Elimination of Leaf Angle Impacts on Plant Reflectance Spectra Using Fusion of Hyperspectral Images and 3D Point Clouds
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Samples
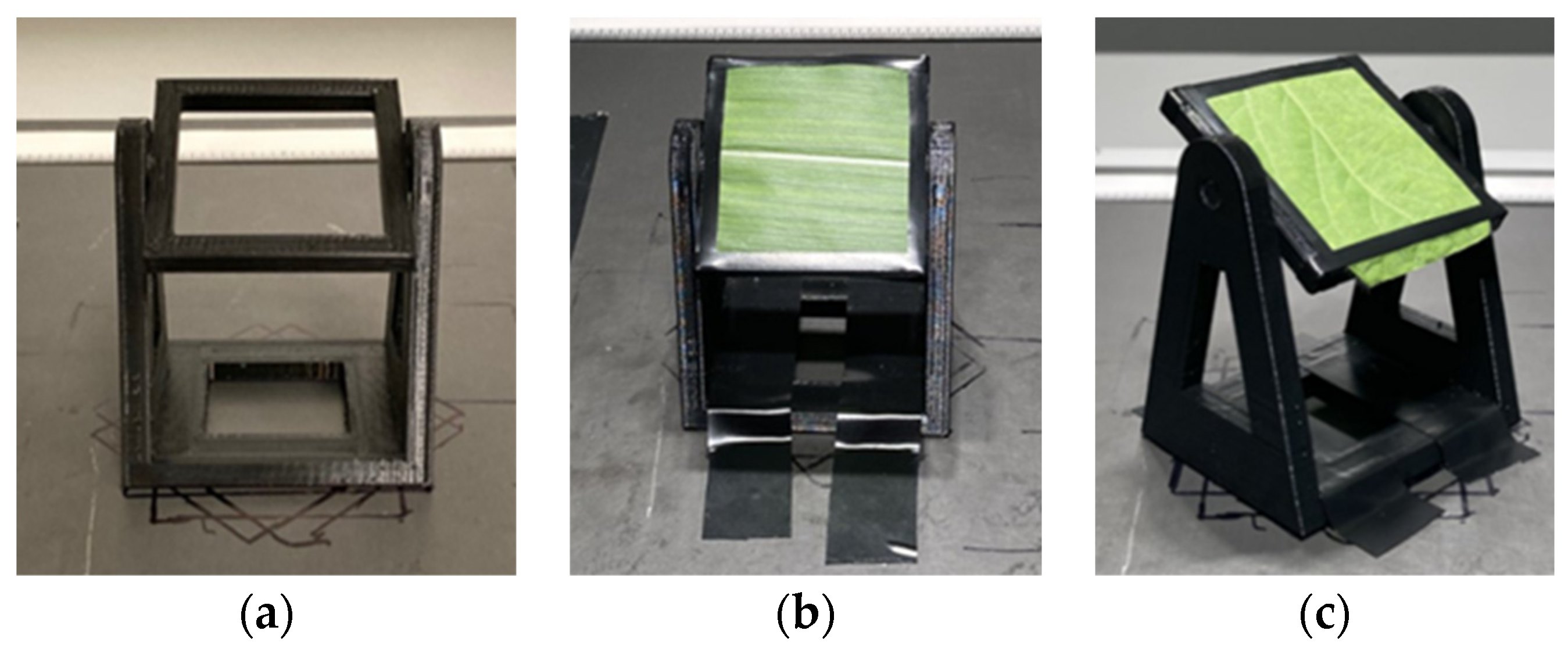
2.2. Image Acquisition of Leaf Samples at Different Tilt Angles and Orientations
2.3. Image Processing of Leaf Samples from Different Tilt Angles and Orientations
2.4. Fusion of Hyperspectral Images and 3D Point Clouds
2.5. Application of 3D Calibration on Plant Leaves
3. Results and Discussion
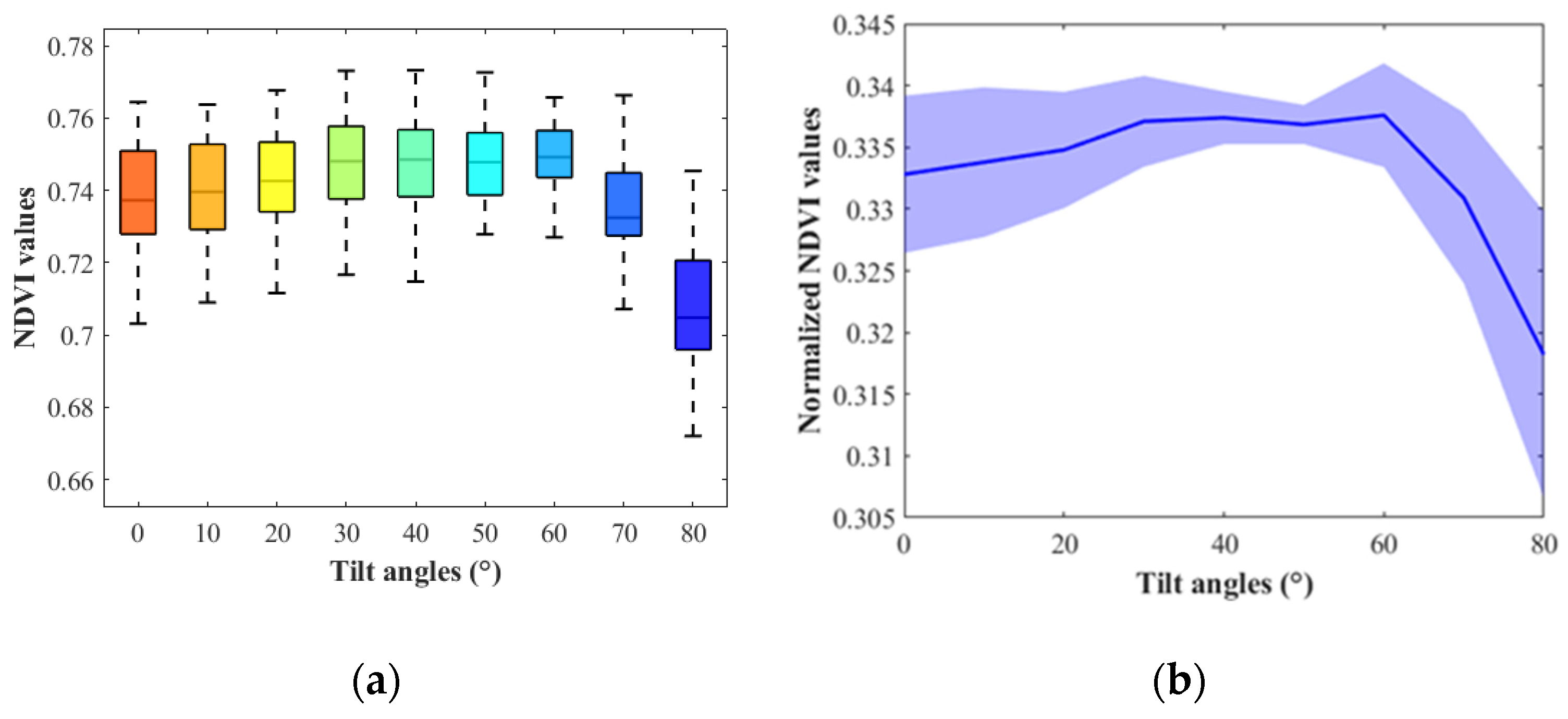
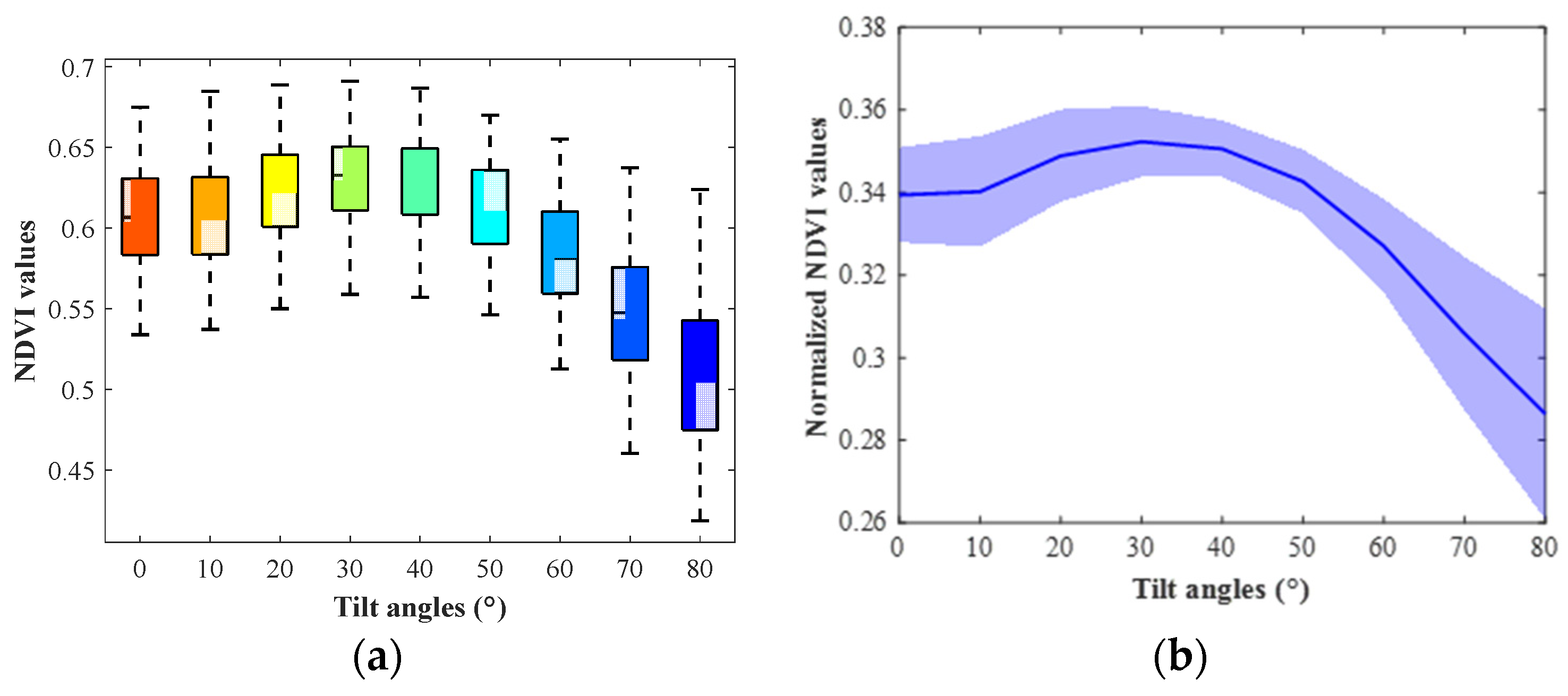
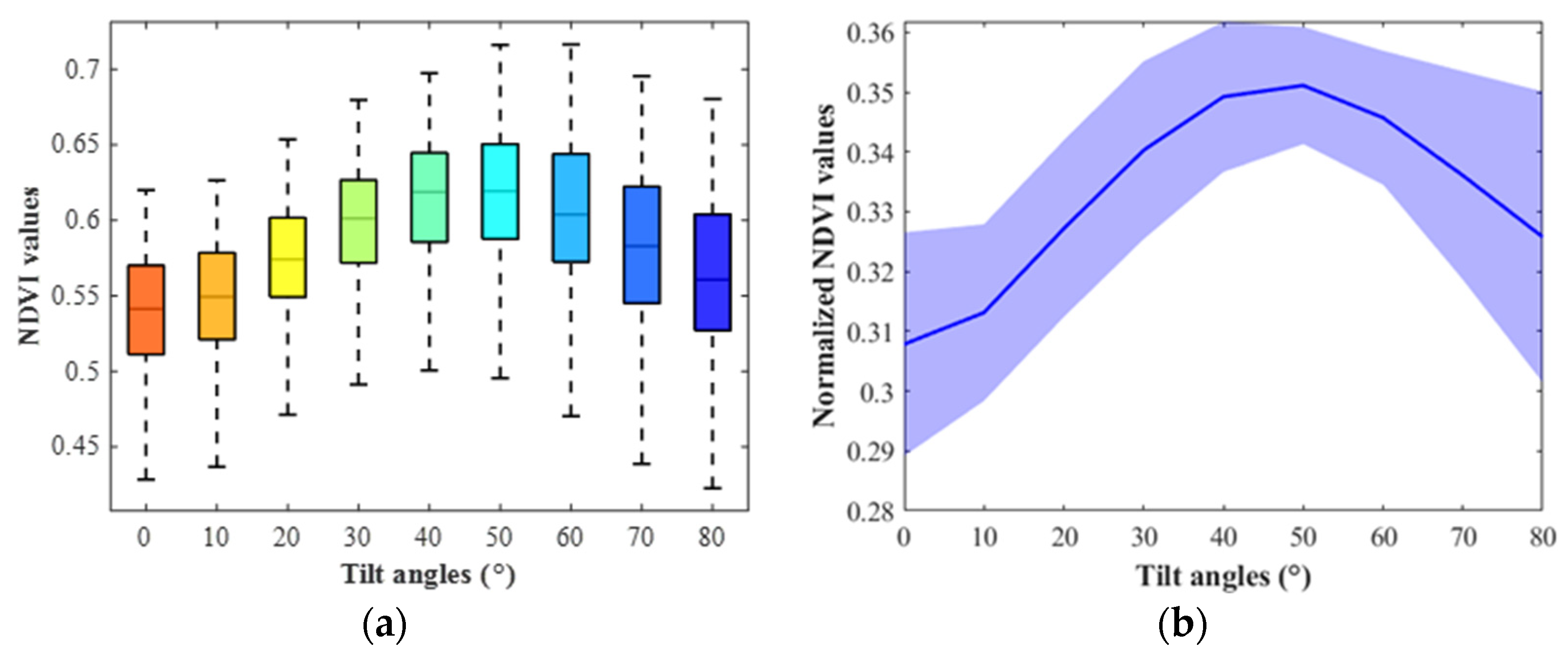
3.1. Soybean NDVI Changing Trend
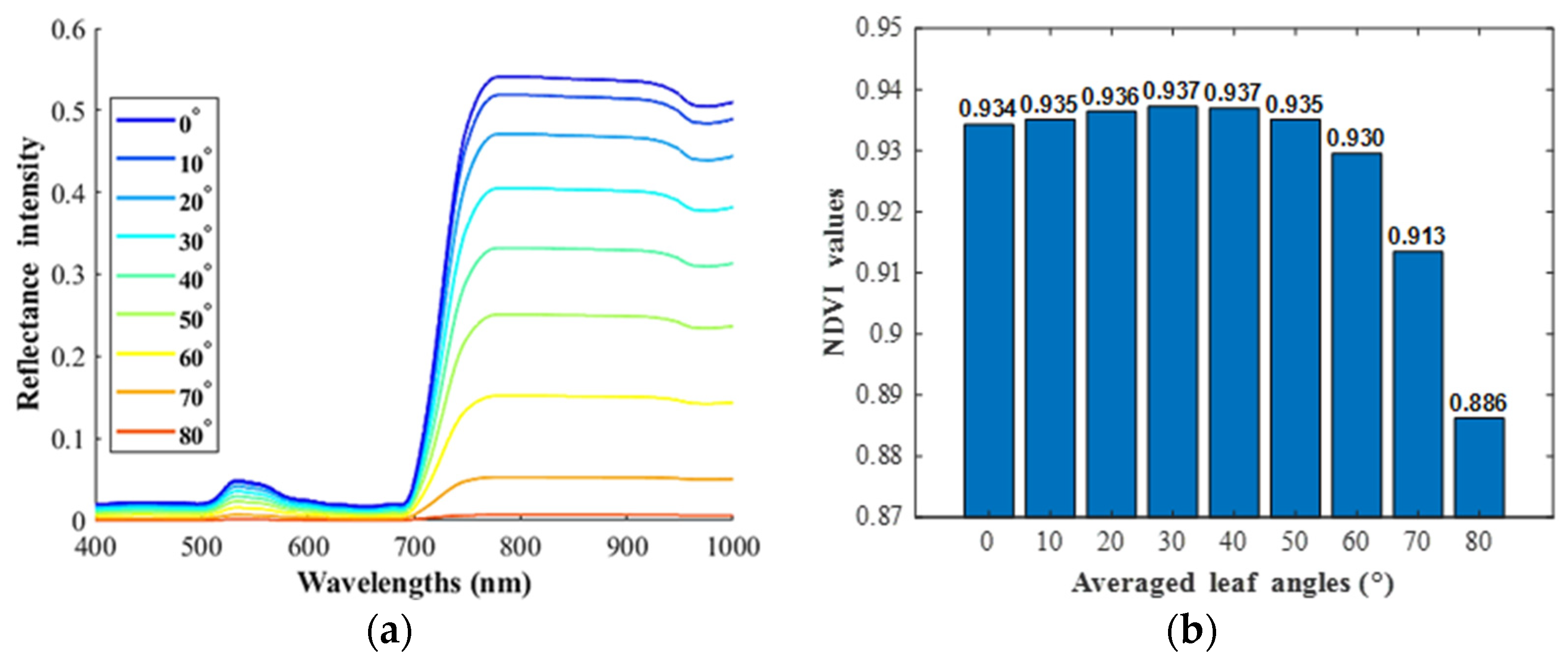
3.2. Corn NDVI Changing Trend and Comparison with PROSAIL Outputs
3.3. SVR Modelling for Soybean and Corn
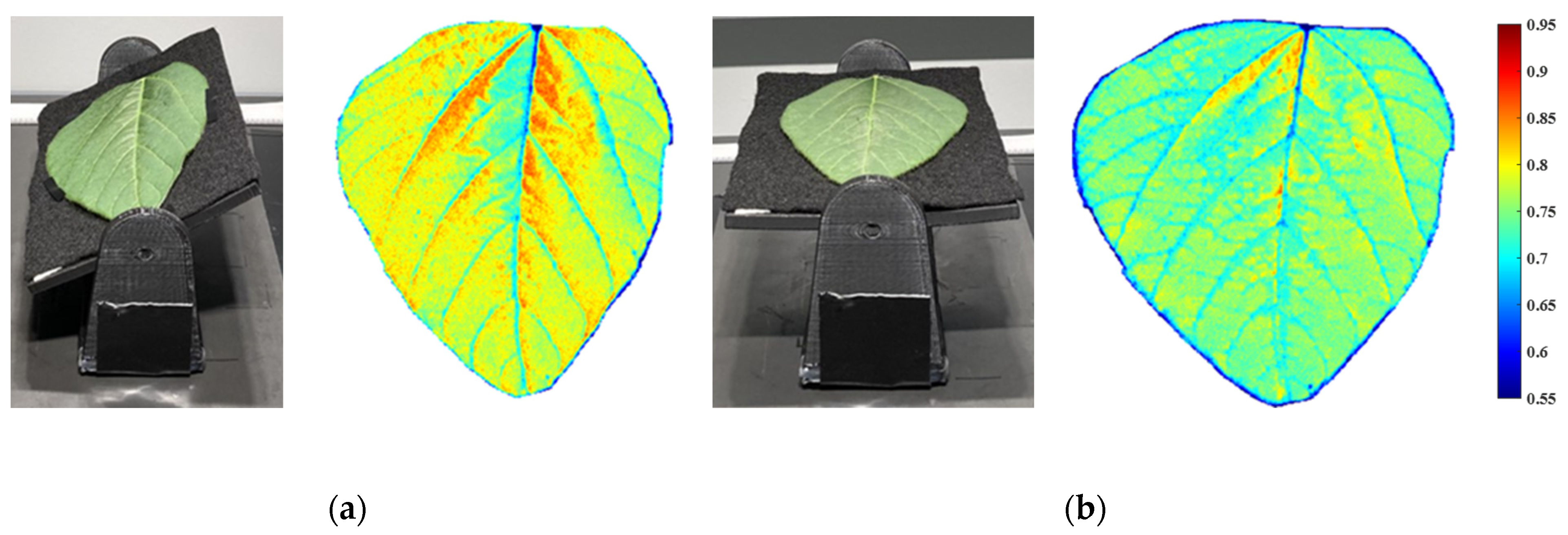
3.4. Fusion of Hyperspectral Images and 3D Point Clouds
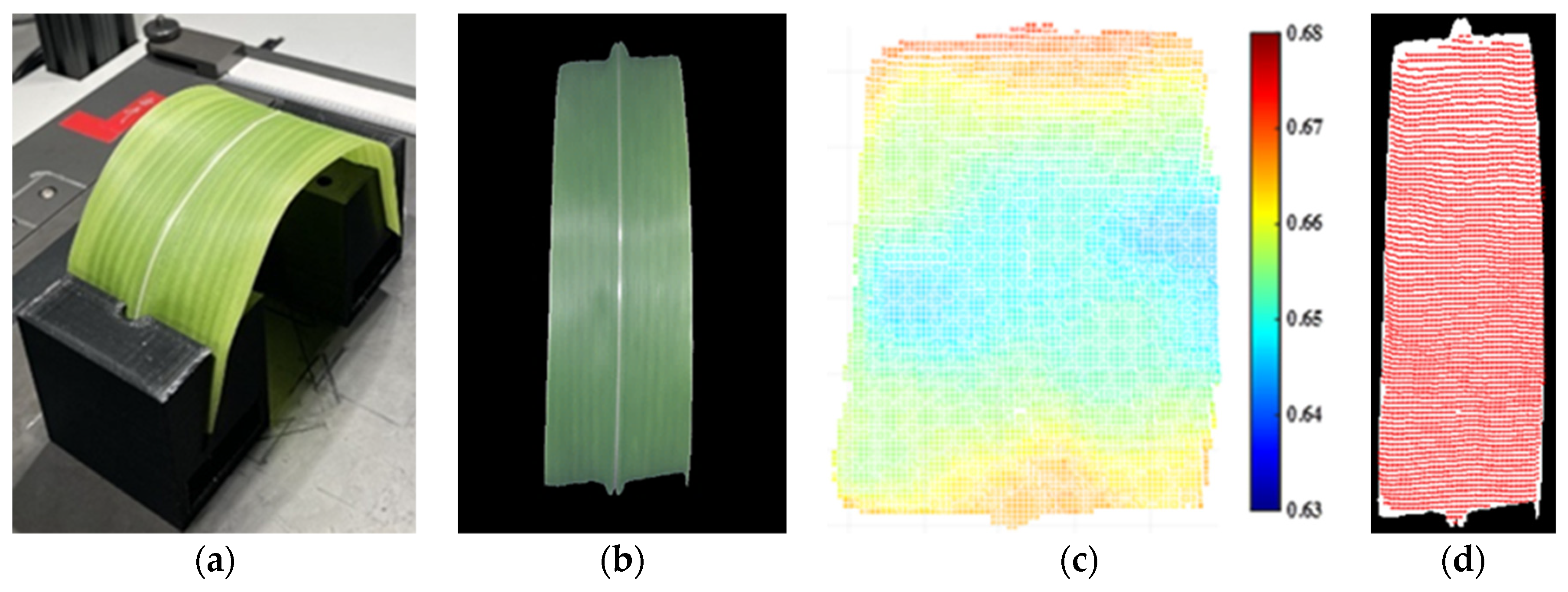
3.5. Application of 3D Calibration on Soybean and Corn Leaves
3.5.1. Soybean Leaf Calibration
3.5.2. Corn Leaf Calibration
3.5.3. Summary of 3D Calibration on Soybean and Corn Leaves
3.5.4. Future Opportunities and Limitations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dhondt, S.; Wuyts, N.; Inzé, D. Cell to whole-plant phenotyping: The best is yet to come. Trends Plant Sci. 2013, 18, 428–439. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Q.; Huang, D. A review of imaging techniques for plant phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef]
- Zhou, J.; Khot, L.R.; Boydston, R.A.; Miklas, P.N.; Porter, L. Low altitude remote sensing technologies for crop stress monitoring: A case study on spatial and temporal monitoring of irrigated pinto bean. Precis. Agric. 2018, 19, 555–569. [Google Scholar] [CrossRef]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef]
- Xiong, X.; Yu, L.; Yang, W.; Liu, M.; Jiang, N.; Wu, D.; Chen, G.; Xiong, L.; Liu, K.; Liu, Q. A high-throughput stereo-imaging system for quantifying rape leaf traits during the seedling stage. Plant Methods 2017, 13, 7. [Google Scholar] [CrossRef]
- Zhao, Y.R.; Yu, K.Q.; He, Y. Hyperspectral imaging coupled with random frog and calibration models for assessment of total soluble solids in mulberries. J. Anal. Methods Chem. 2015, 2015, 343782. [Google Scholar] [CrossRef]
- Yu, K.Q.; Zhao, Y.R.; Li, X.L.; Shao, Y.N.; Liu, F.; He, Y. Hyperspectral imaging for mapping of total nitrogen spatial distribution in pepper plant. PLoS ONE 2014, 9, e116205. [Google Scholar] [CrossRef]
- Behmann, J.; Mahlein, A.K.; Paulus, S.; Dupuis, J.; Kuhlmann, H.; Oerke, E.C.; Plümer, L. Generation and application of hyperspectral 3D plant models: Methods and challenges. Mach. Vis. Appl. 2016, 27, 611–624. [Google Scholar] [CrossRef]
- Zou, X.; Haikarainen, I.; Haikarainen, I.P.; Mäkelä, P.; Mõttus, M.; Pellikka, P. Effects of crop leaf angle on LAI-sensitive narrow-band vegetation indices derived from imaging spectroscopy. Appl. Sci. 2018, 8, 1435. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Verhoef, W.; Baret, F.; Bacour, C.; Zarco-Tejada, P.J.; Asner, G.P.; François, C.; Ustin, S.L. PROSPECT + SAIL models: A review of use for vegetation characterization. Remote Sens. Environ. 2009, 113, S56–S66. [Google Scholar] [CrossRef]
- Salas, E.A.L.; Henebry, G.M. A new approach for the analysis of hyperspectral data: Theory and sensitivity analysis of the Moment Distance Method. Remote Sens. 2013, 6, 20–41. [Google Scholar] [CrossRef]
- Zhang, L.; Maki, H.; Ma, D.; Sánchez-Gallego, J.A.; Mickelbart, M.V.; Wang, L.; Rehman, T.U.; Jin, J. Optimized angles of the swing hyperspectral imaging system for single corn plant. Comput. Electron. Agric. 2019, 156, 349–359. [Google Scholar] [CrossRef]
- Duncan, W.G. Leaf angles, leaf area, and canopy photosynthesis. Crop Sci. 1971, 11, 482–485. [Google Scholar] [CrossRef]
- Maddonni, G.A.; Otegui, M.E. Leaf area, light interception, and crop development in maize. Field Crops Res. 1996, 48, 81–87. [Google Scholar] [CrossRef]
- Gou, L.; Xue, J.; Qi, B.; Ma, B.; Zhang, W. Morphological variation of maize cultivars in response to elevated plant densities. Agron. J. 2017, 109, 1443–1453. [Google Scholar] [CrossRef]
- Hirano, K.; Kawamura, M.; Araki-Nakamura, S.; Fujimoto, H.; Ohmae-Shinohara, K.; Yamaguchi, M.; Fujii, A.; Sasaki, H.; Kasuga, S.; Sazuka, T. Sorghum DW1 positively regulates brassinosteroid signaling by inhibiting the nuclear localization of BRASSINOSTEROID INSENSITIVE 2. Sci. Rep. 2017, 7, 126. [Google Scholar] [CrossRef]
- Zhang, W.; Fu, L.; Men, C.; Yu, J.; Yao, J.; Sheng, J.; Xu, Y.; Wang, Z.; Liu, L.; Yang, J.; et al. Response of brassinosteroids to nitrogen rates and their regulation on rice spikelet degeneration during meiosis. Food Energy Secur. 2020, 9, e201. [Google Scholar] [CrossRef]
- Kao, W.Y.; Forseth, I.N. Diurnal leaf movement, chlorophyll fluorescence and carbon assimilation in soybean grown under different nitrogen and water availabilities. Plant Cell Environ. 1992, 15, 703–710. [Google Scholar] [CrossRef]
- Rosa, L.M.; Dillenburg, L.R.; Forseth, I.N. Responses of soybean leaf angle, photosynthesis and stomatal conductance to leaf and soil water potential. Ann. Bot. 1991, 67, 51–58. [Google Scholar] [CrossRef]
- Bousquet, L.; Lachérade, S.; Jacquemoud, S.; Moya, I. Leaf BRDF measurements and model for specular and diffuse components differentiation. Remote Sens. Environ. 2005, 98, 201–211. [Google Scholar] [CrossRef]
- Granier, C.; Aguirrezabal, L.; Chenu, K.; Cookson, S.J.; Dauzat, M.; Hamard, P.; Thioux, J.-J.; Rolland, G.; Bouchier-Combaud, S.; Lebaudy, A.; et al. PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol. 2006, 169, 623–635. [Google Scholar] [CrossRef]
- An, N.; Palmer, C.M.; Baker, R.L.; Markelz, R.C.; Ta, J.; Covington, M.F.; Maloof, J.N.; Welch, S.M.; Weinig, C. Plant high-throughput phenotyping using photogrammetry and imaging techniques to measure leaf length and rosette area. Comput. Electron. Agric. 2016, 127, 376–394. [Google Scholar] [CrossRef]
- Schaepman-Strub, G.; Schaepman, M.E.; Painter, T.H.; Dangel, S.; Martonchik, J.V. Reflectance quantities in optical remote sensing—Definitions and case studies. Remote Sens. Environ. 2006, 103, 27–42. [Google Scholar] [CrossRef]
- Mishra, P.; Lohumi, S.; Khan, H.A.; Nordon, A. Close-range hyperspectral imaging of whole plants for digital phenotyping: Recent applications and illumination correction approaches. Comput. Electron. Agric. 2020, 178, 105780. [Google Scholar] [CrossRef]
- Jay, S.; Bendoula, R.; Hadoux, X.; Féret, J.B.; Gorretta, N. A physically-based model for retrieving foliar biochemistry and leaf orientation using close-range imaging spectroscopy. Remote Sens. Environ. 2016, 177, 220–236. [Google Scholar] [CrossRef]
- Wiegand, C.L.; Richardson, A.J.; Escobar, D.E.; Gerbermann, A.H. Vegetation indices in crop assessments. Remote Sens. Environ. 1991, 35, 105–119. [Google Scholar] [CrossRef]
- Bannari, A.; Morin, D.; Bonn, F.; Huete, A. A review of vegetation indices. Remote Sens. Rev. 1995, 13, 95–120. [Google Scholar] [CrossRef]
- Behmann, J.; Acebron, K.; Emin, D.; Bennertz, S.; Matsubara, S.; Thomas, S.; Bohnenkamp, D.; Kuska, M.T.; Jussila, J.; Salo, H.; et al. Specim IQ: Evaluation of a new, miniaturized handheld hyperspectral camera and its application for plant phenotyping and disease detection. Sensors 2018, 18, 441. [Google Scholar] [CrossRef]
- Cherkassky, V.; Ma, Y. Practical selection of SVM parameters and noise estimation for SVM regression. Neural Netw. 2004, 17, 113–126. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; Hoefsloot, H.C.; Smit, S.; Vis, D.J.; Smilde, A.K.; van Velzen, E.J.; van Duijnhoven, J.P.M.; van Dorsten, F.A. Assessment of PLSDA cross validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef]
- Gupta, R.; Hartley, R.I. Linear pushbroom cameras. IEEE Trans. Pattern Anal. Mach. Intell. 1997, 19, 963–975. [Google Scholar] [CrossRef]
- Buyer, A.; Schubert, W. Extraction of discontinuity orientations in point clouds. In Proceedings of the ISRM International Symposium—EUROCK 2016, Ürgüp, Turkey, 29–31 August 2016. [Google Scholar]
- Ma, D.; Rehman, T.U.; Zhang, L.; Maki, H.; Tuinstra, M.R.; Jin, J. Modeling of diurnal changing patterns in airborne crop remote sensing images. Remote Sens. 2021, 13, 1719. [Google Scholar] [CrossRef]
- Ma, D.; Rehman, T.U.; Zhang, L.; Maki, H.; Tuinstra, M.R.; Jin, J. Modeling of Environmental Impacts on Aerial Hyperspectral Images for Corn Plant Phenotyping. Remote Sens. 2021, 13, 2520. [Google Scholar] [CrossRef]
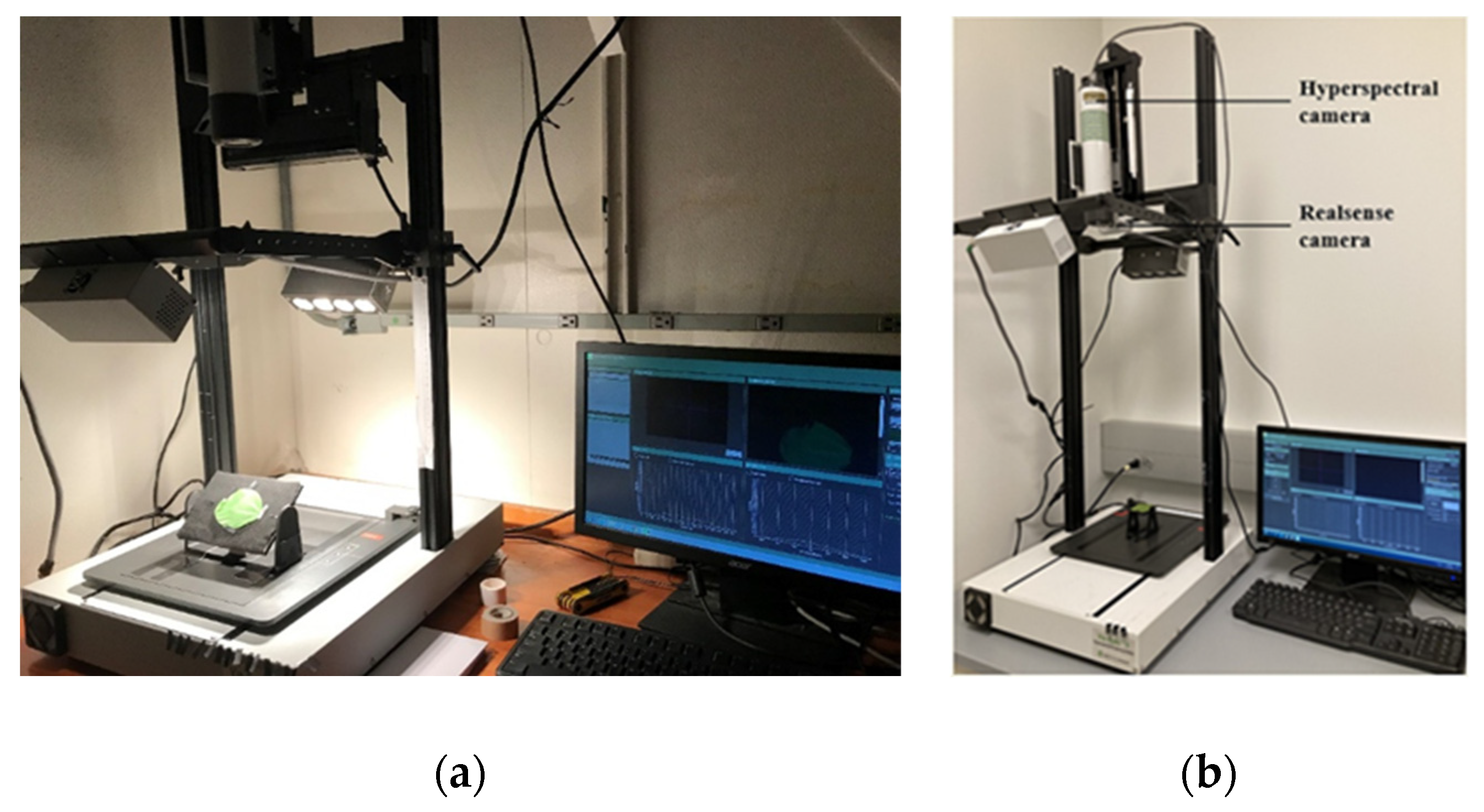





| Camera Types | Parameters | Corresponding Settings |
|---|---|---|
| Hyperspectral camera | Camera model | acA 780–75 gm |
| Spectrograph | Specim® V10H | |
| Camera sensor type | Progressive scan CCD | |
| Camera gain mode | 12-bit | |
| Camera focal length (mm) | Default lens focal length | |
| Camera spectral range (nm) | 362–1043 | |
| Camera field of view (°) | 22.1 | |
| Image spectral dimension (bands) | 582 | |
| Camera numerical aperture | F/1.4 | |
| Image spatial resolution (pixels) | 782 | |
| Image integration time (ms) | 16 | |
| Scanning frame rate (fps) | 60 | |
| Scanning speed (mm/s) | 6.35 | |
| RealSense camera | Depth module | Intel® RealSense™ Depth module D435 |
| Baseline (mm) | 50 | |
| Left/right imagers type | Wide | |
| Depth FOV HD (°) | H:87 ± 3/V:58 ± 1/D:95 ± 3 | |
| Depth FOV VGA (°) | H:75 ± 3/V:62 ± 1/D:89 ± 3 | |
| IR projector | Wide | |
| IR projector FOV (°) | H:90/V:63/D:99 | |
| Color sensor | OV2740 | |
| Color camera FOV (°) | H:69 ± 1/V:42 ± 1/D:77 ± 1 | |
| Image dimensions | 720 × 1280 × 3 |
| Source | SS | df | MS | F | Prob > F |
|---|---|---|---|---|---|
| Columns | 0.8195 | 8 | 0.1024 | 82.8310 | 0 |
| Rows | 0.0394 | 7 | 0.0056 | 4.5568 | 6.006 × 10−5 |
| Interaction | 0.0535 | 56 | 9.5507 × 10−4 | 0.7722 | 0.8846 |
| Error | 0.6233 | 504 | 0.0012 | ||
| Total | 1.5358 | 575 |
| Source | SS | df | MS | F | Prob > F |
|---|---|---|---|---|---|
| Columns | 1.1583 | 8 | 0.1448 | 74.1286 | 0 |
| Rows | 0.5349 | 23 | 0.0764 | 39.1212 | 0 |
| Interaction | 0.1927 | 184 | 0.0034 | 1.7621 | 5.0877 × 10−4 |
| Error | 3.2345 | 1656 | 0.0020 | ||
| Total | 5.1204 | 1727 |
| Original Coordinates (Pixels) | Estimated Coordinates (Pixels) | |||
|---|---|---|---|---|
| Points | Rows | Columns | Rows | Columns |
| 1 | 435 | 122 | 434 | 121 |
| 2 | 435 | 393 | 437 | 394 |
| 3 | 437 | 667 | 436 | 666 |
| 4 | 1097 | 667 | 1100 | 668 |
| 5 | 1755 | 667 | 1753 | 666 |
| 6 | 1755 | 429 | 1757 | 430 |
| 7 | 1752 | 119 | 1751 | 119 |
| 8 | 1092 | 122 | 1094 | 122 |
| 9 | 962 | 312 | 961 | 311 |
| 10 | 964 | 484 | 963 | 483 |
| 11 | 1251 | 479 | 1250 | 478 |
| 12 | 1246 | 312 | 1245 | 311 |
| 13 | 1152 | 395 | 1152 | 397 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Jin, J.; Wang, L.; Rehman, T.U.; Gee, M.T., Jr. Elimination of Leaf Angle Impacts on Plant Reflectance Spectra Using Fusion of Hyperspectral Images and 3D Point Clouds. Sensors 2023, 23, 44. https://doi.org/10.3390/s23010044
Zhang L, Jin J, Wang L, Rehman TU, Gee MT Jr. Elimination of Leaf Angle Impacts on Plant Reflectance Spectra Using Fusion of Hyperspectral Images and 3D Point Clouds. Sensors. 2023; 23(1):44. https://doi.org/10.3390/s23010044
Chicago/Turabian StyleZhang, Libo, Jian Jin, Liangju Wang, Tanzeel U. Rehman, and Mark T. Gee, Jr. 2023. "Elimination of Leaf Angle Impacts on Plant Reflectance Spectra Using Fusion of Hyperspectral Images and 3D Point Clouds" Sensors 23, no. 1: 44. https://doi.org/10.3390/s23010044
APA StyleZhang, L., Jin, J., Wang, L., Rehman, T. U., & Gee, M. T., Jr. (2023). Elimination of Leaf Angle Impacts on Plant Reflectance Spectra Using Fusion of Hyperspectral Images and 3D Point Clouds. Sensors, 23(1), 44. https://doi.org/10.3390/s23010044






