Ensembles of Convolutional Neural Networks and Transformers for Polyp Segmentation
Abstract
1. Introduction
- An up-to-date review of ensembles for polyp segmentation. Actually, to the best of our knowledge, no earlier review on the topic is available in the open literature.
2. Related Work
- Data augmentation [22] increases the size of the training set by adding synthetic samples. Such samples can be created in many ways: the most-common approach in computer vision is to generate new images by simply altering existing ones, for instance by flipping, cropping, or rotating them. However, other approaches are possible, including the generation of completely artificial images [23].
- Ensemble techniques [24] increase accuracy by combining the responses of different classifiers (per-pixel classifiers, in the case of semantic segmentation). As in the case of data augmentation, many different solutions have been proposed to combine the answers and to build the classifiers themselves.
- Three [31,32,33] exhibit results that no longer represent the state-of-the-art for popular datasets, the more so considering that such results were obtained with more benevolent experimental protocols (e.g., ensembles trained and tested on the same dataset) than the one [10] currently adopted by several researchers, including ourselves.
| Paper | Ensemble Size | Deep Labv3 | Deep Labv3+ | Eff- UNet | FPN | HarDNet- MSEG | Mask R-CNN | MultiRes UNet | nnU-Net | PSPNet | Polyp- PVT | SegNet | U-Net | Unet++ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Guo2019 [20] | 3 | 🗸 | 🗸 | 🗸 | ||||||||||
| Kang2019 [31] | 2 | 🗸 | ||||||||||||
| Nguyen2019 [34] | 3 | 🗸 | ||||||||||||
| Shrestha2020 [32] | 2 | 🗸 | ||||||||||||
| ThuHong2020 [35] | 2 | 🗸 | ||||||||||||
| Hong2021 [33] | 5 | 🗸 | ||||||||||||
| Lumini2021a [27] | 14 | 🗸 | ||||||||||||
| Lumini2021b [28] | 14 | 🗸 | ||||||||||||
| Nanni2021 [29] | 2, 10, 20, 30, 60 | 🗸 | 🗸 | |||||||||||
| Thambawita2021 [36] | 3, 7 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | ||||||||
| Tomar2021 [25] | 4 | 🗸 | ||||||||||||
| Nanni2022a [37] | 2, 10, 14 | 🗸 | 🗸 | 🗸 | ||||||||||
| Nanni2022b [30] | 12, 14, 15, 16, 20 | 🗸 | 🗸 | |||||||||||
| Tran2022 [26] | N/A | 🗸 | 🗸 | |||||||||||
| Nanni2023 [38] | 2, 4, 10, 20, 30 | 🗸 | 🗸 |
| Paper | ColDB | ClinDB | CVC-T | EDD 2020 | Polyp-Gen | ETIS | Hyper-Kvasir | Kvasir | MediEval 2020 |
|---|---|---|---|---|---|---|---|---|---|
| Guo2019 [20] | 🗸 | 🗸 | 🗸 | ||||||
| Kang2019 [31] | 🗸 | 🗸 | 🗸 | ||||||
| Nguyen2019 [34] | 🗸 | 🗸 | 🗸 | ||||||
| Shrestha2020 [32] | 🗸 | 🗸 | |||||||
| ThuHong2020 [35] | 🗸 | 🗸 | 🗸 | ||||||
| Hong2021 [33] | 🗸 | ||||||||
| Lumini2021a [27] | 🗸 | ||||||||
| Lumini2021b [28] | 🗸 | ||||||||
| Nanni2021 [29] | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | ||||
| Thambawita2021 [36] | 🗸 | 🗸 | |||||||
| Tomar2021 [25] | 🗸 | ||||||||
| Nanni2022a [37] | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | ||||
| Nanni2022b [30] | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | ||||
| Tran2022 [26] | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | ||||
| Nanni2023 [38] | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 |
| Paper | Accuracy | Dice | F2 | FPS | IoU | Precision | Recall | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|---|
| Guo2019 [20] | 🗸 | 🗸 | |||||||
| Kang2019 [31] | 🗸 | 🗸 | 🗸 | ||||||
| Nguyen2019 [34] | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | |||
| Shrestha2020 [32] | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | ||
| ThuHong2020 [35] | 🗸 | 🗸 | 🗸 | 🗸 | |||||
| Hong2021 [33] | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | ||
| Lumini2021a [27] | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | |||
| Lumini2021b [28] | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | |||
| Nanni2021 [29] | 🗸 | 🗸 | |||||||
| Thambawita2021 [36] | 🗸 | 🗸 | 🗸 | 🗸 | |||||
| Tomar2021 [25] | |||||||||
| Nanni2022a [37] | 🗸 | 🗸 | |||||||
| Nanni2022b [30] | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | 🗸 | |||
| Tran2022 [26] | 🗸 | ||||||||
| Nanni2023 [38] | 🗸 | 🗸 |
| Paper | Kvasir | ClinDB | ColDB | ETIS | CVC-T | PolypGen | MediEval 2020 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IoU | Dice | IoU | Dice | IoU | Dice | IoU | Dice | IoU | Dice | IoU | Dice | IoU | Dice | |
| Guo2019 [20] | 0.967 | 0.983 | 0.962 | 0.980 | 0.970 | 0.985 | ||||||||
| Kang2019 [31] | 0.695 | 0.661 | ||||||||||||
| Nguyen2019 [34] | 0.891 | 0.896 | ||||||||||||
| Shrestha2020 [32] | 0.760 | 0.838 | 0.755 | 0.832 | ||||||||||
| ThuHong2020 [35] | 0.798 | 0.891 | 0.702 | 0.823 | ||||||||||
| Hong2021 [33] | 0.571 | 0.619 | ||||||||||||
| Lumini2021a [27] | 0.820 | 0.885 | ||||||||||||
| Lumini2021b [28] | 0.825 | 0.888 | ||||||||||||
| Nanni2021 [29] | 0.872 | 0.919 | 0.886 | 0.931 | 0.701 | 0.776 | 0.663 | 0.743 | 0.831 | 0.901 | ||||
| Thambawita2021 [36] | 0.840 | |||||||||||||
| Nanni2022a [37] | 0.874 | 0.920 | 0.894 | 0.937 | 0.751 | 0.826 | 0.717 | 0.787 | 0.842 | 0.904 | ||||
| Nanni2022b [30] | 0.870 | 0.918 | 0.884 | 0.929 | 0.695 | 0.768 | 0.644 | 0.727 | 0.833 | 0.904 | ||||
| Nanni2023 [38] | 0.871 | 0.920 | 0.903 | 0.947 | 0.720 | 0.787 | 0.688 | 0.756 | 0.846 | 0.909 | ||||
3. Methods
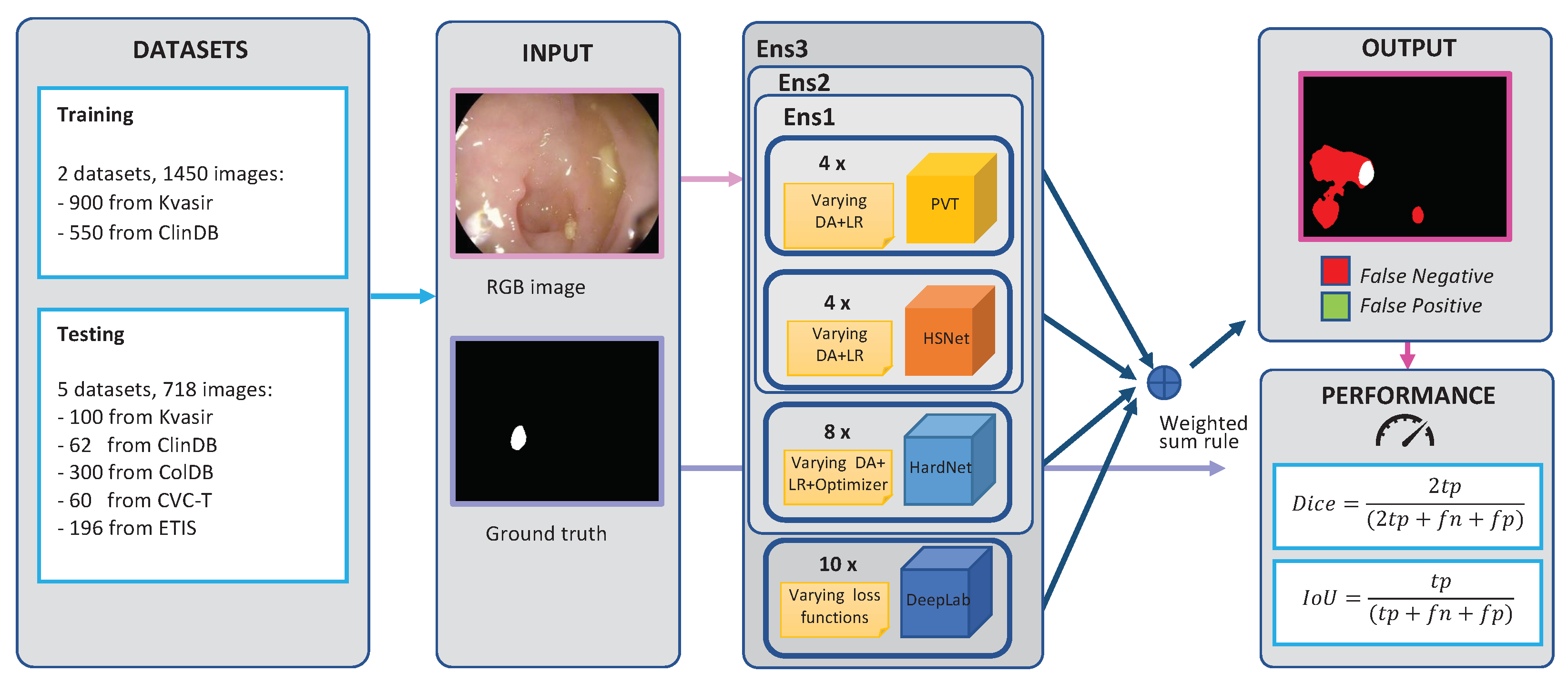
3.1. Structure of the Ensemble
3.2. Performance Metrics and Loss Functions
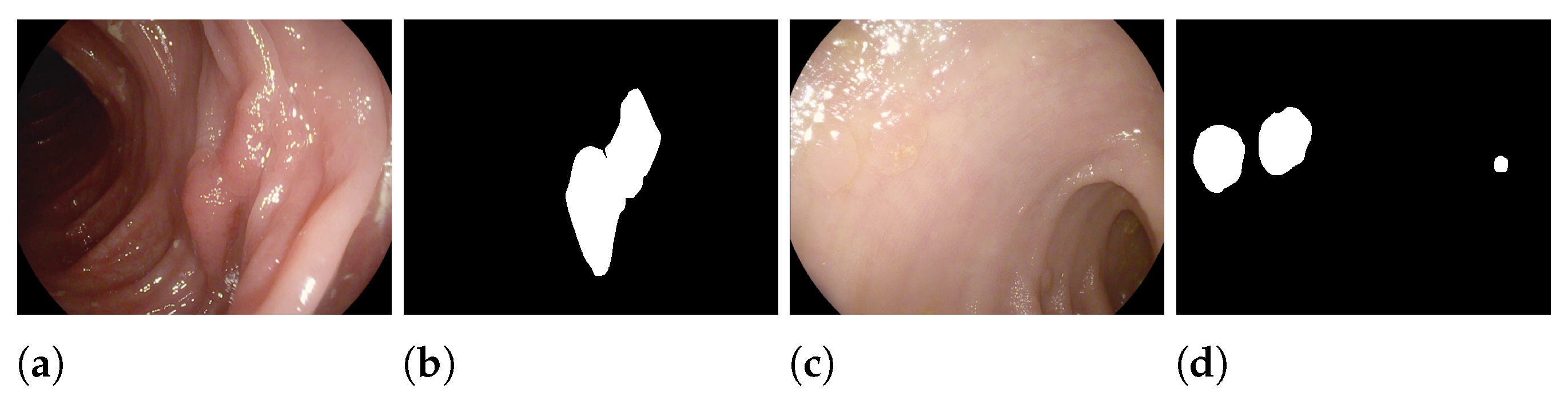
3.3. Datasets and Data Augmentations
- The Kvasir-SEG [56] dataset (“Kvasir”) contains medical images that have been labeled and verified by doctors. The images depict different parts of the digestive system and show both healthy and diseased tissue. The dataset includes images at different resolutions (from 720 × 576 up to 1920 × 1072 pixels) and is organized into folders based on the content of the images. Some of the images also include a small picture-in-picture showing the position of the endoscope inside the body.
- CVC-ColonDB [7] (“ColDB”) is a dataset of 300 images that aims to include a wide range of appearances for polyps. The goal is to provide as much diversity as possible in the dataset.
- CVC-T (sometimes called “Endo”) is the test set of a larger dataset named CVC-EndoSceneStill [47].
- CVC-ClinicDB [39] (“ClinDB”) contains 612 images from 31 videos of colonoscopy procedures. The images have been manually labeled by experts to identify the regions covered by the polyps, and ground truth information is also provided for light reflections. The images are 576 × 768 pixels in size.
- “DA1”: a basic strategy that includes two image flips (up/down and left/right) and a 90-degree counterclockwise rotation; therefore, three synthetic images were created for each original image.
- “DA2”: a sophisticated strategy that creates synthetic images in 13 different ways, including the application of motion blur and shadows to the original images.
3.4. Overview of the Experiments
- (learning rate “a”);
- decaying to after 30 epochs (learning rate “b”).
4. Experimental Results
4.1. Ablation Studies
- “SGD”: stochastic gradient descent.
- “Adam”: Adam.
- “SGD + Adam”: both.
4.2. Proposed Ensembles and Comparison with State-of-the-Art Models
- “Ens1”: an ensemble of 4 Polyp-PVT networks with the segmentation masks obtained with our approach and trained with all possible combinations of Data Augmentations 1 and 2 and Learning Rates a and b (Table 6, last row), plus 4 HSNet networks with the segmentation masks obtained with our approach and trained with all possible combinations of Data Augmentations 1 and 2 and Learning Rates a and b (Table 7, last row).
- “Ens2”: like Ens1, plus eight HarDNet-MSEG networks trained with all possible combinations of the SGD and Adam optimizers, Data Augmentations 1 and 2, and Learning Rates a and b (Table 5, last row).
- “Ens3”: like Ens2, plus the best ensemble introduced in [37] based on DeepLabv3+.
| Method | Kvasir | ClinDB | ColDB | ETIS | CVC-T | Avg | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IoU | Dice | IoU | Dice | IoU | Dice | IoU | Dice | IoU | Dice | IoU | Dice | |
| Ens1 | 0.886 | 0.930 | 0.892 | 0.936 | 0.764 | 0.839 | 0.738 | 0.812 | 0.841 | 0.904 | 0.824 | 0.884 |
| Ens2 | 0.883 | 0.927 | 0.894 | 0.938 | 0.759 | 0.832 | 0.750 | 0.822 | 0.841 | 0.904 | 0.825 | 0.885 |
| Ens3 | 0.882 | 0.928 | 0.894 | 0.937 | 0.752 | 0.824 | 0.760 | 0.830 | 0.840 | 0.905 | 0.826 | 0.885 |
| HarDNet-MSEG [12] | 0.857 | 0.912 | 0.882 | 0.932 | 0.66 | 0.731 | 0.613 | 0.677 | 0.821 | 0.887 | 0.767 | 0.828 |
| Polyp-PVT [13] | 0.864 | 0.917 | 0.889 | 0.937 | 0.727 | 0.808 | 0.706 | 0.787 | 0.833 | 0.9 | 0.804 | 0.869 |
| HSNet [14] | 0.877 | 0.926 | 0.905 | 0.948 | 0.735 | 0.81 | 0.734 | 0.808 | 0.839 | 0.903 | 0.818 | 0.879 |
| [37] | 0.874 | 0.920 | 0.894 | 0.937 | 0.751 | 0.826 | 0.717 | 0.787 | 0.842 | 0.904 | 0.816 | 0.875 |
| PraNet (from [12]) | 0.84 | 0.898 | 0.849 | 0.899 | 0.64 | 0.709 | 0.567 | 0.628 | 0.797 | 0.871 | 0.739 | 0.801 |
| SFA (from [12]) | 0.611 | 0.723 | 0.607 | 0.7 | 0.347 | 0.469 | 0.217 | 0.297 | 0.329 | 0.467 | 0.422 | 0.531 |
| U-Net++ (from [12]) | 0.743 | 0.821 | 0.729 | 0.794 | 0.41 | 0.483 | 0.344 | 0.401 | 0.624 | 0.707 | 0.57 | 0.641 |
| U-Net (from [12]) | 0.746 | 0.818 | 0.755 | 0.823 | 0.444 | 0.512 | 0.335 | 0.398 | 0.627 | 0.71 | 0.581 | 0.652 |
| Eloss101-Mix + FH [38] | 0.871 | 0.920 | 0.903 | 0.947 | 0.720 | 0.787 | 0.688 | 0.756 | 0.846 | 0.909 | 0.806 | 0.864 |
| MIA-Net [58] | 0.876 | 0.926 | 0.899 | 0.942 | 0.739 | 0.816 | 0.725 | 0.8 | 0.835 | 0.9 | 0.815 | 0.877 |
| P2T [59] | 0.849 | 0.905 | 0.873 | 0.923 | 0.68 | 0.761 | 0.631 | 0.7 | 0.805 | 0.879 | 0.768 | 0.834 |
| DBMF [60] | 0.886 | 0.932 | 0.886 | 0.933 | 0.73 | 0.803 | 0.711 | 0.79 | 0.859 | 0.919 | 0.814 | 0.875 |
| SETR [61] | 0.854 | 0.911 | 0.885 | 0.934 | 0.69 | 0.773 | 0.646 | 0.726 | 0.814 | 0.889 | 0.778 | 0.847 |
| TransUnet [62] | 0.857 | 0.913 | 0.887 | 0.935 | 0.699 | 0.781 | 0.66 | 0.731 | 0.824 | 0.893 | 0.785 | 0.851 |
| TransFuse [11] | 0.87 | 0.92 | 0.897 | 0.942 | 0.706 | 0.781 | 0.663 | 0.737 | 0.826 | 0.894 | 0.792 | 0.855 |
| UACANet [63] | 0.859 | 0.912 | 0.88 | 0.926 | 0.678 | 0.751 | 0.678 | 0.751 | 0.849 | 0.91 | 0.789 | 0.85 |
| SANet [64] | 0.847 | 0.904 | 0.859 | 0.916 | 0.67 | 0.753 | 0.654 | 0.75 | 0.815 | 0.888 | 0.769 | 0.842 |
| MSNet [65] | 0.862 | 0.907 | 0.879 | 0.921 | 0.678 | 0.755 | 0.664 | 0.719 | 0.807 | 0.869 | 0.778 | 0.834 |
| SwinE-Net [66] | 0.87 | 0.92 | 0.892 | 0.938 | 0.725 | 0.804 | 0.687 | 0.758 | 0.842 | 0.906 | 0.803 | 0.865 |
| AMNet [67] | 0.865 | 0.912 | 0.888 | 0.936 | 0.69 | 0.762 | 0.679 | 0.756 | - | - | - | - |
5. Conclusions
- A fusion of different convolutional and transformer topologies can achieve state-of-the-art performance;
- Applying different approaches to the learning rate strategy is a feasible method to build a set of segmentation networks;
- A better way to add the transformers (Polyp-PVT and HSNet) in an ensemble is to use the proposed approach for creating the final segmentation mask.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Sauer, A.G.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [PubMed]
- Valderrama-Treviño, A.; Hazzel, E.; Flores, C.; Herrera, M. Colorectal cancer: A review. Artic. Int. J. Res. Med. Sci. 2017, 5, 4667–4676. [Google Scholar] [CrossRef]
- Wieszczy, P.; Regula, J.; Kaminski, M.F. Adenoma detection rate and risk of colorectal cancer. Best Pract. Res. Clin. Gastroenterol. 2017, 31, 441–446. [Google Scholar] [CrossRef]
- Silva, J.; Histace, A.; Romain, O.; Dray, X.; Granado, B. Toward embedded detection of polyps in WCE images for early diagnosis of colorectal cancer. Int. J. Comput. Assist. Radiol. Surg. 2014, 9, 283–293. [Google Scholar] [CrossRef]
- Mamonov, A.V.; Figueiredo, I.N.; Figueiredo, P.N.; Richard Tsai, Y.H. Automated polyp detection in colon capsule endoscopy. IEEE Trans. Med. Imaging 2014, 33, 1488–1502. [Google Scholar] [CrossRef] [PubMed]
- Tajbakhsh, N.; Gurudu, S.R.; Liang, J. Automated polyp detection in colonoscopy videos using shape and context information. IEEE Trans. Med. Imaging 2016, 35, 630–644. [Google Scholar] [CrossRef] [PubMed]
- Bernal, J.; Sánchez, J.; Vilariño, F. Towards automatic polyp detection with a polyp appearance model. Pattern Recognit. 2012, 45, 3166–3182. [Google Scholar] [CrossRef]
- Minaee, S.; Boykov, Y.; Porikli, F.; Plaza, A.; Kehtarnavaz, N.; Terzopoulos, D. Image Segmentation Using Deep Learning: A Survey. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44, 3523–3542. [Google Scholar] [CrossRef]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Johansen, D.; Lange, T.D.; Halvorsen, P.; Johansen, H.D. ResUNet++: An Advanced Architecture for Medical Image Segmentation. In Proceedings of the 2019 IEEE International Symposium on Multimedia (ISM), San Diego, CA, USA, 9–11 December 2019; pp. 225–230. [Google Scholar] [CrossRef]
- Fan, D.P.; Ji, G.P.; Zhou, T.; Chen, G.; Fu, H.; Shen, J.; Shao, L. PraNet: Parallel Reverse Attention Network for Polyp Segmentation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2020; Martel, A.L., Abolmaesumi, P., Stoyanov, D., Mateus, D., Zuluaga, M.A., Zhou, S.K., Racoceanu, D., Joskowicz, L., Eds.; Springer: Cham, Switzerland, 2020; pp. 263–273. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, H.; Hu, Q. TransFuse: Fusing Transformers and CNNs for Medical Image Segmentation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021; de Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Essert, C., Eds.; Springer: Cham, Switzerland, 2021; pp. 14–24. [Google Scholar] [CrossRef]
- Huang, C.H.; Wu, H.Y.; Lin, Y.L. HarDNet-MSEG: A Simple Encoder-Decoder Polyp Segmentation Neural Network that Achieves over 0.9 Mean Dice and 86 FPS. arXiv 2021, arXiv:2101.07172. [Google Scholar] [CrossRef]
- Dong, B.; Wang, W.; Fan, D.P.; Li, J.; Fu, H.; Shao, L. Polyp-PVT: Polyp Segmentation with Pyramid Vision Transformers. arXiv 2021, arXiv:2108.06932. [Google Scholar] [CrossRef]
- Zhang, W.; Fu, C.; Zheng, Y.; Zhang, F.; Zhao, Y.; Sham, C.W. HSNet: A hybrid semantic network for polyp segmentation. Comput. Biol. Med. 2022, 150, 106173. [Google Scholar] [CrossRef] [PubMed]
- Cornelio, C.; Donini, M.; Loreggia, A.; Pini, M.S.; Rossi, F. Voting with random classifiers (VORACE): Theoretical and experimental analysis. Auton. Agent 2021, 35, 2. [Google Scholar] [CrossRef]
- Pacal, I.; Karaboga, D.; Basturk, A.; Akay, B.; Nalbantoglu, U. A comprehensive review of deep learning in colon cancer. Comput. Biol. Med. 2020, 126, 104003. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhuang, P.; Sun, H.H.; Li, G.; Kwong, S.; Li, C. Underwater Image Enhancement via Minimal Color Loss and Locally Adaptive Contrast Enhancement. IEEE Trans. Image Process. 2022, 31, 3997–4010. [Google Scholar] [CrossRef]
- Nisha, J.; Gopi, V.P.; Palanisamy, P. Automated colorectal polyp detection based on image enhancement and dual-path CNN architecture. Biomed. Signal Process. Control 2022, 73, 103465. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Guo, X.; Zhang, N.; Guo, J.; Zhang, H.; Hao, Y.; Hang, J. Automated polyp segmentation for colonoscopy images: A method based on convolutional neural networks and ensemble learning. Med. Phys. 2019, 46, 5666–5676. [Google Scholar] [CrossRef]
- Ji, G.P.; Xiao, G.; Chou, Y.C.; Fan, D.P.; Zhao, K.; Chen, G.; Van Gool, L. Video polyp segmentation: A deep learning perspective. Mach. Intell. Res. 2022, 19, 531–549. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Singh, N.K.; Raza, K. Medical image generation using generative adversarial networks: A review. In Health Informatics: A Computational Perspective in Healthcare; Springer: Singapore, 2021; pp. 77–96. [Google Scholar]
- Dong, X.; Yu, Z.; Cao, W.; Shi, Y.; Ma, Q. A survey on ensemble learning. Front. Comput. Sci. 2020, 14, 241–258. [Google Scholar] [CrossRef]
- Tomar, N.K.; Ibtehaz, N.; Jha, D.; Halvorsen, P.; Ali, S. Improving Generalizability in Polyp Segmentation using Ensemble Convolutional Neural Network. In Proceedings of the 3rd International Workshop and Challenge on Computer Vision in Endoscopy (EndoCV 2021) Co-Located with the 18th IEEE International Symposium on Biomedical Imaging (ISBI 2021), Nice, France, 13 April 2021; Volume 2886, pp. 49–58. [Google Scholar]
- Tran, T.N.; Isensee, F.; Krämer, L.; Yamlahi, A.; Adler, T.; Godau, P.; Tizabi, M.; Maier-Hein, L. Heterogeneous Model Ensemble For Automatic Polyp Segmentation In Endoscopic Video Sequences. In Proceedings of the 4th International Workshop and Challenge on Computer Vision in Endoscopy (EndoCV 2022) Co-Located with the 19th IEEE International Symposium on Biomedical Imaging (ISBI 2022), Kolkata, India, 28–31 March 2022; Volume 3148, pp. 20–24. [Google Scholar]
- Lumini, A.; Nanni, L.; Maguolo, G. Deep ensembles based on Stochastic Activation Selection for Polyp Segmentation. arXiv 2021, arXiv:2104.00850. [Google Scholar]
- Lumini, A.; Nanni, L.; Maguolo, G. Deep Ensembles Based on Stochastic Activations for Semantic Segmentation. Signals 2021, 2, 820–833. [Google Scholar] [CrossRef]
- Nanni, L.; Cuza, D.; Lumini, A.; Loreggia, A.; Brahnam, S. Deep ensembles in bioimage segmentation. arXiv 2021, arXiv:2112.12955. [Google Scholar] [CrossRef]
- Nanni, L.; Cuza, D.; Lumini, A.; Brahnam, S. Data augmentation for deep ensembles in polyp segmentation. In Computational Intelligence Based Solutions for Vision Systems; IOP Publishing: Bristol, UK, 2022; pp. 8-1–8-22. [Google Scholar] [CrossRef]
- Kang, J.; Gwak, J. Ensemble of Instance Segmentation Models for Polyp Segmentation in Colonoscopy Images. IEEE Access 2019, 7, 26440–26447, Erratum in IEEE Access 2020, 8, 100010–100012. [Google Scholar] [CrossRef]
- Shrestha, S.; Khanal, B.; Ali, S. Ensemble U-Net Model for Efficient Polyp Segmentation. In Proceedings of the MediaEval 2020 Workshop, Online, 14–15 December 2020; Volume 2882. [Google Scholar]
- Hong, A.; Lee, G.; Lee, H.; Seo, J.; Yeo, D. Deep Learning Model Generalization with Ensemble in Endoscopic Images. In Proceedings of the 3rd International Workshop and Challenge on Computer Vision in Endoscopy (EndoCV 2021) Co-Located with the 18th IEEE International Symposium on Biomedical Imaging (ISBI 2021), Nice, France, 13 April 2021; Volume 2886, pp. 80–89. [Google Scholar]
- Nguyen, N.Q.; Lee, S.W. Robust Boundary Segmentation in Medical Images Using a Consecutive Deep Encoder-Decoder Network. IEEE Access 2019, 7, 33795–33808. [Google Scholar] [CrossRef]
- Thu Hong, L.T.; Chi Thanh, N.; Long, T.Q. Polyp Segmentation in Colonoscopy Images Using Ensembles of U-Nets with EfficientNet and Asymmetric Similarity Loss Function. In Proceedings of the 2020 RIVF International Conference on Computing and Communication Technologies (RIVF), Ho Chi Minh City, Vietnam, 14–15 October 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Thambawita, V.; Hicks, S.; Halvorsen, P.; Riegler, M. DivergentNets: Medical Image Segmentation by Network Ensemble. In Proceedings of the 3rd International Workshop and Challenge on Computer Vision in Endoscopy (EndoCV 2021) Co-Located with the 18th IEEE International Symposium on Biomedical Imaging (ISBI 2021), Nice, France, 13 April 2021; Volume 2886, pp. 27–38. [Google Scholar]
- Nanni, L.; Lumini, A.; Loreggia, A.; Formaggio, A.; Cuza, D. An Empirical Study on Ensemble of Segmentation Approaches. Signals 2022, 3, 341–358. [Google Scholar] [CrossRef]
- Nanni, L.; Cuza, D.; Lumini, A.; Loreggia, A.; Brahman, S. Polyp Segmentation with Deep Ensembles and Data Augmentation. In Artificial Intelligence and Machine Learning for Healthcare; Vol. 1: Image and Data Analytics; Springer International Publishing: Cham, Switzerland, 2023; pp. 133–153. [Google Scholar] [CrossRef]
- Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; Gil, D.; Rodríguez, C.; Vilariño, F. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput. Med. Imaging Graph. 2015, 43, 99–111. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. In Computer Vision—ECCV 2018; Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y., Eds.; Springer: Cham, Switzerland, 2018; pp. 833–851. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; Chaudhuri, K., Salakhutdinov, R., Eds.; PMLR: Cambridge, MA, USA; Volume 97, pp. 6105–6114. [Google Scholar]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Halvorsen, P.; de Lange, T.; Johansen, D.; Johansen, H.D. Kvasir-SEG: A Segmented Polyp Dataset. In MultiMedia Modeling; Ro, Y.M., Cheng, W.H., Kim, J., Chu, W.T., Cui, P., Choi, J.W., Hu, M.C., De Neve, W., Eds.; Springer: Cham, Switzerland, 2020; pp. 451–462. [Google Scholar] [CrossRef]
- Hosseinzadeh Kassani, S.; Hosseinzadeh Kassani, P.; Wesolowski, M.J.; Schneider, K.A.; Deters, R. Automatic Polyp Segmentation Using Convolutional Neural Networks. In Advances in Artificial Intelligence; Goutte, C., Zhu, X., Eds.; Springer: Cham, Switzerland, 2020; pp. 290–301. [Google Scholar] [CrossRef]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Stoyanov, D., Taylor, Z., Carneiro, G., Syeda-Mahmood, T., Martel, A., Maier-Hein, L., Tavares, J.M.R., Bradley, A., Papa, J.P., Belagiannis, V., et al., Eds.; Springer: Cham, Switzerland, 2018; pp. 3–11. [Google Scholar] [CrossRef]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature Pyramid Networks for Object Detection. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 936–944. [Google Scholar] [CrossRef]
- Vázquez, D.; Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; López, A.M.; Romero, A.; Drozdzal, M.; Courville, A. A benchmark for endoluminal scene segmentation of colonoscopy images. J. Healthc. Eng. 2017, 2017, 4037190. [Google Scholar] [CrossRef] [PubMed]
- Baheti, B.; Innani, S.; Gajre, S.; Talbar, S. Eff-UNet: A Novel Architecture for Semantic Segmentation in Unstructured Environment. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Seattle, WA, USA, 14–19 June 2020; pp. 1473–1481. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Tao, A.; Sapra, K.; Catanzaro, B. Hierarchical Multi-Scale Attention for Semantic Segmentation. arXiv 2020, arXiv:2005.10821. [Google Scholar] [CrossRef]
- Jadon, S. A survey of loss functions for semantic segmentation. arXiv 2020, arXiv:2006.14822. [Google Scholar]
- Sudre, C.H.; Li, W.; Vercauteren, T.; Ourselin, S.; Cardoso, M.J. Generalised Dice Overlap as a Deep Learning Loss Function for Highly Unbalanced Segmentations. arXiv 2017, arXiv:1707.03237. [Google Scholar]
- Rahman, M.A.; Wang, Y. Optimizing intersection-over-union in deep neural networks for image segmentation. In International Symposium on Visual Computing; Springer: Cham, Switzerland, 2016; pp. 234–244. [Google Scholar]
- Cho, Y.J. Weighted Intersection over Union (wIoU): A New Evaluation Metric for Image Segmentation. arXiv 2021, arXiv:2107.09858. [Google Scholar]
- Aurelio, Y.S.; de Almeida, G.M.; de Castro, C.L.; Braga, A.P. Learning from imbalanced datasets with weighted cross-entropy function. Neural Process. Lett. 2019, 50, 1937–1949. [Google Scholar] [CrossRef]
- Pogorelov, K.; Randel, K.R.; Griwodz, C.; Eskeland, S.L.; de Lange, T.; Johansen, D.; Spampinato, C.; Dang-Nguyen, D.T.; Lux, M.; Schmidt, P.T.; et al. Kvasir: A multi-class image dataset for computer aided gastrointestinal disease detection. In Proceedings of the 8th ACM on Multimedia Systems Conference, Taipei, Taiwan, 20–23 June 2017; pp. 164–169. [Google Scholar]
- Shen, T.; Xu, H. Medical image segmentation based on Transformer and HarDNet structures. IEEE Access 2023, 11, 16621–16630. [Google Scholar] [CrossRef]
- Li, W.; Zhao, Y.; Li, F.; Wang, L. MIA-Net: Multi-information aggregation network combining transformers and convolutional feature learning for polyp segmentation. Knowl.-Based Syst. 2022, 247, 108824. [Google Scholar] [CrossRef]
- Wu, Y.H.; Liu, Y.; Zhan, X.; Cheng, M.M. P2T: Pyramid Pooling Transformer for Scene Understanding. IEEE Trans. Pattern Anal. Mach. Intell. 2022; early access. [Google Scholar] [CrossRef]
- Liu, F.; Hua, Z.; Li, J.; Fan, L. DBMF: Dual Branch Multiscale Feature Fusion Network for polyp segmentation. Comput. Biol. Med. 2022, 151, 106304. [Google Scholar] [CrossRef]
- Zheng, S.; Lu, J.; Zhao, H.; Zhu, X.; Luo, Z.; Wang, Y.; Fu, Y.; Feng, J.; Xiang, T.; Torr, P.H.; et al. Rethinking Semantic Segmentation from a Sequence-to-Sequence Perspective with Transformers. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021; pp. 6877–6886. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar] [CrossRef]
- Kim, T.; Lee, H.; Kim, D. UACANet: Uncertainty Augmented Context Attention for Polyp Segmentation. In Proceedings of the 29th ACM International Conference on Multimedia—MM ’21, Virtual, China, 20–24 October 2021; Association for Computing Machinery: New York, NY, USA, 2021; pp. 2167–2175. [Google Scholar] [CrossRef]
- Wei, J.; Hu, Y.; Zhang, R.; Li, Z.; Zhou, S.K.; Cui, S. Shallow Attention Network for Polyp Segmentation. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2021; Volume 12901. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, L.; Lu, H. Automatic Polyp Segmentation via Multi-scale Subtraction Network. arXiv 2021, arXiv:2108.05082. [Google Scholar] [CrossRef]
- Park, K.B.; Lee, J.Y. SwinE-Net: Hybrid deep learning approach to novel polyp segmentation using convolutional neural network and Swin Transformer. J. Comput. Des. Eng. 2022, 9, 616–632. [Google Scholar] [CrossRef]
- Song, P.; Li, J.; Fan, H. Attention based multi-scale parallel network for polyp segmentation. Comput. Biol. Med. 2022, 146, 105476. [Google Scholar] [CrossRef]


| OPT | DA | LR | Kvasir | ClinDB | ColDB | ETIS | CVC-T | Avg |
|---|---|---|---|---|---|---|---|---|
| SGD | 1 | a | 0.893 | 0.875 | 0.745 | 0.667 | 0.882 | 0.812 |
| b | 0.862 | 0.794 | 0.705 | 0.628 | 0.873 | 0.772 | ||
| SGD | 2 | a | 0.914 | 0.944 | 0.747 | 0.727 | 0.901 | 0.847 |
| b | 0.871 | 0.875 | 0.702 | 0.650 | 0.885 | 0.797 | ||
| Adam | 1 | a | 0.906 | 0.924 | 0.751 | 0.716 | 0.903 | 0.840 |
| b | 0.910 | 0.910 | 0.748 | 0.702 | 0.884 | 0.831 | ||
| Adam | 2 | a | 0.896 | 0.927 | 0.778 | 0.774 | 0.893 | 0.854 |
| b | 0.895 | 0.916 | 0.758 | 0.716 | 0.885 | 0.834 | ||
| SGD + Adam | 1 + 2 | a | 0.918 | 0.947 | 0.778 | 0.756 | 0.909 | 0.862 |
| b | 0.907 | 0.928 | 0.775 | 0.770 | 0.899 | 0.856 | ||
| a + b | 0.915 | 0.931 | 0.785 | 0.781 | 0.904 | 0.863 |
| DA | LR | SM | Kvasir | ClinDB | ColDB | ETIS | CVC-T | Avg |
|---|---|---|---|---|---|---|---|---|
| 1 | a | No | 0.911 | 0.926 | 0.788 | 0.773 | 0.871 | 0.854 |
| 1 | b | No | 0.924 | 0.924 | 0.793 | 0.800 | 0.877 | 0.864 |
| 2 | a | No | 0.910 | 0.923 | 0.804 | 0.749 | 0.891 | 0.855 |
| 2 | b | No | 0.917 | 0.921 | 0.794 | 0.763 | 0.891 | 0.857 |
| 1 + 2 | a | No | 0.918 | 0.926 | 0.803 | 0.755 | 0.873 | 0.855 |
| 1 + 2 | a | Yes | 0.919 | 0.930 | 0.809 | 0.765 | 0.884 | 0.861 |
| 1 + 2 | b | No | 0.931 | 0.920 | 0.792 | 0.776 | 0.876 | 0.859 |
| 1 + 2 | b | Yes | 0.931 | 0.921 | 0.798 | 0.791 | 0.882 | 0.865 |
| 1 + 2 | a + b | No | 0.926 | 0.932 | 0.821 | 0.800 | 0.891 | 0.874 |
| 1 + 2 | a + b | Yes | 0.926 | 0.933 | 0.824 | 0.808 | 0.895 | 0.877 |
| DA | LR | SM | Kvasir | ClinDB | ColDB | ETIS | CVC-T | Avg |
|---|---|---|---|---|---|---|---|---|
| 1 | a | No | 0.923 | 0.921 | 0.789 | 0.733 | 0.898 | 0.853 |
| 1 | b | No | 0.930 | 0.934 | 0.821 | 0.783 | 0.901 | 0.873 |
| 2 | a | No | 0.909 | 0.944 | 0.806 | 0.750 | 0.901 | 0.862 |
| 2 | b | No | 0.913 | 0.947 | 0.816 | 0.783 | 0.903 | 0.872 |
| 1 + 2 | a | No | 0.923 | 0.925 | 0.794 | 0.703 | 0.891 | 0.847 |
| 1 + 2 | a | Yes | 0.922 | 0.929 | 0.808 | 0.746 | 0.903 | 0.862 |
| 1 + 2 | b | No | 0.931 | 0.938 | 0.821 | 0.775 | 0.896 | 0.872 |
| 1 + 2 | b | Yes | 0.929 | 0.943 | 0.822 | 0.783 | 0.903 | 0.876 |
| 1 + 2 | a + b | No | 0.928 | 0.943 | 0.829 | 0.779 | 0.902 | 0.876 |
| 1 + 2 | a + b | Yes | 0.928 | 0.945 | 0.828 | 0.791 | 0.905 | 0.879 |
| Method | Minmax | Avgmax | Avgmin | Maxmin | |||
|---|---|---|---|---|---|---|---|
| HarDNet-MSEG | P | 2.51 | 71.84 | −6.53 | −11.45 | 2.57% | 2.57% |
| Polyp-PVT | 26.22 | 70.15 | −21.06 | −30.04 | 1.62% | 1.09% | |
| 19.53 | 38.55 | −20.61 | −29.47 | ||||
| HSNet | 24.21 | 67.81 | −38.49 | −69.06 | 2.92% | 0.85% | |
| 33.19 | 87.04 | −42.63 | −84.26 | ||||
| 31.31 | 122.01 | −63.13 | −123.45 | ||||
| 36.47 | 160.11 | −95.23 | −165.40 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nanni, L.; Fantozzi, C.; Loreggia, A.; Lumini, A. Ensembles of Convolutional Neural Networks and Transformers for Polyp Segmentation. Sensors 2023, 23, 4688. https://doi.org/10.3390/s23104688
Nanni L, Fantozzi C, Loreggia A, Lumini A. Ensembles of Convolutional Neural Networks and Transformers for Polyp Segmentation. Sensors. 2023; 23(10):4688. https://doi.org/10.3390/s23104688
Chicago/Turabian StyleNanni, Loris, Carlo Fantozzi, Andrea Loreggia, and Alessandra Lumini. 2023. "Ensembles of Convolutional Neural Networks and Transformers for Polyp Segmentation" Sensors 23, no. 10: 4688. https://doi.org/10.3390/s23104688
APA StyleNanni, L., Fantozzi, C., Loreggia, A., & Lumini, A. (2023). Ensembles of Convolutional Neural Networks and Transformers for Polyp Segmentation. Sensors, 23(10), 4688. https://doi.org/10.3390/s23104688








