2.1. CLIP
The contrastive language-image pre-training (CLIP) (2021) [
21] model is a pretrained neural network released by OpenAI in early 2021 to match images and text, which can be said to be a classic work in the field of recent multimodal research. The model directly uses a large amount of Internet data for pretraining and achieved the best performance in many tasks. The CLIP model uses 400 million image–text pairs collected by OpenAI, encodes the text and images separately, and is trained using metric learning to improve the similarities between the images and text.
Since the introduction of CLIP, there have been many derivative works, such as image-guided text generatIon with CLIP (MAGIC) (2022) [
22] by Yixuan Su et al., where the downstream tasks generate image captions and stories by combining images and text. Patashnik et al.’s (2021) [
23], proposed text-driven manipulation of a style-based generator architecture for generative adversarial networks (styleGAN) imagery (Styleclip) combined StyleGAN and CLIP using three methods.The first was a text-guided latent optimization wherein the CLIP model was used as a loss network, which is a general approach but requires a few minutes of optimization for operation on images. The second was a latent residual mapper trained for a specific text prompt: given a starting point in the latent space (input image to operate on), the mapper produces a local step. The third was a method for mapping text prompts to input independent (global) directions in StyleGAN’s style space to provide control over the manipulation strength and decoupling. Khaatak et al. (2023) [
24] proposed multi-modal Prompt learning (MaPLe), inspired by CLIP, to achieve prompt learning on two modalities simultaneously further enhancing the multimodal alignment ability of the model on downstream tasks.
The main model of the present study was also designed using the CLIP model and has two parts for pretraining and testing.
2.2. Global Model Introduction
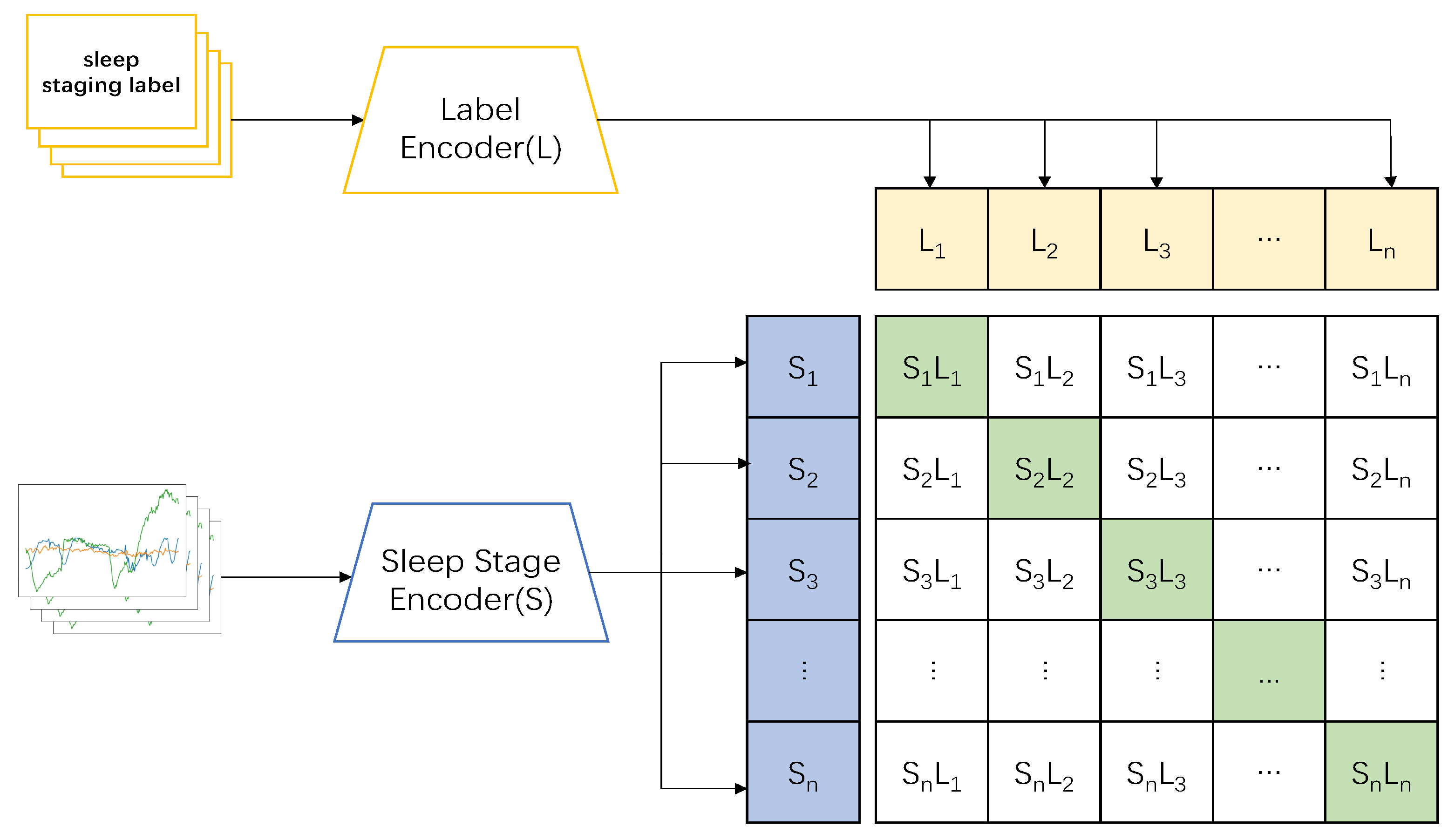
Based on the original structure of the model, we understood CLIP as the pictures and labels of the target classification signals in natural language, and we carried out a series of procedures, such as feature comparison and pairing, to classify the target signals.
By analogy, we believed that the expected sleep-staging tasks could be completed by applying the sleep staging label directly as an input marker signal to the dataset and processing the extracted single-channel EEG signals for feature comparison and matching. Accordingly, we designed the following model framework for sleep staging.
Thus, we designed a CLIP model based on the preconditioning model to complete the sleep-staging tasks. In the pretraining part, we applied the sleep stage data
and sleep label data
as inputs; the two sets of data were then imported into the corresponding encoders to obtain features
, and
, respectively, following which the model combined the two encoders to train the correct pairing of sleep and label data:
(show in
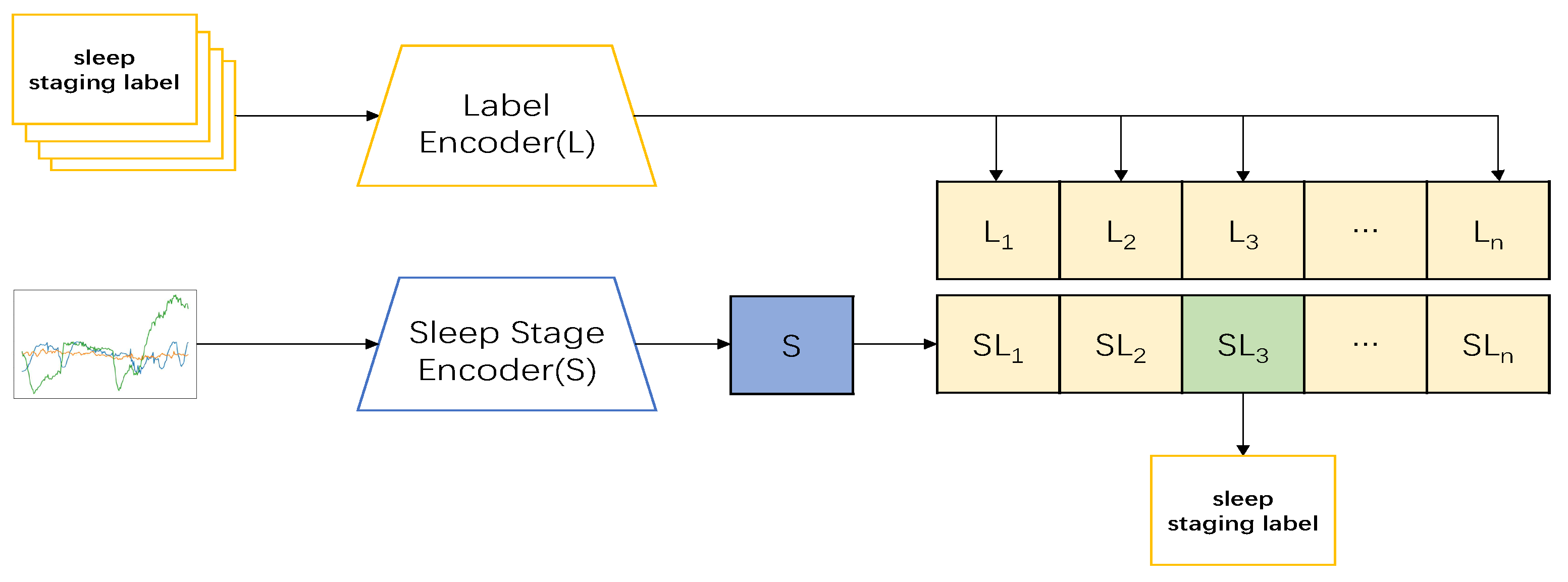
Figure 1). In the testing part, the datum
was input, and its feature
was obtained through the encoder. The feature was then input into the model and predicted with the label data in the model to obtain
; the sleep-staging result of the input data with the highest predicted value was then taken as the output (show in
Figure 2).The pseudocode of the whole model processing process will be shown in
Table 2.
2.3. Encoder
The method of extracting sleep signal features for sleep-staging tasks was the most important component of this work, and also the most important improvement to the CLIP model. The encoder was divided into two parts: a sleep-stage encoder for feature extraction from sleep signals and a label encoder for processing sleep signal labels.
Owing to its efficiency and speed, the label encoder module selected an autoregressive model as the transformer, which was primarily composed of multi-head attention (
) contiguous modules followed by a multilayer perception (
) module. The MLP block consists of two fully connected layers with a nonlinear rectified linear unit (ReLU) function and dropout in between. In this study, the transformer model used prenormalized residual linkages to generate more stable gradients.Then,
K identical layers were stacked to generate the final features. Next, a tag
was added to the input, the state of which was the feature vector of the final output. During operation, the transformer first applied the label data
to the linear projection
layer, which mapped the features to the hidden dimension
. The output of this preprojection was then sent to the transformer, and the final feature vector was appended to
so that the input features became the center
, where the subscript 0 represented the input to the first layer. Next,
was passed through the transformer layer as follows:
Finally, the context vector was rewired from the final output and used as input to the cosine similarity function.
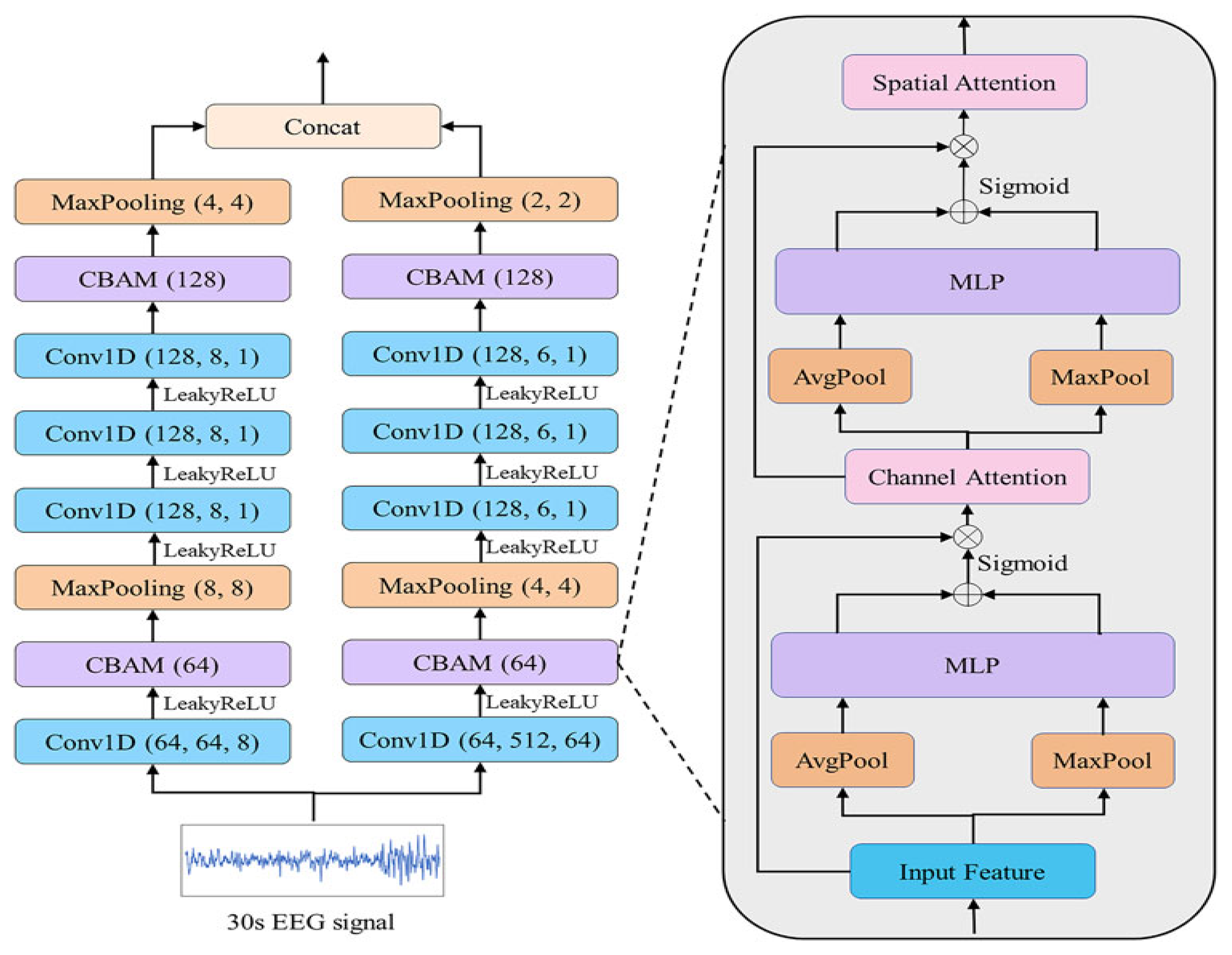
In the sleep-stage encoder, we used the EEG feature extraction module of Wang et al.’s DynamicSleepNet model [
25], the structure of which is shown in
Figure 3. The encoder was designed according to the characteristics of the EEG signals in each sleep period. The
wave (4–8 Hz), which is stable in the relaxed state with eyes closed, was an effective feature for distinguishing the W stage. The
wave (8–13 Hz), which is common in the late stage of N1, was also an effective feature for distinguishing the N1 stage. At a sampling rate of 100 Hz, each convolution window captured 0.64 s (F = 1/T) of information, which meant that relatively high-frequency information, such as
, was captured. The
wave was complete. The main waveform of the N3 period was the
wave (0.5–4 Hz), which contained relatively low-frequency information; a large convolution kernel of 512 was designed for this feature, and each convolution window captured 5.12 s of information, which meant that we completely extracted the
wave and other low-frequency information.
First, the EEG epochs were input into the Conv1D layer to obtain the initial features F as , where ; n is the total number of features; d is the length of ; and the 1D convolution layer (Conv1D) is the convolution operation in the effective feature extraction module (EFEM).
Next, the acquired features were input into the channel-attention module in the CBAM module. The features
F were then compressed to obtain
and
by the average (AvgPool) and maximum pool (MaxPool). The average and maximum pool steps were used to compress channel information so that
was compressed into
and
. These two sets of features were then input into a shared
network, which used the sigmoid activation function to generate the channel attention features
.
where
is the sigmoid activation function, and
and
are the pooling and
layers, respectively. Then,
was used to scale the features
F as follows:
where ⊗ refers to pointwise multiplication. The channel attention module processes the features
F to obtain
, which has the same dimensions as
F. The obtained
wass next input into the average and maximum pooling layers to obtain
and
, respectively. These two features are connected and the convolution operation is applied to them; then, the sigmoid activation function was used to generate the spatial attention feature map
as follows:
where
refers to the convolution operation with kernel size 7. Finally, the features
are scaled by Ms as follows:
where
was the last set of features after CBAM block processing, and the most essential features as well as the most important parts of each feature were adaptively selected for classification.
Finally, was input into later modules to obtain the output EEG features . These output features S were imported into the cosine similarity value calculation module.
2.4. Cosine Similarity Value and Loss Function
Next, to predict the sleep data and labels better, we computed the cosine similarities between their features as follows:
Here, and represent the vector lengths of and , respectively.
After obtaining the pairwise occurrences
, the overall arrangement was in the form of the matrix columns in
Figure 2. The diagonal pairs were the positive samples that we wanted, while the other pairs were the negative samples. Following the basic idea of contrastive learning, the distances among the positive samples were as large as possible from the distances between the positive and negative samples, so that the model could learn the features among the positive samples better. Here, we trained the model on cross-entropy, and the loss functions for the sleep data
S and label data
L are below:
Here,
represents the true probability distribution of
L and
S, while
represents the assumed probability distributions of
L, and
S. Accordingly, the overall loss of the model is given as follows: