Fetal Electrocardiogram Extraction from the Mother’s Abdominal Signal Using the Ensemble Kalman Filter
Abstract
:1. Introduction
2. Theory and Methods
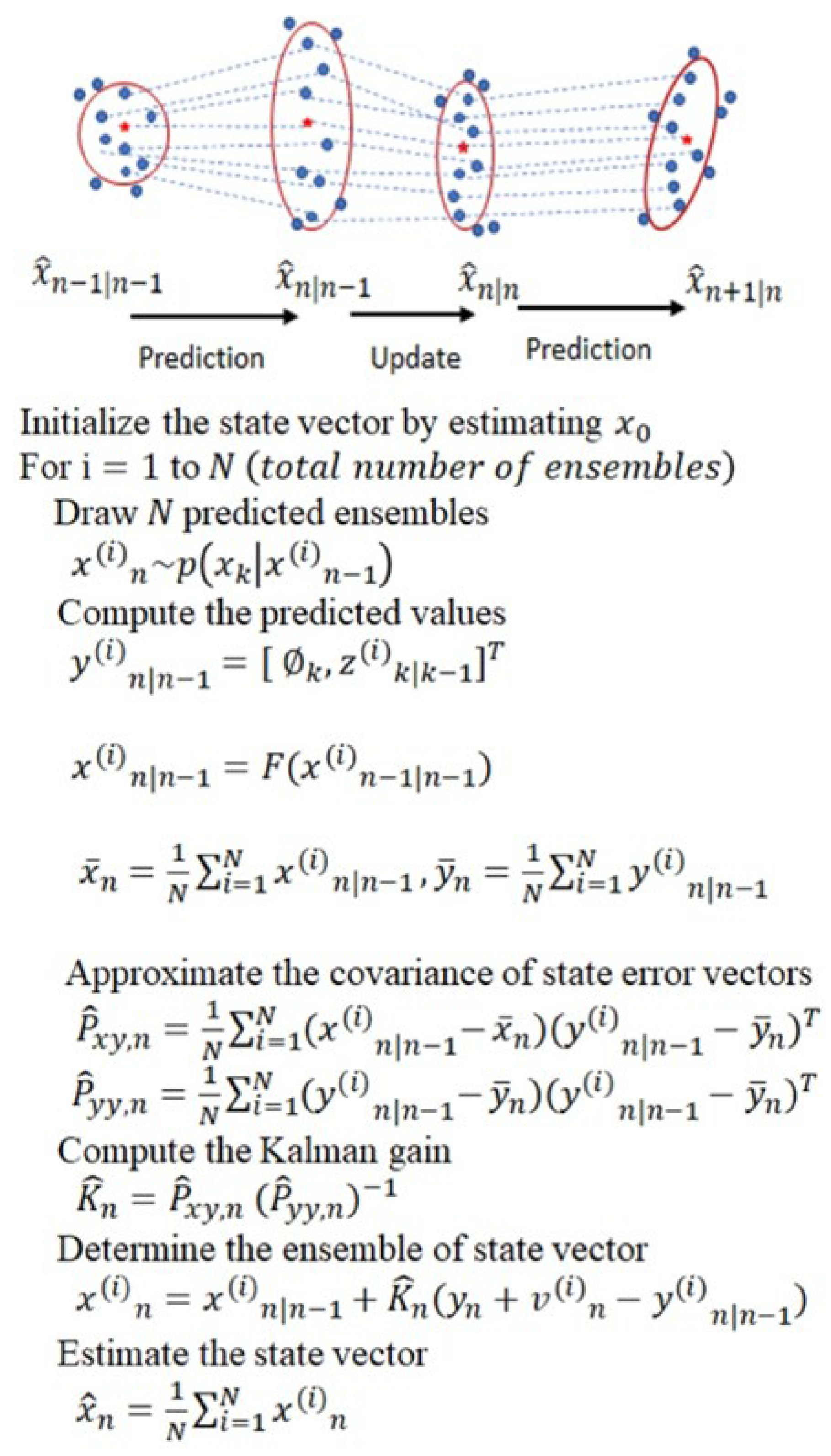
2.1. Ensemble Kalman Filter (EnKF)
2.2. State-Space Model of a Synthetic ECG
2.3. EnKF Based fECG Extraction Algorithm
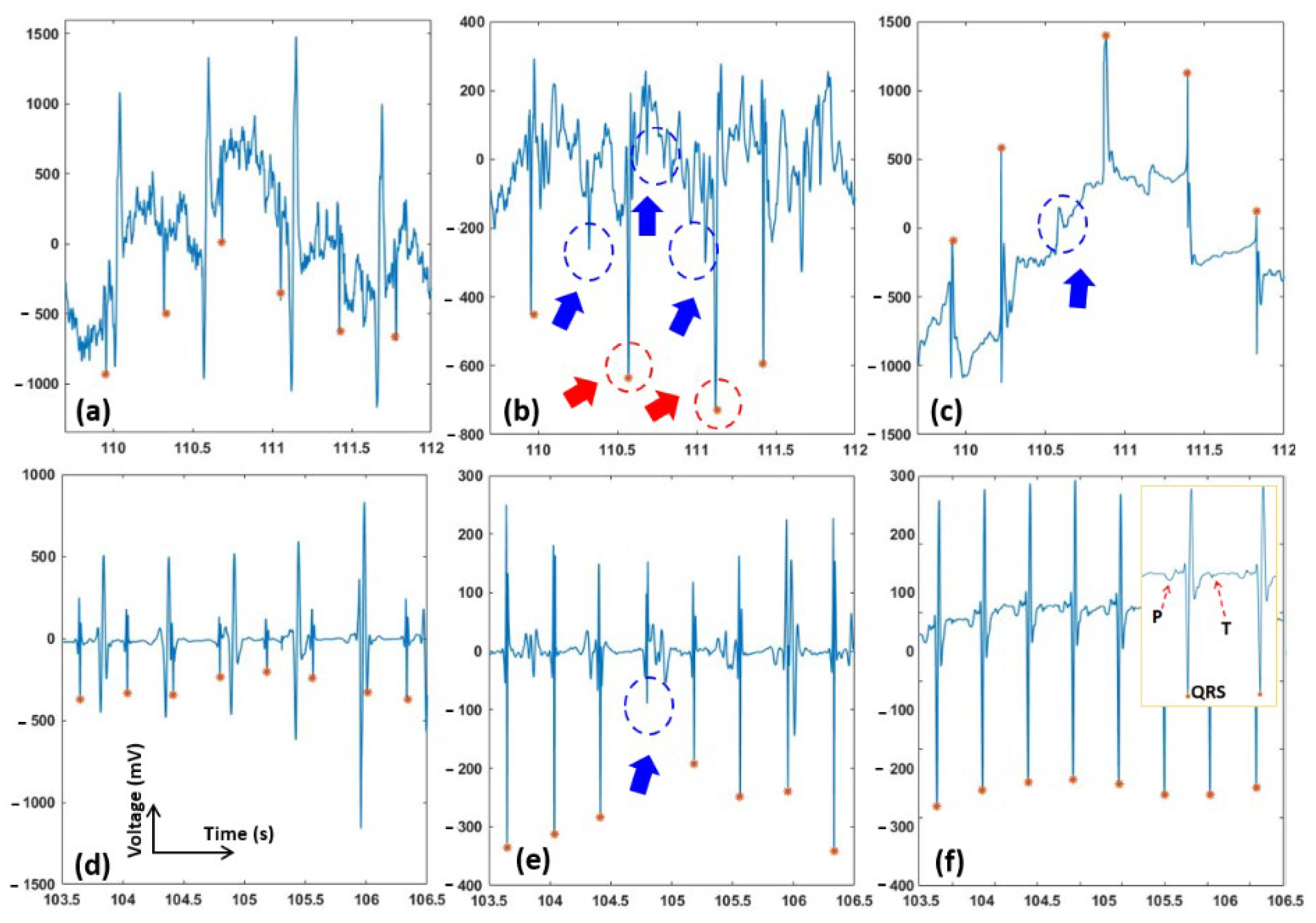
2.4. Data for Testing
2.4.1. Challenge Databank
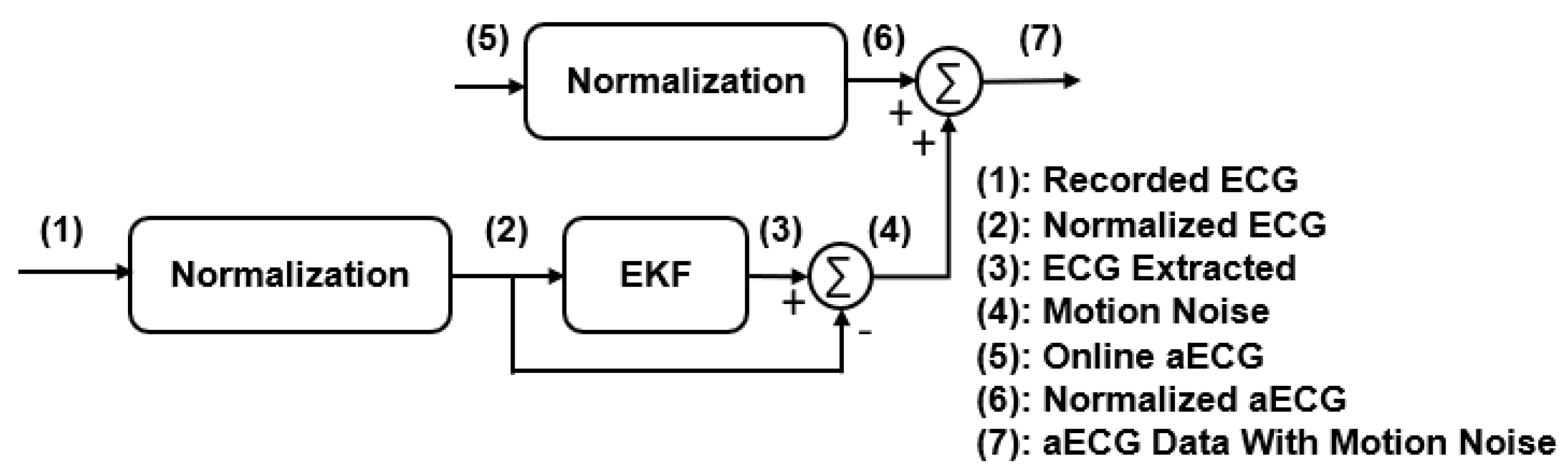
2.4.2. Modified Signals with Motional Artifacts Added
2.4.3. Our Human Data
2.5. Comparison Criteria
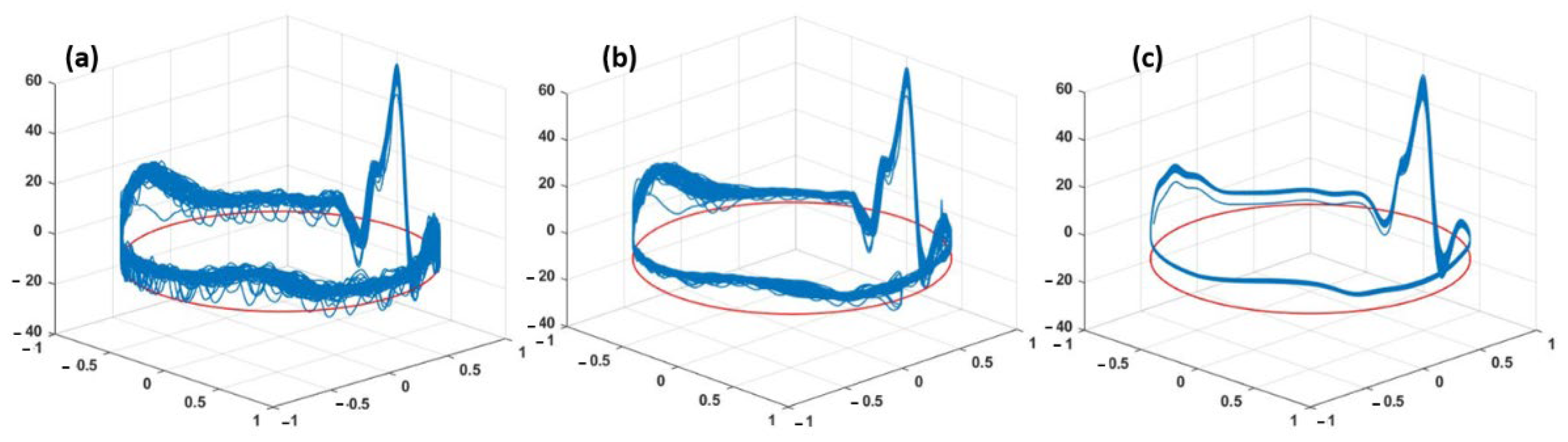
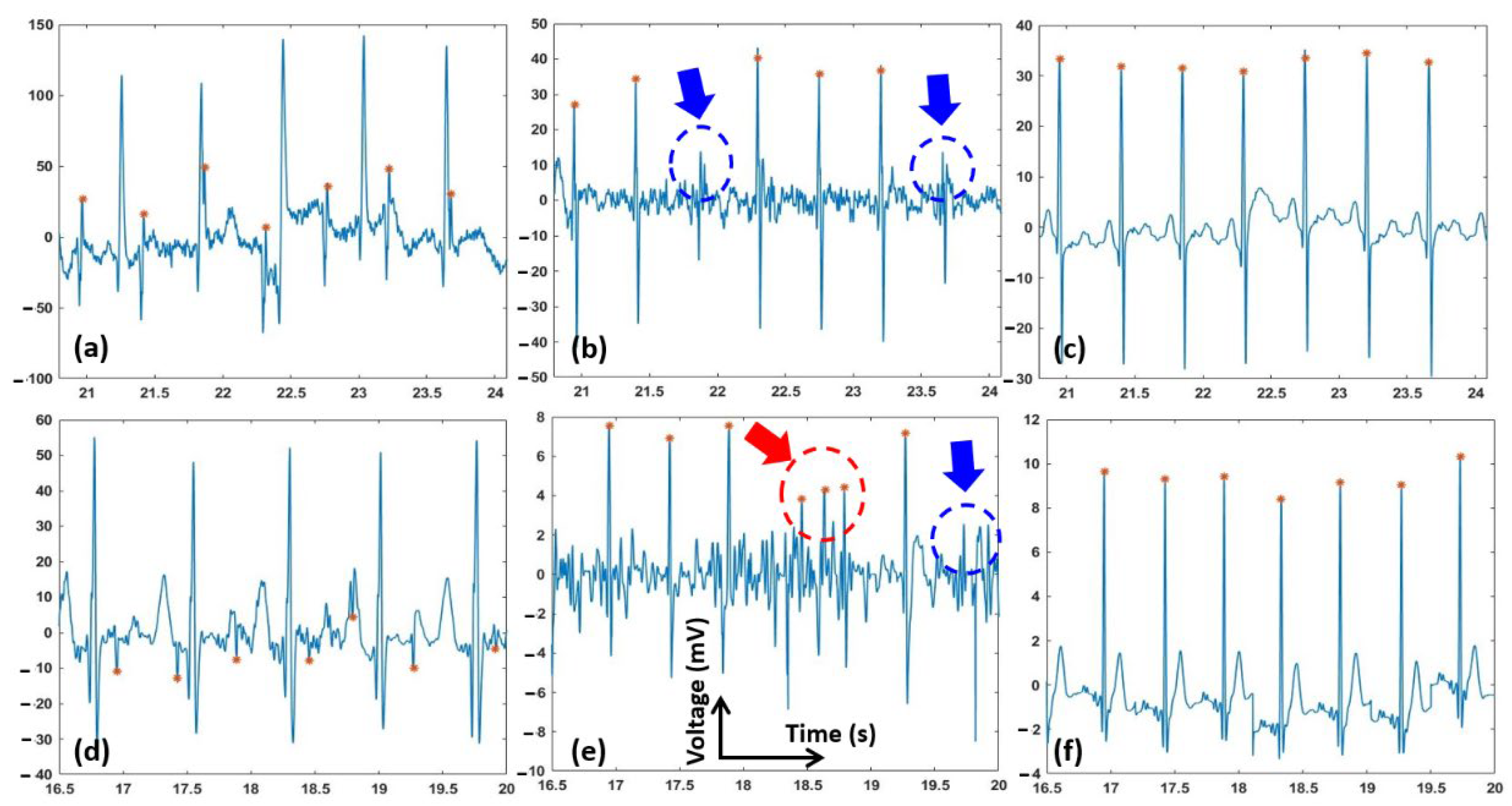
3. Results
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gregory, E.C. Trends in Fetal and Perinatal Mortality in the United States, 2006–2012; US Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Health Statistics: Hyattsville, MD, USA, 2014.
- Grivell, R.M.; Alfirevic, Z.; Gyte, G.M.; Devane, D. Antenatal cardiotocography for fetal assessment. Cochrane Database Syst. Rev. 2015, 9, CD007863. [Google Scholar] [CrossRef] [PubMed]
- Zambrano, L.D.; Ellington, S.; Strid, P.; Galang, R.R.; Oduyebo, T.; Tong, V.T.; Woodworth, K.R.; Nahabedian III, J.F.; Azziz-Baumgartner, E.; Gilboa, S.M. Update: Characteristics of symptomatic women of reproductive age with laboratory-confirmed SARS-CoV-2 infection by pregnancy status—United States, January 22–October 3, 2020. Morb. Mortal. Wkly. Rep. 2020, 69, 1641. [Google Scholar] [CrossRef] [PubMed]
- Rados, C. FDA Cautions against Ultrasound “Keepsake” Images. FDA Consumer Magazine. 2015; Volume 11. Available online: www.fda.gov/fdac/features/2004/104_images.html (accessed on 1 March 2022).
- Jaros, R.; Kahankova, R.; Martinek, R.; Nedoma, J.; Fajkus, M.; Slanina, Z. Fetal phonocardiography signal processing from abdominal records by non-adaptive methods. In Photonics Applications in Astronomy, Communications, Industry, and High-Energy Physics Experiments 2018; International Society for Optics and Photonics: Bellingham, WA, USA, 2018; Volume 10808, p. 108083E. [Google Scholar]
- Martinek, R.; Kahankova, R.; Jezewski, J.; Jaros, R.; Mohylova, J.; Fajkus, M.; Nedoma, J.; Janku, P.; Nazeran, H. Comparative effectiveness of ICA and PCA in extraction of fetal ECG from abdominal signals: Toward non-invasive fetal monitoring. Front. Physiol. 2018, 9, 648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sameni, R.; Jutten, C.; Shamsollahi, M.B. Multichannel electrocardiogram decomposition using periodic component analysis. IEEE Trans. Biomed. Eng. 2008, 55, 1935–1940. [Google Scholar] [CrossRef] [Green Version]
- Taha, L.Y.; Abdel-Raheem, E. Extraction of fetal electrocardiogram signals using blind source extraction based parallel linear predictor filter. In Proceedings of the 2018 IEEE International Symposium on Signal Processing and Information Technology (ISSPIT), Louisville, KY, USA, 6–8 December 2018; pp. 208–211. [Google Scholar]
- Taha, L.Y.; Abdel-Raheem, E. Efficient blind source extraction of noisy mixture utilising a class of parallel linear predictor filters. IET Signal. Process. 2018, 12, 1009–1016. [Google Scholar] [CrossRef]
- Cardoso, J.-F. Blind signal separation: Statistical principles. Proc. IEEE 1998, 86, 2009–2025. [Google Scholar] [CrossRef] [Green Version]
- Hyvärinen, A.; Oja, E. Independent component analysis: Algorithms and applications. Neural Netw. 2000, 13, 411–430. [Google Scholar] [CrossRef] [Green Version]
- Le, T.; Fortunato, J.; Maritato, N.; Cho, Y.; Nguyen, Q.-D.; Ghirmai, T.; Lau, M.P.; Han, H.-D.; Nguyen, C.K.; Nguyen, V.C. Home-based mobile fetal/maternal electrocardiogram acquisition and extraction with cloud assistance. In Proceedings of the 2019 IEEE MTT-S International Microwave Biomedical Conference (IMBioC), Nanjing, China, 6–8 May 2019; pp. 1–4. [Google Scholar]
- Yamamoto, U.; Nakamura, Y.; Yokouchi, H.; Hiroyasu, T. Improving the accuracy of the method for removing motion artifacts from fNIRS data using ICA and an accelerometer. In Proceedings of the 2014 World Automation Congress (WAC), Waikoloa, HI, USA, 3–7 August 2014; pp. 131–136. [Google Scholar]
- Yu, Q.; Yan, H.; Song, L.; Guo, W.; Liu, H.; Si, J.; Zhao, Y. Automatic identifying of maternal ECG source when applying ICA in fetal ECG extraction. Biocybern. Biomed. Eng. 2018, 38, 448–455. [Google Scholar] [CrossRef]
- Ungureanu, M.; Bergmans, J.W.; Oei, S.G.; Strungaru, R. Fetal ECG Extraction during Labor Using an Adaptive Maternal Beat Subtraction Technique. Biomed. Eng. 2007, 52, 56–60. [Google Scholar] [CrossRef]
- Varanini, M.; Tartarisco, G.; Billeci, L.; Macerata, A.; Pioggia, G.; Balocchi, R. An efficient unsupervised fetal QRS complex detection from abdominal maternal ECG. Physiol. Meas. 2014, 35, 1607. [Google Scholar] [CrossRef]
- Zaunseder, S.; Andreotti, F.; Cruz, M.; Stepan, H.; Schmieder, C.; Malberg, H.; Jank, A.; Wessel, N. Fetal QRS detection by means of Kalman filtering and using the event synchronous canceller. In Proceedings of the 7th International Workshop on Biosignal Interpretation, Como, Italy, 2–4 July 2012; pp. 2–4. [Google Scholar]
- Lee, K.J.; Lee, B. Sequential total variation denoising for the extraction of fetal ECG from single-channel maternal abdominal ECG. Sensors 2016, 16, 1020. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Chen, D.; Sun, G. Detection of fetal ECG R wave from single-lead abdominal ECG using a combination of RR time-series smoothing and template-matching approach. IEEE Access 2019, 7, 66633–66643. [Google Scholar] [CrossRef]
- Behar, J.; Oster, J.; Clifford, G.D. Combining and benchmarking methods of foetal ECG extraction without maternal or scalp electrode data. Physiol. Meas. 2014, 35, 1569. [Google Scholar] [CrossRef] [Green Version]
- Matonia, A.; Jezewski, J.; Horoba, K.; Gacek, A.; Labaj, P. The maternal ECG suppression algorithm for efficient extraction of the fetal ECG from abdominal signal. In Proceedings of the 2006 International Conference of the IEEE Engineering in Medicine and Biology Society, New York, NY, USA, 30 August–3 September 2006; pp. 3106–3109. [Google Scholar]
- Fotiadou, E.; Van Laar, J.O.; Oei, S.G.; Vullings, R. Enhancement of low-quality fetal electrocardiogram based on time-sequenced adaptive filtering. Med. Biol. Eng. Comput. 2018, 56, 2313–2323. [Google Scholar] [CrossRef] [Green Version]
- Kaleem, A.M.; Kokate, R.D. An efficient adaptive filter for fetal ECG extraction using neural network. J. Intell. Syst. 2019, 28, 589–600. [Google Scholar] [CrossRef]
- Martinek, R.; Kahankova, R.; Nazeran, H.; Konecny, J.; Jezewski, J.; Janku, P.; Bilik, P.; Zidek, J.; Nedoma, J.; Fajkus, M. Non-invasive fetal monitoring: A maternal surface ECG electrode placement-based novel approach for optimization of adaptive filter control parameters using the LMS and RLS algorithms. Sensors 2017, 17, 1154. [Google Scholar] [CrossRef]
- Wu, S.; Shen, Y.; Zhou, Z.; Lin, L.; Zeng, Y.; Gao, X. Research of fetal ECG extraction using wavelet analysis and adaptive filtering. Comput. Biol. Med. 2013, 43, 1622–1627. [Google Scholar] [CrossRef]
- Andreotti, F.; Riedl, M.; Himmelsbach, T.; Wedekind, D.; Zaunseder, S.; Wessel, N.; Malberg, H. Maternal signal estimation by Kalman filtering and template adaptation for fetal heart rate extraction. In Proceedings of the Computing in Cardiology 2013, Zaragoza, Spain, 22–25 September 2013; pp. 193–196. [Google Scholar]
- Suganthy, M.; Manjula, S. Enhancement of SNR in fetal ECG signal extraction using combined SWT and WLSR in parallel EKF. Clust. Comput. 2019, 22, 3875–3881. [Google Scholar] [CrossRef]
- Vullings, R.; De Vries, B.; Bergmans, J.W. An adaptive Kalman filter for ECG signal enhancement. IEEE Trans. Biomed. Eng. 2010, 58, 1094–1103. [Google Scholar] [CrossRef] [Green Version]
- Martinek, R.; Kahankova, R.; Nedoma, J.; Fajkus, M.; Cholevova, K. Fetal ECG preprocessing using wavelet transform. In Proceedings of the 10th International Conference on Computer Modeling and Simulation, Sydney, Australia, 8–10 January 2018; pp. 39–43. [Google Scholar]
- Jothi, S.H.; Prabha, K.H. Fetal electrocardiogram extraction using adaptive neuro-fuzzy inference systems and undecimated wavelet transform. IETE J. Res. 2012, 58, 469–475. [Google Scholar] [CrossRef]
- Sharma, M.; Ritchie, P.; Ghirmai, T.; Cao, H.; Lau, M.P. Unobtrusive acquisition and extraction of fetal and maternal ECG in the home setting. In Proceedings of the 2017 IEEE SENSORS, Glasgow, UK, 29 October–1 November 2017; pp. 1–3. [Google Scholar]
- Sarafan, S.; Le, T.; Naderi, A.M.; Nguyen, Q.-D.; Kuo, B.T.-Y.; Ghirmai, T.; Han, H.-D.; Lau, M.P.; Cao, H. Investigation of methods to extract fetal electrocardiogram from the mother’s abdominal signal in practical scenarios. Technologies 2020, 8, 33. [Google Scholar] [CrossRef]
- Jagannath, D.; Selvakumar, A.I. Issues and research on foetal electrocardiogram signal elicitation. Biomed. Signal. Process. Control 2014, 10, 224–244. [Google Scholar] [CrossRef]
- Keller, J.; Hendricks Franssen, H.J.; Marquart, G. Comparing seven variants of the ensemble Kalman filter: How many synthetic experiments are needed? Water Resour. Res. 2018, 54, 6299–6318. [Google Scholar] [CrossRef] [Green Version]
- Anderson, B.D.; Moore, J.B. Optimal Filtering. Courier Corporation; Dover Publication: Mineola, NY, USA, 2012. [Google Scholar]
- Katzfuss, M.; Stroud, J.R.; Wikle, C.K. Understanding the ensemble Kalman filter. Am. Stat. 2016, 70, 350–357. [Google Scholar] [CrossRef]
- McSharry, P.E.; Clifford, G.D.; Tarassenko, L.; Smith, L.A. A dynamical model for generating synthetic electrocardiogram signals. IEEE Trans. Biomed. Eng. 2003, 50, 289–294. [Google Scholar] [CrossRef] [Green Version]
- Sameni, R.; Shamsollahi, M.B.; Jutten, C.; Clifford, G.D. A nonlinear Bayesian filtering framework for ECG denoising. IEEE Trans. Biomed. Eng. 2007, 54, 2172–2185. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Fritsche, C.; Yin, F.; Gunnarsson, F.; Gustafsson, F. Sequential Monte Carlo methods and theoretical bounds for proximity report based indoor positioning. IEEE Trans. Veh. Technol. 2018, 67, 5372–5386. [Google Scholar] [CrossRef]
- Cao, H.; Yu, F.; Zhao, Y.; Zhang, X.; Tai, J.; Lee, J.; Darehzereshki, A.; Bersohn, M.; Lien, C.-L.; Chi, N.C. Wearable multi-channel microelectrode membranes for elucidating electrophysiological phenotypes of injured myocardium. Integr. Biol. 2014, 6, 789–795. [Google Scholar] [CrossRef] [Green Version]
- Silva, I.; Behar, J.; Sameni, R.; Zhu, T.; Oster, J.; Clifford, G.D.; Moody, G.B. Noninvasive fetal ECG: The PhysioNet/computing in cardiology challenge 2013. In Proceedings of the Computing in Cardiology 2013, Zaragoza, Spain, 22–25 September 2013; pp. 149–152. [Google Scholar]
- Sarafan, S.; Le, T.; Ellington, F.; Zhang, Z.; Lau, M.P.; Ghirmai, T.; Hameed, A.; Cao, H. Development of a Home-based Fetal Electrocardiogram (ECG) Monitoring System. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Guadalajara, Mexico, 1–5 November 2021; pp. 7116–7119. [Google Scholar]
- Niknazar, M.; Rivet, B.; Jutten, C. Fetal ECG extraction by extended state Kalman filtering based on single-channel recordings. IEEE Trans. Biomed. Eng. 2012, 60, 1345–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohl, T.; Müller, A.; Tchatcheva, K.; Achenbach, S.; Gembruch, U. Fetal transesophageal echocardiography: Clinical introduction as a monitoring tool during cardiac intervention in a human fetus. Ultrasound Obstet. Gynecol. Off. J. Int. Soc. Ultrasound Obstet. Gynecol. 2005, 26, 780–785. [Google Scholar] [CrossRef]
- Rychik, J.; Ayres, N.; Cuneo, B.; Gotteiner, N.; Hornberger, L.; Spevak, P.J.; Van Der Veld, M. American Society of Echocardiography guidelines and standards for performance of the fetal echocardiogram. J. Am. Soc. Echocardiogr. 2004, 17, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Corsten-Janssen, N.; Bouman, K.; Diphoorn, J.C.; Scheper, A.J.; Kinds, R.; El Mecky, J.; Breet, H.; Verheij, J.B.; Suijkerbuijk, R.; Duin, L.K. A prospective study on rapid exome sequencing as a diagnostic test for multiple congenital anomalies on fetal ultrasound. Prenat. Diagn. 2020, 40, 1300–1309. [Google Scholar] [CrossRef] [PubMed]
- Vo, K.; Naeini, E.K.; Naderi, A.; Jilani, D.; Rahmani, A.M.; Dutt, N.; Cao, H. P2E-WGAN: ECG waveform synthesis from PPG with conditional wasserstein generative adversarial networks. In Proceedings of the 36th Annual ACM Symposium on Applied Computing, online. 22–26 March 2021; pp. 1030–1036. [Google Scholar]

| Average F1 (%) | Average SE (%) | Average PPE (%) | ||
|---|---|---|---|---|
| Online Data without Motion Noise | EKF | 88.90 ± 5 | 86.73 ± 5.5 | 91.16 ± 4.6 |
| EnKF | 97.25 ± 2.4 | 96.91 ± 0.5 | 97.59 ± 3.8 | |
| Online Data with Motion Noise | EKF | 78 ± 6.58 | 75.38 ± 7.4 | 80.80 ± 5.1 |
| EnKF | 89.04 ± 3 | 88.2 ± 1.7 | 89.9 ± 4.5 | |
| Our Clinical Data | EKF | 82.3 ± 5.5 | 100 ± 0.1 | 71.4 ± 6.4 |
| EnKF | 94.3 ± 1.2 | 89.2 ± 1.5 | 100 ± 0.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarafan, S.; Le, T.; Lau, M.P.H.; Hameed, A.; Ghirmai, T.; Cao, H. Fetal Electrocardiogram Extraction from the Mother’s Abdominal Signal Using the Ensemble Kalman Filter. Sensors 2022, 22, 2788. https://doi.org/10.3390/s22072788
Sarafan S, Le T, Lau MPH, Hameed A, Ghirmai T, Cao H. Fetal Electrocardiogram Extraction from the Mother’s Abdominal Signal Using the Ensemble Kalman Filter. Sensors. 2022; 22(7):2788. https://doi.org/10.3390/s22072788
Chicago/Turabian StyleSarafan, Sadaf, Tai Le, Michael P. H. Lau, Afshan Hameed, Tadesse Ghirmai, and Hung Cao. 2022. "Fetal Electrocardiogram Extraction from the Mother’s Abdominal Signal Using the Ensemble Kalman Filter" Sensors 22, no. 7: 2788. https://doi.org/10.3390/s22072788
APA StyleSarafan, S., Le, T., Lau, M. P. H., Hameed, A., Ghirmai, T., & Cao, H. (2022). Fetal Electrocardiogram Extraction from the Mother’s Abdominal Signal Using the Ensemble Kalman Filter. Sensors, 22(7), 2788. https://doi.org/10.3390/s22072788








