Deep 3D Neural Network for Brain Structures Segmentation Using Self-Attention Modules in MRI Images
Abstract
:1. Introduction
2. Material and Methods
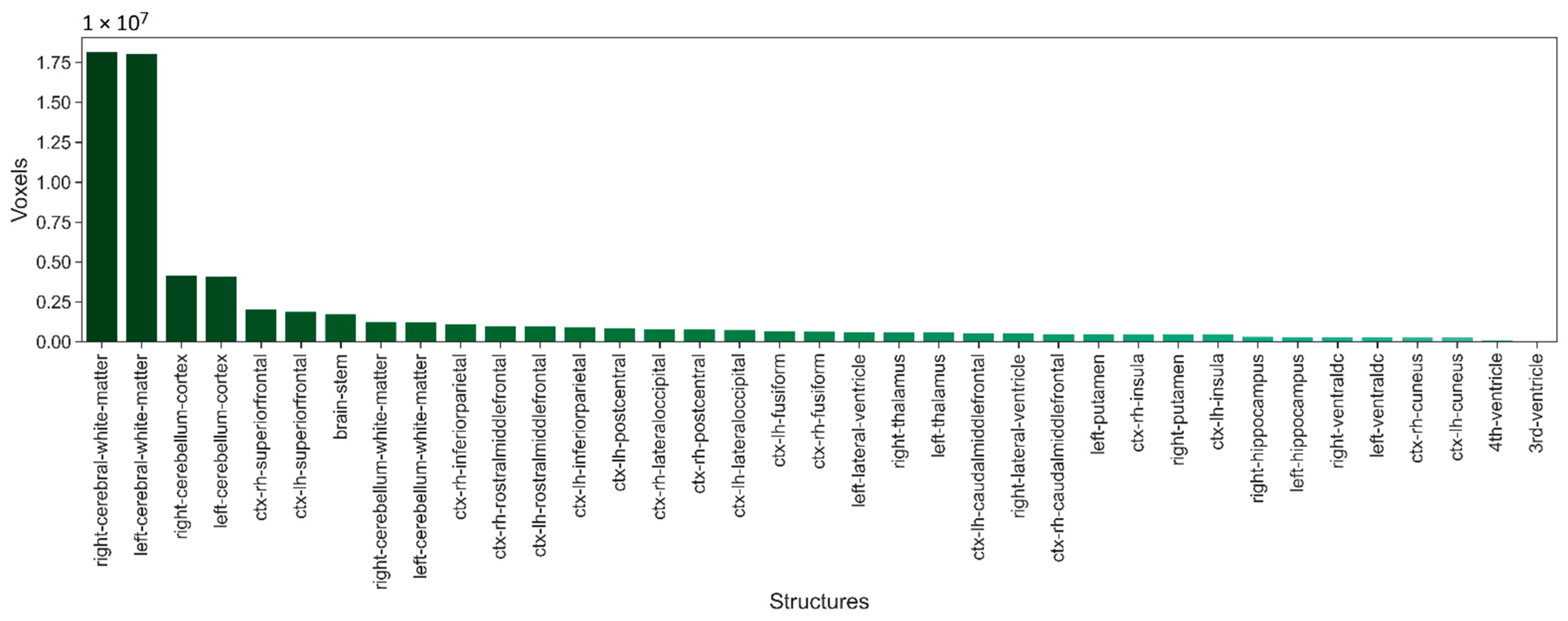
2.1. Dataset
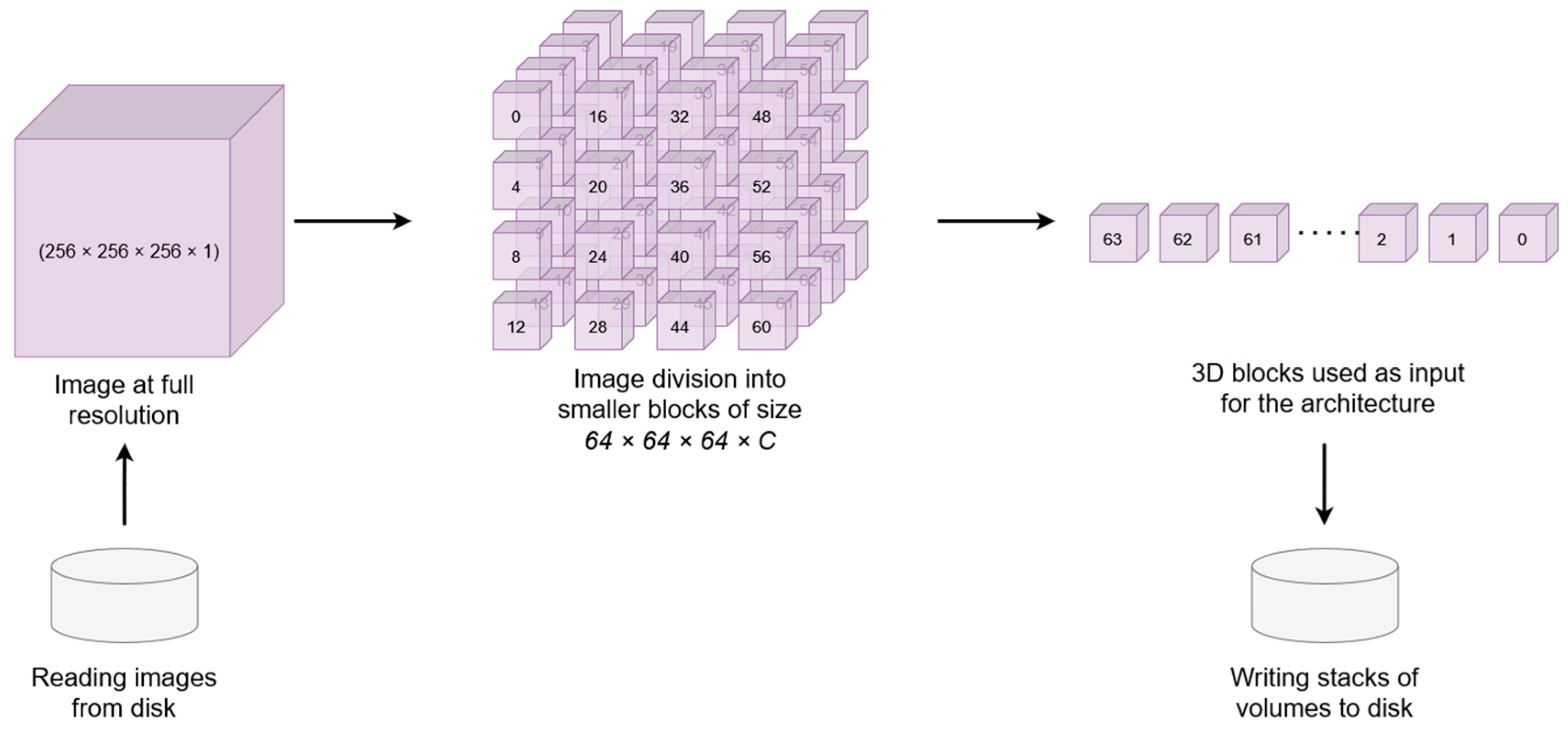
2.2. Data Preprocesing
2.3. Computational Resources
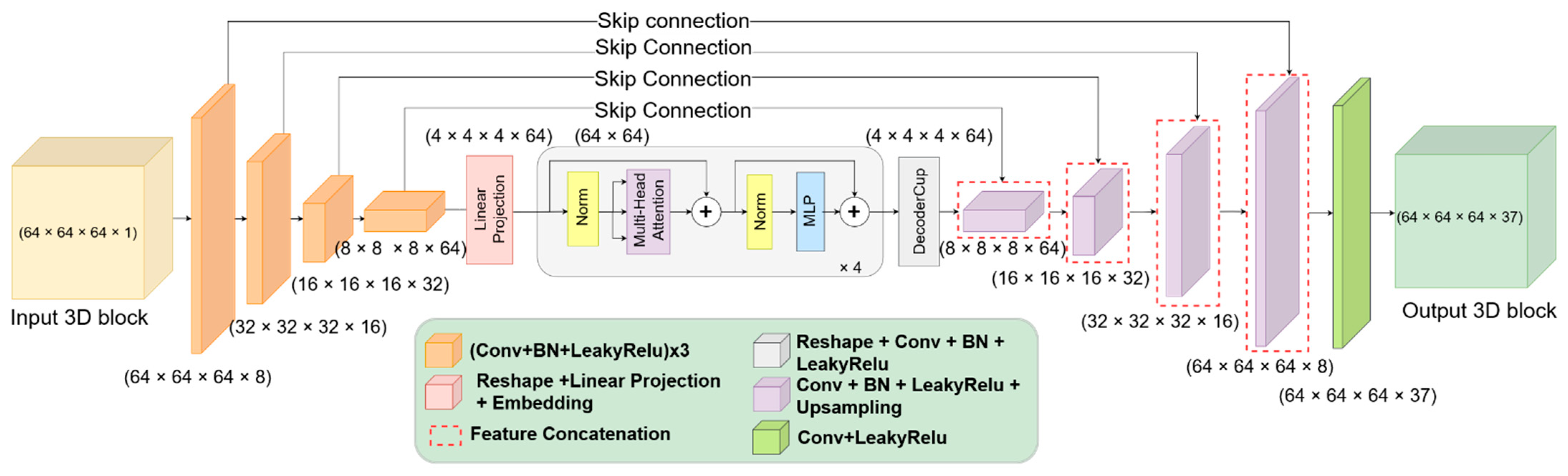
2.4. Model for Brain Structures Segmentation from MRI
2.5. Self-Attention Mechanism for Brain Segmentation
2.6. Loss Functions and Class Weights
2.7. Metrics
2.8. Training Process
3. Results
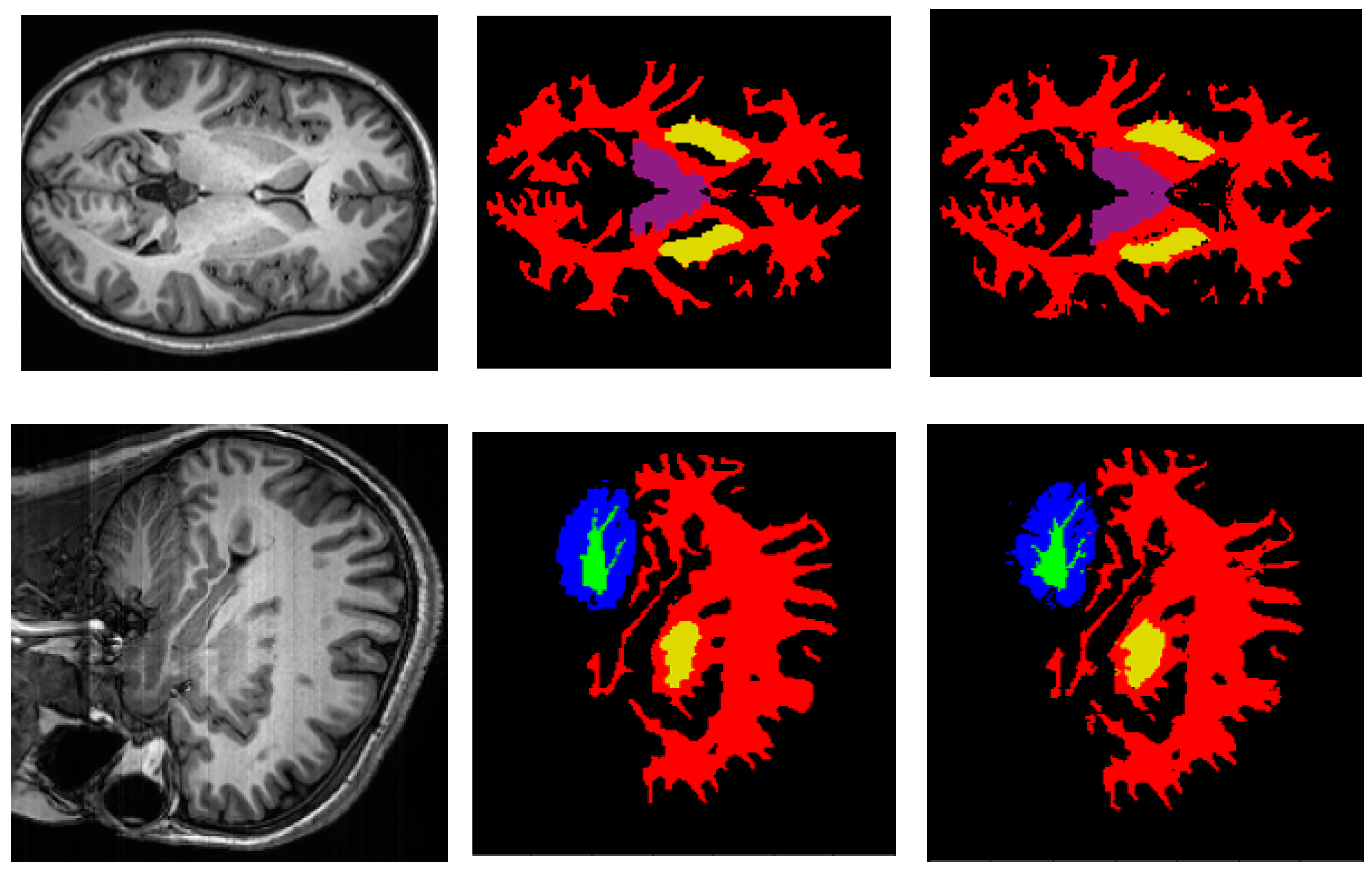
3.1. Performance of Proposed Deep Neural Network Architecture
3.2. Patch Resolution Size Determination
3.3. Comparison with Other Methods
4. Discussion and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Miller-Thomas, M.M.; Benzinger, T.L.S. Neurologic Applications of PET/MR Imaging. Magn. Reson. Imaging Clin. N. Am. 2017, 25, 297–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mier, W.; Mier, D. Advantages in functional imaging of the brain. Front. Hum. Neurosci. 2015, 9, 249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neeb, H.; Zilles, K.; Shah, N.J. Fully-automated detection of cerebral water content changes: Study of age- and gender-related H2O patterns with quantitative MRI. NeuroImage 2006, 29, 910–922. [Google Scholar] [CrossRef] [PubMed]
- Shenton, M.E.; Dickey, C.C.; Frumin, M.; McCarley, R.W. A review of MRI findings in schizophrenia. Schizophr. Res. 2001, 49, 1–52. [Google Scholar] [CrossRef] [Green Version]
- Widmann, G.; Henninger, B.; Kremser, C.; Jaschke, W. MRI Sequences in Head & Neck Radiology—State of the Art. ROFO Fortschr. Geb. Rontgenstr. Nuklearmed. 2017, 189, 413–422. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Yang, L.T.; Zhang, Q.; Armstrong, D.; Deen, M.J. Convolutional neural networks for medical image analysis: State-of-the-art, comparisons, improvement and perspectives. Neurocomputing 2021, 444, 92–110. [Google Scholar] [CrossRef]
- Xie, X.; Niu, J.; Liu, X.; Chen, Z.; Tang, S.; Yu, S. A survey on incorporating domain knowledge into deep learning for medical image analysis. Med. Image Anal. 2021, 69, 101985. [Google Scholar] [CrossRef]
- Ilhan, U.; Ilhan, A. Brain tumor segmentation based on a new threshold approach. Procedia Comput. Sci. 2017, 120, 580–587. [Google Scholar] [CrossRef]
- Deng, W.; Xiao, W.; Deng, H.; Liu, J. MRI brain tumor segmentation with region growing method based on the gradients and variances along and inside of the boundary curve. In Proceedings of the 2010 3rd International Conference on Biomedical Engineering and Informatics, Yantai, China, 16–18 October 2010; Volume 1, pp. 393–396. [Google Scholar]
- Ashburner, J.; Friston, K.J. Unified segmentation. NeuroImage 2005, 26, 839–851. [Google Scholar] [CrossRef]
- Liu, J.; Guo, L. A New Brain MRI Image Segmentation Strategy Based on K-means Clustering and SVM. In Proceedings of the 2015 7th International Conference on Intelligent Human-Machine Systems and Cybernetics, Hangzhou, China, 26–27 August 2015; Volume 2, pp. 270–273. [Google Scholar]
- Zhao, X.; Zhao, X.-M. Deep learning of brain magnetic resonance images: A brief review. Methods 2021, 192, 131–140. [Google Scholar] [CrossRef]
- Despotović, I.; Goossens, B.; Philips, W. MRI Segmentation of the Human Brain: Challenges, Methods, and Applications. Comput. Math. Methods Med. 2015, 2015, e450341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischl, B.; Salat, D.H.; Busa, E.; Albert, M.; Dieterich, M.; Haselgrove, C.; van der Kouwe, A.; Killiany, R.; Kennedy, D.; Klaveness, S.; et al. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron 2002, 33, 341–355. [Google Scholar] [CrossRef] [Green Version]
- Shattuck, D.W.; Leahy, R.M. BrainSuite: An automated cortical surface identification tool. Med. Image Anal. 2002, 6, 129–142. [Google Scholar] [CrossRef]
- Jenkinson, M.; Beckmann, C.F.; Behrens, T.E.J.; Woolrich, M.W.; Smith, S.M. FSL. NeuroImage 2012, 62, 782–790. [Google Scholar] [CrossRef] [Green Version]
- Yushkevich, P.A.; Gao, Y.; Gerig, G. ITK-SNAP: An interactive tool for semi-automatic segmentation of multi-modality biomedical images. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 3342–3345. [Google Scholar]
- Pieper, S.; Halle, M.; Kikinis, R. 3D Slicer. In Proceedings of the 2004 2nd IEEE International Symposium on Biomedical Imaging: Nano to Macro (IEEE Cat No. 04EX821), Arlington, VA, USA, 15–18 April 2004; Volume 1, pp. 632–635. [Google Scholar]
- Please Help Sustain the Horos Project—Horos Project. Available online: https://horosproject.org/download-donation/ (accessed on 24 August 2021).
- Qin, C.; Wu, Y.; Liao, W.; Zeng, J.; Liang, S.; Zhang, X. Improved U-Net3+ with stage residual for brain tumor segmentation. BMC Med. Imaging 2022, 22, 14. [Google Scholar] [CrossRef]
- Sun, J.; Peng, Y.; Guo, Y.; Li, D. Segmentation of the multimodal brain tumor image used the multi-pathway architecture method based on 3D FCN. Neurocomputing 2021, 423, 34–45. [Google Scholar] [CrossRef]
- Dai, W.; Woo, B.; Liu, S.; Marques, M.; Tang, F.; Crozier, S.; Engstrom, C.; Chandra, S. Can3d: Fast 3d Knee Mri Segmentation Via Compact Context Aggregation. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 1505–1508. [Google Scholar]
- Deepthi Murthy, T.S.; Sadashivappa, G. Brain tumor segmentation using thresholding, morphological operations and extraction of features of tumor. In Proceedings of the 2014 International Conference on Advances in Electronics Computers and Communications, Bangalore, India, 10–11 October 2014; pp. 1–6. [Google Scholar]
- Polakowski, W.E.; Cournoyer, D.A.; Rogers, S.K.; DeSimio, M.P.; Ruck, D.W.; Hoffmeister, J.W.; Raines, R.A. Computer-aided breast cancer detection and diagnosis of masses using difference of Gaussians and derivative-based feature saliency. IEEE Trans. Med. Imaging 1997, 16, 811–819. [Google Scholar] [CrossRef]
- Wani, M.A.; Batchelor, B.G. Edge-region-based segmentation of range images. IEEE Trans. Pattern Anal. Mach. Intell. 1994, 16, 314–319. [Google Scholar] [CrossRef]
- Wu, J.; Ye, F.; Ma, J.-L.; Sun, X.-P.; Xu, J.; Cui, Z.-M. The Segmentation and Visualization of Human Organs Based on Adaptive Region Growing Method. In Proceedings of the 2008 IEEE 8th International Conference on Computer and Information Technology Workshops, Washington, WA, USA, 8–11 July 2008; pp. 439–443. [Google Scholar]
- Passat, N.; Ronse, C.; Baruthio, J.; Armspach, J.-P.; Maillot, C.; Jahn, C. Region-growing segmentation of brain vessels: An atlas-based automatic approach. J. Magn. Reson. Imaging JMRI 2005, 21, 715–725. [Google Scholar] [CrossRef]
- Gibbs, P.; Buckley, D.L.; Blackband, S.J.; Horsman, A. Tumour volume determination from MR images by morphological segmentation. Phys. Med. Biol. 1996, 41, 2437–2446. [Google Scholar] [CrossRef]
- Pohlman, S.; Powell, K.A.; Obuchowski, N.A.; Chilcote, W.A.; Grundfest-Broniatowski, S. Quantitative classification of breast tumors in digitized mammograms. Med. Phys. 1996, 23, 1337–1345. [Google Scholar] [CrossRef] [PubMed]
- Maksoud, E.A.A.; Elmogy, M.; Al-Awadi, R.M. MRI Brain Tumor Segmentation System Based on Hybrid Clustering Techniques. In Proceedings of the Advanced Machine Learning Technologies and Applications, Cairo, Egypt, 28–30 November 2014; Hassanien, A.E., Tolba, M.F., Taher Azar, A., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2014; pp. 401–412. [Google Scholar]
- Artaechevarria, X.; Munoz-Barrutia, A.; Ortiz-de-Solorzano, C. Combination Strategies in Multi-Atlas Image Segmentation: Application to Brain MR Data. IEEE Trans. Med. Imaging 2009, 28, 1266–1277. [Google Scholar] [CrossRef] [PubMed]
- Coupé, P.; Manjón, J.V.; Fonov, V.; Pruessner, J.; Robles, M.; Collins, D.L. Patch-based segmentation using expert priors: Application to hippocampus and ventricle segmentation. NeuroImage 2011, 54, 940–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Suh, J.W.; Das, S.R.; Pluta, J.B.; Craige, C.; Yushkevich, P.A. Multi-Atlas Segmentation with Joint Label Fusion. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 611–623. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Wang, Q.; Zhang, D.; Nie, F.; Huang, H.; Shen, D. A generative probability model of joint label fusion for multi-atlas based brain segmentation. Med. Image Anal. 2014, 18, 881–890. [Google Scholar] [CrossRef] [Green Version]
- Kass, M.; Witkin, A.; Terzopoulos, D. Snakes: Active contour models. Int. J. Comput. Vis. 1988, 1, 321–331. [Google Scholar] [CrossRef]
- Wu, Z.; Guo, Y.; Park, S.H.; Gao, Y.; Dong, P.; Lee, S.-W.; Shen, D. Robust brain ROI segmentation by deformation regression and deformable shape model. Med. Image Anal. 2018, 43, 198–213. [Google Scholar] [CrossRef]
- Rajendran, A.; Dhanasekaran, R. Fuzzy Clustering and Deformable Model for Tumor Segmentation on MRI Brain Image: A Combined Approach. Procedia Eng. 2012, 30, 327–333. [Google Scholar] [CrossRef] [Green Version]
- Khotanlou, H.; Atif, J.; Colliot, O.; Bloch, I. 3D Brain Tumor Segmentation Using Fuzzy Classification and Deformable Models. In Proceedings of the Fuzzy Logic and Applications, Crema, Italy, 15–17 September 2005; Bloch, I., Petrosino, A., Tettamanzi, A.G.B., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 312–318. [Google Scholar]
- Le, W.T.; Maleki, F.; Romero, F.P.; Forghani, R.; Kadoury, S. Overview of Machine Learning: Part 2: Deep Learning for Medical Image Analysis. Neuroimaging Clin. N. Am. 2020, 30, 417–431. [Google Scholar] [CrossRef]
- Liu, X.; Wang, H.; Li, Z.; Qin, L. Deep learning in ECG diagnosis: A review. Knowl. Based Syst. 2021, 227, 107187. [Google Scholar] [CrossRef]
- Nogales, A.; García-Tejedor, Á.J.; Monge, D.; Vara, J.S.; Antón, C. A survey of deep learning models in medical therapeutic areas. Artif. Intell. Med. 2021, 112, 102020. [Google Scholar] [CrossRef] [PubMed]
- Tng, S.S.; Le, N.Q.K.; Yeh, H.-Y.; Chua, M.C.H. Improved Prediction Model of Protein Lysine Crotonylation Sites Using Bidirectional Recurrent Neural Networks. J. Proteome Res. 2022, 21, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Le, N.Q.K.; Ho, Q.-T. Deep transformers and convolutional neural network in identifying DNA N6-methyladenine sites in cross-species genomes. Methods 2021. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Wang, Z.; Kuen, J.; Ma, L.; Shahroudy, A.; Shuai, B.; Liu, T.; Wang, X.; Wang, G.; Cai, J.; et al. Recent advances in convolutional neural networks. Pattern Recognit. 2018, 77, 354–377. [Google Scholar] [CrossRef] [Green Version]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 53. [Google Scholar] [CrossRef]
- Bank, D.; Koenigstein, N.; Giryes, R. Autoencoders. arXiv 2020, arXiv:2003.05991. [Google Scholar]
- Montúfar, G. Restricted Boltzmann Machines: Introduction and Review. arXiv 2018, arXiv:1806.07066. [Google Scholar]
- Sherstinsky, A. Fundamentals of Recurrent Neural Network (RNN) and Long Short-Term Memory (LSTM) Network. arXiv 2018, arXiv:1808.03314. [Google Scholar] [CrossRef] [Green Version]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Backpropagation Applied to Handwritten Zip Code Recognition. Neural Comput. 1989, 1, 541–551. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. arXiv 2017, arXiv:1706.03762. [Google Scholar]
- Fukushima, K. Neocognitron: A self-organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol. Cybern. 1980, 36, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Nie, D.; Wang, L.; Gao, Y.; Shen, D. Fully convolutional networks for multi-modality isointense infant brain image segmentation. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016. [Google Scholar] [CrossRef] [Green Version]
- Bao, S.; Chung, A.C.S. Multi-scale structured CNN with label consistency for brain MR image segmentation. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2018, 6, 113–117. [Google Scholar] [CrossRef]
- Henschel, L.; Conjeti, S.; Estrada, S.; Diers, K.; Fischl, B.; Reuter, M. FastSurfer—A fast and accurate deep learning based neuroimaging pipeline. NeuroImage 2020, 219, 117012. [Google Scholar] [CrossRef] [PubMed]
- Brosch, T.; Tang, L.Y.W.; Yoo, Y.; Li, D.K.B.; Traboulsee, A.; Tam, R. Deep 3D Convolutional Encoder Networks With Shortcuts for Multiscale Feature Integration Applied to Multiple Sclerosis Lesion Segmentation. IEEE Trans. Med. Imaging 2016, 35, 1229–1239. [Google Scholar] [CrossRef]
- Valverde, S.; Cabezas, M.; Roura, E.; González-Villà, S.; Pareto, D.; Vilanova, J.; Ramió-Torrentà, L.; Rovira, À.; Oliver, A.; Lladó, X. Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach. NeuroImage 2017, 155, 159–168. [Google Scholar] [CrossRef] [Green Version]
- Gabr, R.E.; Coronado, I.; Robinson, M.; Sujit, S.J.; Datta, S.; Sun, X.; Allen, W.J.; Lublin, F.D.; Wolinsky, J.S.; Narayana, P.A. Brain and lesion segmentation in multiple sclerosis using fully convolutional neural networks: A large-scale study. Mult. Scler. Houndmills Basingstoke Engl. 2020, 26, 1217–1226. [Google Scholar] [CrossRef]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.-M.; Larochelle, H. Brain tumor segmentation with Deep Neural Networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Havaei, M.; Dutil, F.; Pal, C.; Larochelle, H.; Jodoin, P.-M. A Convolutional Neural Network Approach to Brain Tumor Segmentation. In Proceedings of the Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Munich, Germany, 5 October 2015; Crimi, A., Menze, B., Maier, O., Reyes, M., Handels, H., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; pp. 195–208. [Google Scholar]
- McKinley, R.; Jungo, A.; Wiest, R.; Reyes, M. Pooling-Free Fully Convolutional Networks with Dense Skip Connections for Semantic Segmentation, with Application to Brain Tumor Segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 169–177. [Google Scholar]
- Chen, L.; Bentley, P.; Rueckert, D. Fully automatic acute ischemic lesion segmentation in DWI using convolutional neural networks. NeuroImage Clin. 2017, 15, 633–643. [Google Scholar] [CrossRef]
- Akkus, Z.; Ali, I.; Sedlar, J.; Kline, T.L.; Agrawal, J.P.; Parney, I.F.; Giannini, C.; Erickson, B.J. Predicting 1p19q Chromosomal Deletion of Low-Grade Gliomas from MR Images using Deep Learning. arXiv 2016, arXiv:1611.06939. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Munich, Germany, 5–9 October 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Kumar, P.; Nagar, P.; Arora, C.; Gupta, A. U-Segnet: Fully Convolutional Neural Network Based Automated Brain Tissue Segmentation Tool. In Proceedings of the 2018 25th IEEE International Conference on Image Processing (ICIP), Athens, Greece, 7–10 October 2018; pp. 3503–3507. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Trans. Med. Imaging 2020, 39, 1856–1867. [Google Scholar] [CrossRef] [Green Version]
- Ibtehaz, N.; Rahman, M.S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Kwabena Patrick, M.; Felix Adekoya, A.; Abra Mighty, A.; Edward, B.Y. Capsule Networks—A survey. J. King Saud Univ. Comput. Inf. Sci. 2019, 34, 1295–1310. [Google Scholar] [CrossRef]
- Salehinejad, H.; Baarbe, J.; Sankar, S.; Barfett, J.; Colak, E.; Valaee, S. Recent Advances in Recurrent Neural Networks. arXiv 2018, arXiv:1801.01078. [Google Scholar]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. BERT: Pre-Training of Deep Bidirectional Transformers for Language Understanding. arXiv 2019, arXiv:1810.04805. [Google Scholar]
- Zheng, S.; Lu, J.; Zhao, H.; Zhu, X.; Luo, Z.; Wang, Y.; Fu, Y.; Feng, J.; Xiang, T.; Torr, P.H.S.; et al. Rethinking Semantic Segmentation from a Sequence-to-Sequence Perspective with Transformers. arXiv 2021, arXiv:2012.15840. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Valanarasu, J.M.J.; Oza, P.; Hacihaliloglu, I.; Patel, V.M. Medical Transformer: Gated Axial-Attention for Medical Image Segmentation. arXiv 2021, arXiv:2102.10662. [Google Scholar]
- Klein, A.; Tourville, J. 101 Labeled Brain Images and a Consistent Human Cortical Labeling Protocol. Front. Neurosci. 2012, 6, 171. [Google Scholar] [CrossRef] [Green Version]
- Sugino, T.; Kawase, T.; Onogi, S.; Kin, T.; Saito, N.; Nakajima, Y. Loss Weightings for Improving Imbalanced Brain Structure Segmentation Using Fully Convolutional Networks. Healthcare 2021, 9, 938. [Google Scholar] [CrossRef]
- Sudre, C.H.; Li, W.; Vercauteren, T.; Ourselin, S.; Jorge Cardoso, M. Generalised Dice Overlap as a Deep Learning Loss Function for Highly Unbalanced Segmentations. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Cardoso, M.J., Arbel, T., Carneiro, G., Syeda-Mahmood, T., Tavares, J.M.R.S., Moradi, M., Bradley, A., Greenspan, H., Papa, J.P., Madabhushi, A., et al., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 240–248. [Google Scholar]
- Lin, T.-Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2999–3007. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2021, arXiv:2010.11929. [Google Scholar]
- Wachinger, C.; Reuter, M.; Klein, T. DeepNAT: Deep Convolutional Neural Network for Segmenting Neuroanatomy. NeuroImage 2018, 170, 434–445. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.G.; Conjeti, S.; Navab, N.; Wachinger, C. QuickNAT: A Fully Convolutional Network for Quick and Accurate Segmentation of Neuroanatomy. arXiv 2018, arXiv:1801.04161. [Google Scholar]
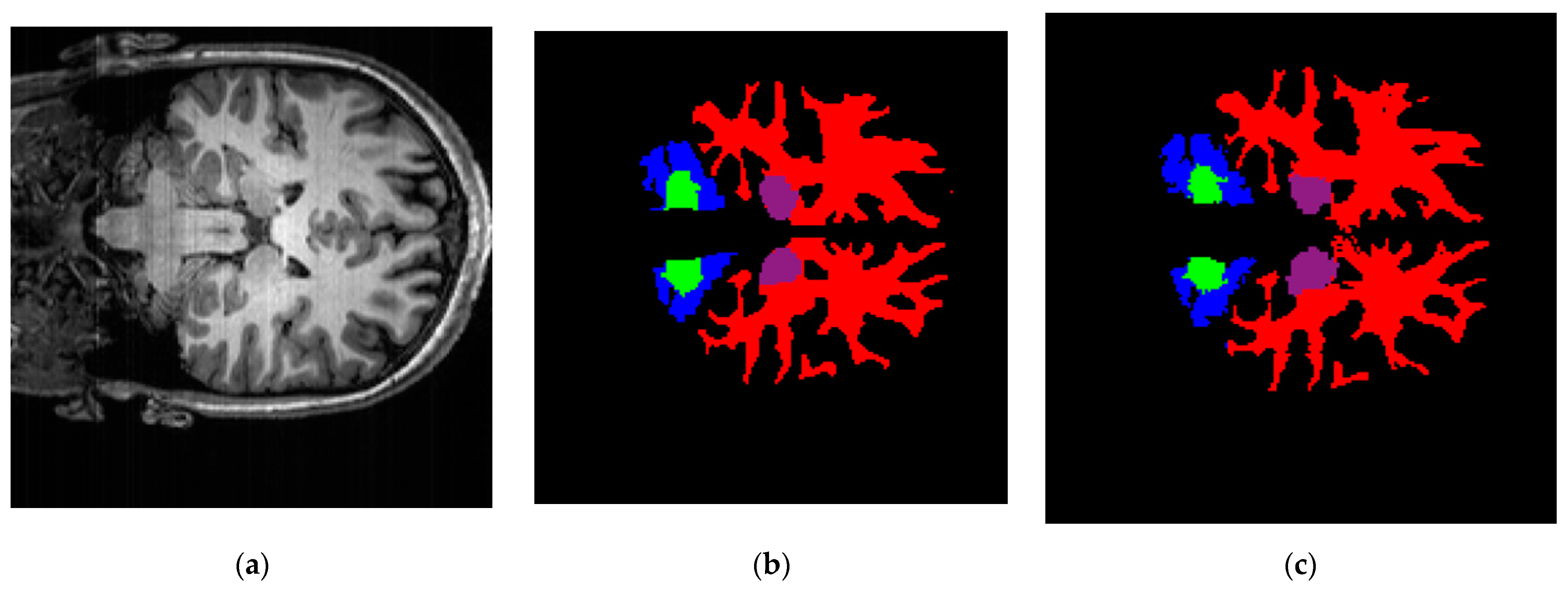

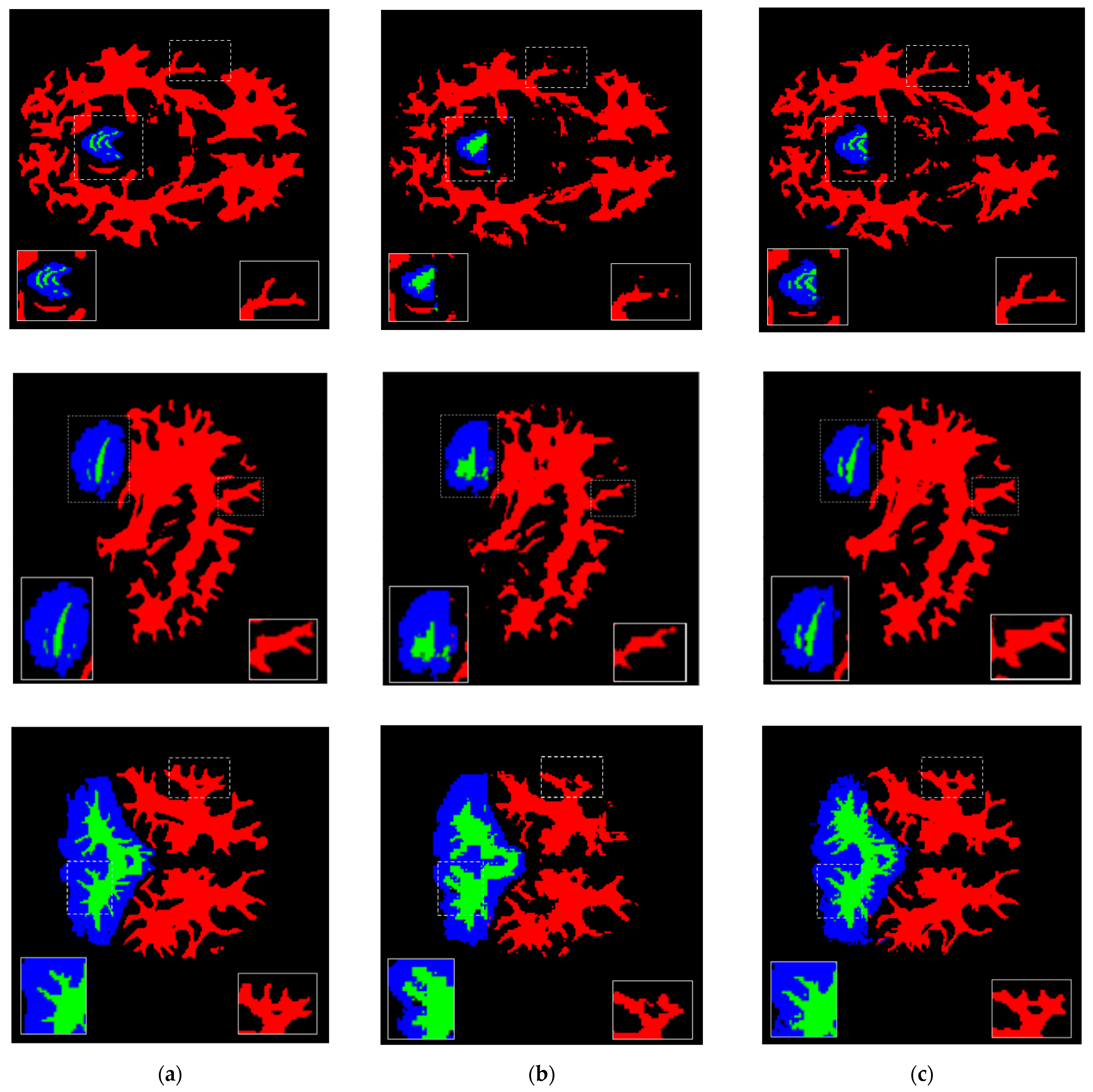
| Brain Structure | Precision | Recall | Dice Score | IoU |
|---|---|---|---|---|
| Left cerebral white matter | 0.95 | 0.91 | 0.93 | 0.86 |
| Right cerebral white matter | 0.97 | 0.89 | 0.93 | 0.86 |
| Left cerebellum white matter | 0.90 | 0.75 | 0.82 | 0.69 |
| Right cerebellum white matter | 0.93 | 0.77 | 0.85 | 0.73 |
| Left cerebellum cortex | 0.87 | 0.82 | 0.84 | 0.73 |
| Right cerebellum cortex | 0.89 | 0.72 | 0.80 | 0.66 |
| Left lateral ventricle | 0.64 | 0.91 | 0.75 | 0.60 |
| Right lateral ventricle | 0.78 | 0.91 | 0.84 | 0.72 |
| Left thalamus | 0.80 | 0.92 | 0.86 | 0.74 |
| Right thalamus | 0.90 | 0.89 | 0.89 | 0.80 |
| Left putamen | 0.85 | 0.84 | 0.85 | 0.73 |
| Right putamen | 0.91 | 0.81 | 0.86 | 0.75 |
| 3rd ventricle | 0.57 | 0.96 | 0.72 | 0.56 |
| 4th ventricle | 0.67 | 0.94 | 0.78 | 0.64 |
| Brain stem | 0.87 | 0.93 | 0.90 | 0.83 |
| Left hippocampus | 0.88 | 0.67 | 0.76 | 0.62 |
| Right hippocampus | 0.89 | 0.80 | 0.84 | 0.73 |
| Left ventral DC | 0.78 | 0.83 | 0.80 | 0.68 |
| Right ventral DC | 0.62 | 0.87 | 0.72 | 0.57 |
| Ctx left caudal middle frontal | 0.84 | 0.43 | 0.57 | 0.40 |
| Ctx right caudal middle frontal | 0.50 | 0.24 | 0.32 | 0.20 |
| Ctx left cuneus | 0.56 | 0.65 | 0.60 | 0.44 |
| Ctx right cuneus | 0.54 | 0.74 | 0.62 | 0.46 |
| Ctx left fusiform | 0.68 | 0.61 | 0.64 | 0.48 |
| Ctx right fusiform | 0.78 | 0.65 | 0.71 | 0.55 |
| Ctx left inferior parietal | 0.64 | 0.54 | 0.58 | 0.42 |
| Ctx right inferior parietal | 0.60 | 0.70 | 0.65 | 0.49 |
| Ctx left lateral occipital | 0.69 | 0.74 | 0.71 | 0.56 |
| Ctx right lateral occipital | 0.73 | 0.69 | 0.71 | 0.56 |
| Ctx left post central | 0.54 | 0.82 | 0.66 | 0.49 |
| Ctx right post central | 0.71 | 0.70 | 0.71 | 0.55 |
| Ctx right rostral middle frontal | 0.57 | 0.81 | 0.67 | 0.50 |
| Ctx left rostral middle frontal | 0.51 | 0.82 | 0.63 | 0.46 |
| Ctx left superior frontal | 0.74 | 0.81 | 0.77 | 0.63 |
| Ctx right superior frontal | 0.77 | 0.79 | 0.78 | 0.65 |
| Ctx left insula | 0.81 | 0.84 | 0.82 | 0.70 |
| Ctx right insula | 0.70 | 0.87 | 0.78 | 0.64 |
| Macro average | 0.75 | 0.78 | 0.75 | 0.63 |
| Weighted average | 0.97 | 0.97 | 0.97 | 0.95 |
| Model | Brain Structures | Mean Dice Score | p-Value |
|---|---|---|---|
| UNet (baseline) | 37 | 0.790 ± 0.0210 | 0.0012850 |
| DenseUNet (finetuned) | 102 | 0.819 ± 0.0110 | 0.0211314 |
| Proposed model | 37 | 0.900 ± 0.0360 | - |
| Model | Segments | Time Per Brain Structure | Mean Dice Score |
|---|---|---|---|
| DeepNAT [78] | 27 | ~133 s (on a Multi-GPU Machine) | 0.906 |
| QuickNAT [79] | 27 | ~0.74 s (on a Multi-GPU Machine) | 0.901 |
| DenseUNet | 102 | 0.64 s (±0.0091 s) (Single GPU Machine) | 0.819 |
| FreeSurfer [79] | ~190 | ~75.8 s | - |
| Proposed model | 37 | 0.038 s (±0.0016 s) (on a Multi-GPU Machine) | 0.903 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laiton-Bonadiez, C.; Sanchez-Torres, G.; Branch-Bedoya, J. Deep 3D Neural Network for Brain Structures Segmentation Using Self-Attention Modules in MRI Images. Sensors 2022, 22, 2559. https://doi.org/10.3390/s22072559
Laiton-Bonadiez C, Sanchez-Torres G, Branch-Bedoya J. Deep 3D Neural Network for Brain Structures Segmentation Using Self-Attention Modules in MRI Images. Sensors. 2022; 22(7):2559. https://doi.org/10.3390/s22072559
Chicago/Turabian StyleLaiton-Bonadiez, Camilo, German Sanchez-Torres, and John Branch-Bedoya. 2022. "Deep 3D Neural Network for Brain Structures Segmentation Using Self-Attention Modules in MRI Images" Sensors 22, no. 7: 2559. https://doi.org/10.3390/s22072559
APA StyleLaiton-Bonadiez, C., Sanchez-Torres, G., & Branch-Bedoya, J. (2022). Deep 3D Neural Network for Brain Structures Segmentation Using Self-Attention Modules in MRI Images. Sensors, 22(7), 2559. https://doi.org/10.3390/s22072559







