C. The correlation between drug-induced sleep endoscopy findings and severity of obstructive sleep apnea. Auris Nasus Larynx 2021, 48, 434–440. [Google Scholar] [CrossRef]
- Kwon, O.E.; Jung, S.Y.; Al-Dilaijan, K.; Min, J.-Y.; Lee, K.H.; Kim, S.W. Is epiglottis surgery necessary for obstructive sleep apnea patients with epiglottis obstruction? Laryngoscope 2019, 129, 2658–2662. [Google Scholar] [CrossRef]
- Li, H.-Y.; Lo, Y.-L.; Wang, C.-J.; Hsin, L.-J.; Lin, W.-N.; Fang, T.-J.; Lee, L.-A. Dynamic drug-induced sleep computed tomography in adults with obstructive sleep apnea. Sci. Rep. 2016, 6, 35849. [Google Scholar] [CrossRef]
- Xia, F.; Sawan, M. Clinical and Research Solutions to Manage Obstructive Sleep Apnea: A Review. Sensors 2021, 21, 1784. [Google Scholar] [CrossRef] [PubMed]
- Edwards, C.; Almeida, O.P.; Ford, A.H. Obstructive sleep apnea and depression: A systematic review and meta-analysis. Maturitas 2020, 142, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Sunter, G.; Omercikoglu, O.H.; Vural, E.; Gunal, D.I.; Agan, K. Risk assessment of obstructive sleep apnea syndrome and other sleep disorders in multiple sclerosis patients. Clin. Neurol. Neurosurg. 2021, 207, 106749. [Google Scholar] [CrossRef] [PubMed]
- Jonas, D.E.; Amick, H.R.; Feltner, C.; Weber, R.P.; Arvanitis, M.; Stine, A.; Lux, L.; Middleton, J.C.; Voisin, C.; Harris, R.P. Screening for Obstructive Sleep Apnea in Adults Us Preventive Services Task Force Recommendation Statement; Report No.: 14-05216-EF-1; Agency for Healthcare Research and Quality (US): Rockville, MD, USA, January 2017.
- Kezirian, E.J.; Hohenhorst, W.; de Vries, N. Drug-induced sleep endoscopy: The VOTE classification. Eur. Arch. Oto-Rhino-Laryngol. 2011, 268, 1233–1236. [Google Scholar] [CrossRef]
- Tutsoy, O. Pharmacological, non-pharmacological policies and mutation: An artificial intelligence based multi-dimensional policy making algorithm for controlling the casualties of the pandemic diseases. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44, 9477–9488. [Google Scholar] [CrossRef]
- Gonzalez, R.C.; Woods, R.E. Digital Image Processing, 4th ed.; Pearson: Harlow, UK, 2017; pp. 184–187. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: London, UK, 2016; pp. 330–372. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 1, 1097–1105. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7 June 2015. [Google Scholar] [CrossRef]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.-Y.; Berg, A.C. SSD: Single shot multibox detector. In Proceedings of the 14th European Conference on Computer Vision—ECCV 2016, Amsterdam, The Netherlands, 11–14 October 2016; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; Volume 9905, pp. 21–37. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, real-time object detection. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. YOLO9000: Better, faster, stronger. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement (Tech Report) 2018. Available online: https://pjreddie.com/media/files/papers/YOLOv3.pdf (accessed on 20 January 2021).
- Bochkovskiy, A.; Wang, C.Y.; Liao, H.Y.M. YOLOv4: Optimal speed and accuracy of object detection. arXiv 2022, arXiv:2004.10934. [Google Scholar] [CrossRef]
- Wang, C.Y.; Bochkovskiy, A.; Liao, H.Y.M. Scaled-YOLOv4: Scaling cross stage partial network. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2021, Nashville, TN, USA, 20–25 June 2021. [Google Scholar] [CrossRef]
- Wang, C.Y.; Bochkovskiy, A.; Liao, H.Y.M. YOLOv7: Trainablebag-of-freebiessetsnewstate-of-the-artforreal-timeobject detectors. arXiv 2022, arXiv:2207.02696. [Google Scholar] [CrossRef]
- EKEN, S. Medical data analysis for different data types. Int. J. Comput. Exp. Sci. Eng. 2020, 6, 138–144. [Google Scholar] [CrossRef]
- Ferrer-Lluis, I.; Castillo-Escario, Y.; Montserrat, J.M.; Jané, R. Enhanced monitoring of sleep position in sleep apnea patients: Smartphone triaxial accelerometry compared with video-validated position from polysomnography. Sensors 2021, 21, 3689. [Google Scholar] [CrossRef] [PubMed]
- Kou, W.; Carlson, D.A.; Baumann, A.J.; Donnan, E.; Luo, Y.; Pandolfino, J.E.; Etemadi, M. A deep-learning-based unsupervised model on esophageal manometry using variational autoencoder. Artif. Intell. Med. 2021, 112, 102006. [Google Scholar] [CrossRef]
- Liu, Y.; Feng, Y.; Li, Y.; Xu, W.; Wang, X.; Han, D. Automatic classification of the obstruction site in obstructive sleep apnea based on snoring sounds. Am. J. Otolaryngol. 2022, 43, 103584. [Google Scholar] [CrossRef]
- Hanif, U.; Kiaer, E.K.; Capasso, R.; Liu, S.Y.; Mignot, E.J.M.; Sorensen, H.B.D.; Jennum, P. Automatic scoring of drug-induced sleep endoscopy for obstructive sleep apnea using deep learning. Sleep Med. 2023, 102, 19–29. [Google Scholar] [CrossRef]
- Heckbert, P.S. Color image quantization for frame buffer display. Comput. Graph. 1982, 16, 297–307. [Google Scholar] [CrossRef]
- Torre, C.; Camacho, M.; Liu, S.-Y.; Huon, L.-K.; Capasso, R. Epiglottis collapse in adult obstructive sleep apnea: A systematic review. Laryngoscope 2016, 126, 515–523. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Chao, Y.; Suzuki, K.; Wu, K. Fast connected-component labeling. Pattern Recognit. 2009, 42, 1977–1987. [Google Scholar] [CrossRef]
- Morera, Á.; Sánchez, Á.; Moreno, A.B.; Sappa, Á.D.; Vélez, J.F. SSD vs. YOLO for detection of outdoor urban advertising panels under multiple variabilities. Sensors 2020, 20, 4587. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-P.; Liaw, J.-J. A novel image measurement algorithm for common mushroom caps based on convolutional neural network. Comput. Electron. Agric. 2020, 171, 105336. [Google Scholar] [CrossRef]
- Su, H.-H.; Pan, H.-W.; Lu, C.-P.; Chuang, J.-J.; Yang, T. Automatic detection method for cancer cell nucleus image based on deep-learning analysis and color layer signature analysis algorithm. Sensors 2020, 20, 4409. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Mark Liao, H.-Y.; Wu, Y.-H.; Chen, P.-Y.; Hsieh, J.-W.; Yeh, I.-H. CSPNet: A new backbone that can enhance learning capability of CNN. In Proceedings of the 2020 IEEE Conference on Computer Vision and Pattern Recognition Workshop (CVPR Workshop), Seattle, WA, USA, 14–19 June 2020. [Google Scholar] [CrossRef]
- Misra, D. Mish: A self regularized non-monotonic activation function. arXiv 2020, arXiv:1908.08681. [Google Scholar] [CrossRef]
- Zheng, Z.; Wang, P.; Liu, W.; Li, J.; Ye, R.; Ren, D. Distance-iou loss: Faster and better learning for bounding box regression. In Proceedings of the AAAI Conference on Artificial Intelligence 2020, New York, NY, USA, 7–12 February 2020. [Google Scholar] [CrossRef]
- LabelImg. Available online: https://github.com/tzutalin/labelImg (accessed on 27 July 2023).



















Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, H.-H.; Lu, C.-P. Development of a Deep Learning-Based Epiglottis Obstruction Ratio Calculation System. Sensors 2023, 23, 7669. https://doi.org/10.3390/s23187669
Su H-H, Lu C-P. Development of a Deep Learning-Based Epiglottis Obstruction Ratio Calculation System. Sensors. 2023; 23(18):7669. https://doi.org/10.3390/s23187669
Chicago/Turabian StyleSu, Hsing-Hao, and Chuan-Pin Lu. 2023. "Development of a Deep Learning-Based Epiglottis Obstruction Ratio Calculation System" Sensors 23, no. 18: 7669. https://doi.org/10.3390/s23187669
APA StyleSu, H.-H., & Lu, C.-P. (2023). Development of a Deep Learning-Based Epiglottis Obstruction Ratio Calculation System. Sensors, 23(18), 7669. https://doi.org/10.3390/s23187669





