Abstract
Magnetic Resonance Imaging (MRI) typically recruits multiple sequences (defined here as “modalities”). As each modality is designed to offer different anatomical and functional clinical information, there are evident disparities in the imaging content across modalities. Inter- and intra-modality affine and non-rigid image registration is an essential medical image analysis process in clinical imaging, as for example before imaging biomarkers need to be derived and clinically evaluated across different MRI modalities, time phases and slices. Although commonly needed in real clinical scenarios, affine and non-rigid image registration is not extensively investigated using a single unsupervised model architecture. In our work, we present an unsupervised deep learning registration methodology that can accurately model affine and non-rigid transformations, simultaneously. Moreover, inverse-consistency is a fundamental inter-modality registration property that is not considered in deep learning registration algorithms. To address inverse consistency, our methodology performs bi-directional cross-modality image synthesis to learn modality-invariant latent representations, and involves two factorised transformation networks (one per each encoder-decoder channel) and an inverse-consistency loss to learn topology-preserving anatomical transformations. Overall, our model (named “FIRE”) shows improved performances against the reference standard baseline method (i.e., Symmetric Normalization implemented using the ANTs toolbox) on multi-modality brain 2D and 3D MRI and intra-modality cardiac 4D MRI data experiments. We focus on explaining model-data components to enhance model explainability in medical image registration. On computational time experiments, we show that the FIRE model performs on a memory-saving mode, as it can inherently learn topology-preserving image registration directly in the training phase. We therefore demonstrate an efficient and versatile registration technique that can have merit in multi-modal image registrations in the clinical setting.
1. Introduction
Clinical decision-making from magnetic resonance imaging (MRI) is based on combining anatomical and functional information across multiple MRI sequences (defined throughout as “modalities”). Multiple imaging biomarkers can be derived across different MR modalities and organ areas. This makes image registration an important MR image analysis process, as it is commonly required to “pair” images from different modalities, time points and slices. Hence, both intra- and inter-modality image registration are essential components in clinical MR image analysis [1], and finds wide use in longitudinal analysis and multi-modal image fusion [2].
Although numerous deep learning (DL) methods have been devised for medical image analysis [2], DL-based image registration tasks have been relatively less explored [3,4]. Among DL-based registration studies, supervised learning-based methods showed promising results. The main disadvantage of supervised learning is that it necessitates laborious and time-consuming manual annotations for model training, even as it is difficult to design generalised frameworks [5,6,7]. Unsupervised DL-based image registration methods gain increasing popularity, as they aim to overcome the need of training datasets with ground truth annotations [3,4]. However, unsupervised learning has been mainly investigated on intra(single)-modal image registration [3,4,8] and on either 3D volumes [9,10,11,12,13] or 2D images [14,15,16,17,18]. Also, previous unsupervised learning methods involve affine registration before training, which is a laborious, time-consuming and computationally expensive task [2,3,4,9,10,11,12,13,14,15,16,17,18].
To date, most previous unsupervised DL methods can perform either affine or non-rigid registration [3,4,9,10,12,13,14,15,16,17,18]. To our knowledge, the only unsupervised DL study that has developed a method for both affine and non-rigid registrations is by de Vos et al. [11]. In this work, DL modelling was performed using two autonomous models in the analysis pipeline to address both affine and non-rigid registrations, whilst also requiring affine transformations before training. Inverse-consistency-based transformation models show improved ability to preserve the contextual and topology information in image registration [19]. Note that a deformable image registration between two images is called inverse consistent, when the correspondence between images is invariant to the order of the choice of the source and target image [19,20,21].
In our work, we demonstrate a bi-directional unsupervised DL model that is capable of performing multi-modal (n-D, where n = 2–4) affine and non-rigid image transformations. As will be discussed in the next sections, we model n-D affine and non-rigid registrations through bi-directional cross-modality synthesis and inverse-consistent spatial transformations. Unlike previous unsupervised learning studies that focus on estimating asymmetric transformations and cannot preserve topology [2,3,4,9,10,11,12,13,14,15,16,17,18], our proposed technique is efficient for both affine and non-rigid image registrations, as demonstrated in multi-modal brain and cardiac image registration experiments.
1.1. Motivation
Unsupervised DL techniques have considerable potential in medical image registration. In this paper we combine up-to-date progress in unsupervised learning with strong prior knowledge about medical image registration: reaching high registration performance, we synchronically perform multi-modal image synthesis and factorised spatial transformations (one per each encoder-decoder channel). This operation allows us to efficiently reach inverse-consistent multi-modal affine and non-rigid image registration of multi-dimensional medical imaging data, making our work efficient and versatile for the medical imaging community.
The FIRE model provides a generalised architecture for registration applications, as the synthesis components of the FIRE model can be customised and re-trained if/when data from additional medical imaging modalities would need to be co-registered. The synthesis factor captures global image modality information, determining how organ topology is rendered in the target image. Between other things, maintaining a representation of the modality characteristics in the synthesis factor of the model provides the ability to potentially model data from multiple modalities. We evaluate this function using the multi-modal MR data of variable anatomical information (see also the “model explainability” section).
Finally, the incorporation of factorised spatial transformations in the FIRE model allows us to learn varied image transformations. We examine whether the FIRE architecture and its associated loss functions can directly output transformation fields for both affine and non-rigid registrations. We demonstrate that the FIRE model architecture efficiently learns to produce inverse-consistent deformations that are intrinsically topology-preserving.
1.2. Overview of the Proposed Approach
Learning image representations and spatial transformations through image synthesis is an area of recent work in medical image analysis [20]. However, there was no previous consideration about the precision of the synthesis process and the versatility of the spatial transformations. This is important in medical image analysis, as both affine and deformable registrations are commonly required for a typical clinical dataset.
Following our previous work [22], we demonstrate a versatile registration method called “FIRE” by explicitly incorporating spatial transformation into our cross-domain bi-directional image synthesis (Figure 1). We show that our method can robustly model 2D/3D and 4D registrations when examined on multi- and intra-modality brain and cardiac MRI, respectively. We provide thorough explanations regarding our model explainability in medical image registration. Moreover, we demonstrate the efficiency of our method on computational time experiments.
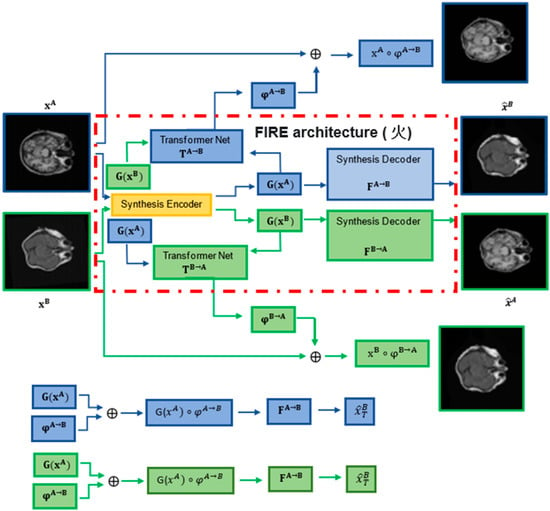
Figure 1.
FIRE model (火, Chinese symbol for “fire”) architecture (red dash-dotted line box). Two synthesis encoders G(xA) and G(xB) extract modality-invariant (latent) representations. Two synthesis decoders FA→B and FB→A map the representations extracted by G(xA) and G(xB) to B and A (images synthesised), respectively (see FA→B and FB→A at the bottom). Finally, two transformation networks TA→B and TB→A model the transformation fields, finalising the FIRE training process.
1.3. Contributions
Our main contributions are as follows:
- With the use of two spatial transformation networks factorised into our cross-domain synthesis, the “火”-shape FIRE model architecture can be trained (through our combined loss functions) end to end, to perform inverse-consistent unsupervised registrations using a single DL network for both inter- and intra-modality registration.
- Incorporating a new affine regularisation loss to eliminate non-rigid transformation where needed, we show that our FIRE model simultaneously learns both affine and non-rigid transformations.
- Overall, the novel FIRE model loss functions and training procedure are designed to allow simultaneous learning for synthesis and registration of 2D, 3D and 4D multi-modal MR data.
- We show that our FIRE model is explainable and provides a comprehensive (affine and non-rigid) framework for future improvements and generalisation.
2. Related Work
In this section we review previous work on registration methods using DL (Section 2.1). We then review inverse-consistent DL transformations (Section 2.2). Finally, we review methods that solve inter-modality registration through image synthesis (Section 2.3).
Since we detail the background and motivation of our work against previous studies in the Introduction section of this paper, here we focus on discussing similarities and differences of our FIRE model against previous methods that are relevant to our application domain.
2.1. Registration Methods Using DL
Early DL registration methods were mostly adaptations of the conventional image alignment scheme, where DL was used to extract convolutional features for guiding correspondence detection across subjects [23], or to learn new similarity metrics across images [24,25].
Reinforcement learning methods have shown promising results, but they are still designed to solve image registration through iterative processes, making them computationally expensive and time-consuming [26,27,28]. As discussed, although DL-supervised learning-based methods have also been promising for image registration, one of their main disadvantages is that they require laborious and time-consuming manual annotations [5,6,7,29].
To overcome the limitations of supervised learning methods, unsupervised learning approaches were recently developed that learn image registration by focusing to minimise the loss between the deformed image and fixed (target) image. The development of a spatial transformer network (STN) in 2015 by Jaderberg et al. has inspired many unsupervised learning methods as it can be inserted into existing DL models [8]. The STN allows explicit manipulation of images within a network and can potentially perform image similarity loss calculations during the training and testing process. Balakrishnan et al. demonstrated Voxelmorph, a 3D medical image registration algorithm using STN within a convolutional neural network (CNN), in which parameters are learned by the local cross-correlation loss function [30,31]. Krebs et al. proposed a probabilistic formulation of the registration problem through unsupervised learning, in which an STN was used within a conditional variational autoencoder [32]. Kim et al. developed CycleMorph, a cycle-consistent DL method for deformable image registration (through an STN component) and demonstrated accurate registration results [33]. Unlike previous techniques [30,31,32,34,35], the CycleMorph model incorporated inverse consistency directly in the training phase [33] to improve model efficiency and robustness.
To date, the only unsupervised DL study that has developed a method for both affine and non-rigid registrations is by de Vos et al. [11]. However, in this work, DL modelling required the incorporation of two autonomous models in the analysis to address both affine and non-rigid registrations, whilst also requiring affine transformations as part of the modelling process (therefore adding complexity and computational cost).
2.2. Inverse-Consistent DL Transformations
Most recent studies are mainly focusing on performing non-rigid deformations [30,31,32,33,34,35,36], whilst requiring computationally expensive and laborious affine transformations in pre-processing using ANTs [30,36,37] or some other software application [20]. Note that affine transformation is inherently inverse-consistent because of the invertibility of the transformation matrix. In the past decade, inverse consistency has mainly been achieved by incorporating diffeomorphic deformation fields [37]. Some of the most popular examples include the diffeomorphic Demons algorithms [38,39].
Several DL diffeomorphic architectures have recently been proposed, such as the Voxelmorph [30,31,34,35], the Quicksilver [38] and the CycleMorph [33]. However, most of these unsupervised diffeomorphic models were tested on intra-modality brain data [30,31,34,35,38]. As already discussed in Section 2.1, the CycleMorph model is still considered the only technique to effectively incorporate inverse consistency directly in the training phase, whilst it was tested on multi-modal (brain MRI and liver CT data) medical imaging data. In our work, we define the output deformation fields with simple inverse-consistency constraints, devising a model that can learn topology-preserving registratrion in an expandable and versatile architecture. It may be important to emphasise that Nielsen et al. recently concluded that the diffeomorphism fails to account for local discontinuities in brain images with pathological conditions [39]. Despite this finding, we should highlight that it is straightforward to integrate diffeomorphic layer components into the FIRE framework if deemed as appropriate (for example, the Voxelmorph velocity field [30,31], or the CycleMorph homeomorphic mapping [33] could potentially be incorporated).
In clinical practice, image registration tasks commonly involve comprehensive image deformations, i.e., a combination of affine and non-rigid registration. In our study, we demonstrate a versatile and efficient DL model that can perform both affine and non-rigid (deformable) registration, and which can learn to produce inverse-consistent and intrinsically topology-preserving image deformations.
2.3. Image Synthesis DL Methods
Image synthesis-based methods have potential in recent DL-based inter-modality registration algorithms. Alongside the recent developments on adversarial cycle-consistent loss [40], most recent methods focus on using CycleGAN for synthesis-based inter-modality registration [20,41]. Wei et al. performed image registration by devising CycleGAN-based image synthesis as a separate processing step prior to registration [41]. Qin et al. proposed a bi-directional unsupervised training framework, which is conceptually close to our model. However, our FIRE model is simultaneously learning synthesis and registration on a memory-saving architecture that requires less hyperparameters when balancing all different losses (see Section 3). Moreover, the FIRE model is evaluated on 2D, 3D and 4D data involving different organs (brain and heart; note that 4D here denotes periodic heart deformations observed across different time phases of cardiac cine-MRI).
3. Methods
Our proposed model can be described as a bi-directional cross-domain image synthesis structure, with two factorised spatial transformation components (one per each encoder-decoder channel) [22].
Each time two images xA and xB are processed, the FIRE model learns φA→B and φB→A transformations, warping xA and xB into xA ◦ φA→B and xB ◦ φB→A, via the transformation networks TA→B and TB→A. In parallel, two synthesis encoders G(xA) and G(xB) extract modality-invariant latent representations through the synthesis decoders FA→B and FB→A, mapping the representations extracted by G(xA) and G(xB) to the synthesed images B and A, respectively.
3.1. Architecture
Our model involves the following components (Figure 1): (a) one synthesis encoder G, which extracts modality-invariant representations G(xA) and G(xB); (b) two synthesis decoders FA→B and FB→A, which map G(xA) and G(xB) to B = FA→B(G(xA)) and A = FB→A(G(xB)) (synthesised images); and (c) two transformation networks, TA→B and TB→A, which model the transformation fields φA→B = TA→B(G(xA), G(xB)) and φB→A = TB→A(G(xB), G(xA)). Note that during training, G(xA) and G(xB) are warped into G(xA) ◦ φA→B and G(xB) ◦ φB→A before being used to generate the synthesised images: = FA→B(G(xA) ◦ φA→B) and = FB→A(G(xB) ◦ φB→A).
3.2. Encoder and Decoder for Synthesis
The structures of the encoder and decoders were inspired by the design of the CycleGAN [41], which includes a set of downsampling and upsampling convolutional layers connected through a series of Resnet blocks.
We modified the architecture of the CycleGAN generator by adding two decoders in the generator and by eliminating the discriminator (instead of having one decoder in the generator followed by a discriminator, as in the original CycleGAN, Figure 1 and Figure 2) [41]. In our model, we did not need to recruit adversarial learning through the discriminator, since by involving two decoders and geometrical mapping via our STN-derived deformations in the cross-domain synthesis learning, our model was designed to match the (pixel intensity and geometrical) distribution of generated images to the data distribution in either target domain.
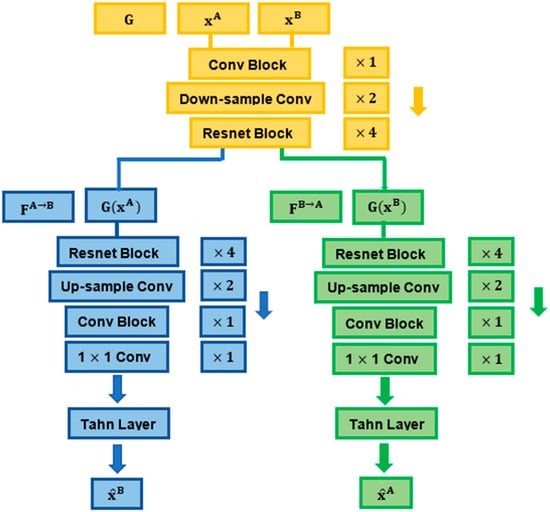
Figure 2.
Details of the encoder and decoders used for the synthesis process.
The encoder G contained one convolutional block (7 × 7, 64) with stride 1, two downsampling convolutional layers (3 × 3, 128; 3 × 3, 256) with stride 2 and four Resnet blocks of the same size (3 × 3, 256) with stride 1. The decoders contained four consecutive Resnet blocks of the same size (3 × 3, 256) with stride 1, two convolutional layers used for upsampling (3 × 3, 128; 3 × 3, 64) with stride 1 and a convolutional block (7 × 7, 3) with stride 1. The synthesised output was finally produced by a 1 × 1 convolutional layer, followed by a Tanh layer [22].
The downsampling layers (in the encoder) encode the images down to an abstract latent representation. In this process, the model learns weights that enable the reducing of the spatial resolution of the feature (image pixel) maps. The output feature maps derived from downsampling are then passed through a series of Resnet blocks with skip connections to further interpret and process their underlying information while addressing the vanishing gradient problem that may occur in large networks [20,41]. Through the upsampling layers, the model learns to transpose (reverse) convolutions, decoding the latent representations back to the size of the ouput image.
3.3. Transformation Network
The transformation networks of our model, TA→B and TB→A, can learn both affine and non-rigid transformations (denoted as φaf and φnr, respectively, Figure 3). Each transformation network consists of a subnetwork for affine transformation followed by a subnetwork for nonrigid transformation, as shown in Figure 3.
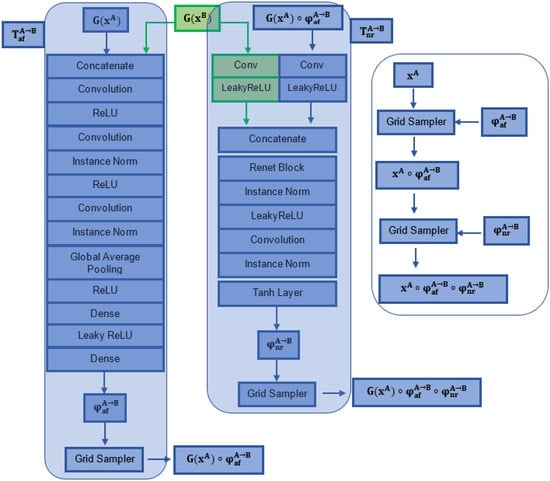
Figure 3.
Affine and non-rigid spatial transformation networks incorporated in the FIRE model.
To derive deeper geometrical inter-relationships from the processed data, we modified the start of the STN structure by adding three convolutional layers with instance normalization and standard LeakyReLU layers (Figure 3). In specific, the affine transformation network Taf uses an initial convolutional layer of size (1 × 8, 7) and stride 2, followed by two “convolutional layer + ReLU + Instance Norm” blocks (the size of the first and second convolutional layers used were 8 × 10, 5 and 16 × 20, 2, respectively, with stride 2). Following the two “convolutional layer + ReLU + Instance Norm” blocks, two Dense (fully connected) layers were recruited to compute the parameters of the affine transformation matrix (2 × 3 for 2D; 3 × 4 for 3D transformations) [8]. Essentially, the input of the affine subnetwork is the concatenated features obtained from the two input images. The convolutional features derived from the third convolutional layer were resampled into a fixed-size feature vector using Global Average Pooling [8,22], followed by two fully connected layers that were used to generate the transformation matrix.
Non-rigid transformation network TnrA→B receives the affinely transformed feature G(xA) ◦ TnrA→B and the untransformed feature G(xB) as inputs, and subsequently processes them in parallel layers (Figure 3). The features extracted are finally concatenated and used as inputs into a ResNet block with instance normalization and LeakyReLU layers. To produce the non-rigid deformation φnrA→B, a final convolutional layer and a Tanh activation function are implemented. The Tanh layer is performed on a normalised coordinate system: a coordinate p ∈ [−1, 1] n exists for an n-D image [22].
As described, we factorised two separate transformation networks for each encode-decoder channel in our FIRE model, which means that the above processes described for the A→B (affine and non-rigid) transformations are identical for the B→A affine and non-rigid transformations as well.
3.4. Training Procedure and Overall Loss Function
Our FIRE model is trained for simultaneously learning mutually inverse registration and synthesis tasks across both “A→B” and “B→A” directions.
The A→B image synthesis process synthesised images and , so that is registered with xA and is identical to the target image xB. The A→B transformation learned by TA→B is applied to the features G(xA) for synthesis purposes, and subsequently to image xA for registration.
The B→A image synthesis process performs backward registration and synthesis. All parameters of the entire network are updated end to end in the training process, and only G and a single transformation network are required to deform the moving image in the testing phase, for either modality input image.
To train the proposed FIRE model, a synthesis loss ( and a registration loss (reg) are recruited. A regularisation process () is introduced to perform topology-preserving deformation and spatial smoothing [22]. Combining these terms, the overall loss function of the proposed model is:
As described in the next subsections, we formulate most of the terms in syn, reg and using root-mean-square (RMS) calculated on normalised inputs. As a result, apart from regularising the smoothness of the non-rigid deformation field, no hyperparameter is required to balance different losses, as detailed below.
3.5. Synthesis Loss
There are four synthesis loss terms involved, each one supporting a different purpose (see Figure 4). First, to perform accurate cross-domain synthesis, the following synthesis accuracy loss was defined (by implementing the RMS error):
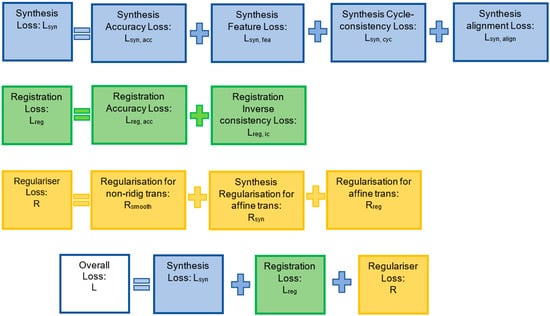
Figure 4.
Graphical description of all losses defined in the FIRE model. Full descriptions and mathematical details are provided in Section 3.5, Section 3.6 and Section 3.7 (in Methods).
Note that for image synthesis, the synthesised images and aim to be identical to the target images xA and xB, respectively, which is defined in Equation (2).
Second, G is designed to learn modality-invariant features. To represent this, we defined the following feature loss, which aims to minimise the RMS error between G(xA) and G(xB) , as well as G(xB) and G(xA) :
Equation (3) was designed to minimise the error between the G outcomes from either modality and the transformations learned in the G outcomes of the other modality.
Third, the cycle-consistency loss used in CycleGAN [20,41] has been proven to be critical for its superior performance in cross-domain image synthesis. For robust cross-modality synthesis performance, a cycle-consistency loss was therefore designed:
Essentially, Equation (4) encourages FB→A (G()) = xA and FA→B (G()) = xB, enforcing these mappings to be reverses of each other (by minimising the error of either mapping procedure to the inputs xA and xB).
Finally, aligning x and is important to transfer the geometric correspondence from either xA or xB (learned through the transformation networks), and to the synthesis process. To this end, the synthesis alignment loss was defined as:
3.6. Registration Loss
To perform synthesis and registration, features extracted by G were transformed and registration was then performed through the following transformations to the input images xA and xB: φA→B = φafA→B ◦ φnrA→B and φB→A = φafB→A ◦ φnrB→A. Because the output of the synthesis process F•→• (G(·)) is aligned with its input, the synthesised image obtained from a transformed xA image should be identical to xB. Based on this, we defined the following registration accuracy loss:
Finally, for inverse-consistent registration, the transformations φA→B and φB→A should be mutually inverse; thus, the following inverse-consistency loss was defined:
This inverse-consistent loss can be seen as a transformation-oriented cycle-consistency loss, which encourages the composition of mutual mappings from the moving image to the fixed image on a bi-directional mode. For example, for xA (moving image) to xB (fixed image) registration, it minimises the error for both and mappings.
The overall entire registration loss was computed as:
In practice, the non-rigid transformation field φnr was calculated on G(x) and linearly resampled before being applied to image x.
3.7. Regularisation
Inspired by conventional image registration, in which regularisation is commonly an integral part [11,22], we also add regularisation terms in both affine and non-rigid transformations. The main novelty in terms of regularisation in our work is that we incorporate separate regularization terms for both affine and non-rigid transformations in our model. At that stage, it is important to note a fundamental difference between affine and non-rigid transformation [11,30,31,32,33]: affine registration recruits global transformation fields, whilst non-rigid registration recruits mainly local transformation fields [11]. Non-rigid registration may contain affine transformations. However, affine registration should not involve non-rigid transformations.
In that context, we first add a bending energy penalty term to reinforce smooth displacements for both non-rigid transformation fields φnrA→B and φnrB→A, as follows:
where ∇ is the Laplacian operator.
Second, because our FIRE model is designed to also calculate affine registrations, we propose to induce a separate regularization term to eliminate non-rigid transformations when affine transformation is present in the data. During image synthesis, the affinely transformed features G(xA) ◦ φafA→B and G(xB) ◦ φafB→A can be used as inputs into the synthesis decoders to obtain FA→B(G(xA) ◦ φaf A→B) and FB→A(G(xB) ◦ φafB→A). The regularisation of the synthesis is then computed as:
Similarly, a regularisation for registration, , is defined using the affinely deformed xA and xB:
The overall regularisation of the FIRE model was [22]:
where λ is the scaling parameter for , which is the only hyperparameter used in the FIRE loss. We should note that to register n-D images, λ = 22n/10N where N is the number of points in the input image. It is important to also emphasise that when non-rigid transformation is present, the model uses all three parameters, and , from Equation (13). When affine registration is present, the model focuses on using the and terms (since the Laplacian operator is not activated during affine registration).
3.8. Optimization
Computing Lreg requires the input of the transformed images into G, which creates a circular computing graph. Furthermore, different networks in the proposed FIRE model show different behaviours in the training process. For example, Taf•→• is more sensitive to changes in L, compared to G and F•→•.
To address this issue, we implemented three Adam optimisers to separately optimise parameters for (a) Taf•→•, (b) Tnr•→• and (c) the synthesis encoder/two decoders. We used a uniform training procedure regardless of the size of the datasets. To optimise speed of convergence, learning rates for training Taf•→• and Tnr•→• and G/F•→• were set to 10−5, 5 × 10−5 and 10−4, respectively. The FIRE model was trained end to end for 144,000 iterations for both datasets (empirical observation 1: for both brain and cardiac data, the total duration of the training phase was approximately 15–18 h on a Tesla P40 GPU with 24 G memory (about 7.5–9 h per dataset); empirical observation 2: for the cardiac data, model convergence was reached in the first 2–3 h, but the model was kept under training for all 144,000 iterations).
4. Experiments
The performance of our proposed model was evaluated using multi- (for inter-modality registration) and single- (for intra-modality registration) modal MR data.
4.1. MRBrainS Data
For multi-modality registration, the training data from the MRBrains13 (http://mrbrains13.isi.uu.nl/, last accessed on 1 February 2022) and the MRBrains18 (https://mrbrains18.isi.uu.nl/, last accessed on 1 February 2022) Challenges were fused. The fused dataset contained multi-modality brain MR data.
In detail, the dataset consisted of 3D T1-weighted, T2-Fluid-attenuated IR (T2-FLAIR) and inversion recovery (IR) data from 12 subjects, acquired using 3T MRI. The 3D T1, T2-FLAIR and IR datasets included 192, 48 and 48 slices per patient, respectively. The 3D T1 and IR data were already co-registered to the T2-FLAIR data. Images across all modalities had a voxel size of 0.958 × 0.958 × 3.000 mm3.
To assess model performance, manual annotations from three brain anatomical structures were used: the brain stem (BS), the cerebellum (Ce) and the white matter (WHM). For the training, validation and testing processes, we used all MRI slices from eight, one and three patients, respectively (Table 1). To perform 3D and 2D registration, all data were resampled to 1.28 mm3 per voxel.

Table 1.
Total number of images for training, validation and testing per brain and cardiac dataset. Note that for the cardiac data, only two images per cardiac slice contained annotations and were evaluated at testing.
In the registration process, 2D and 3D registration was performed between T1 and T2-FLAIR data, and 2D registration between IR and T2-FLAIR data. During training, moderate-to-strong (20–50% change across at least one dimension) affine and non-rigid transformations were randomly applied to the moving and fixed images. In the testing phase, each of the T1 and IR data were randomly transformed 20 times and were subsequently allowed to be registered to the corresponding T2-FLAIR data.
4.2. ACDC Data
To perform intra-modality registration, 4D cardiac cine MRI data from the 2017 ACDC (https://www.creatis.insa-lyon.fr/Challenge/acdc, last accessed on 1 February 2022) Challenge were used. Note that the fourth dimension here describes temporal resolution. The voxel size was between 1.37–1.68 × 1.37–1.68 × 5–8 mm3, and each 4D image has 28–40 phases covering the cardiac cycle.
In specific, the training dataset contained MRI data from 100 patients with different cardiovascular pathologies, with manual annotations of the myocardium and the left ventricle per patient at two cine-MR phases (images) per slice.
To train the FIRE model, we used all MRI phases per slice. To test the model, only the two annotated phases per slice were evaluated. For the training, validation and testing process, MRI data from 60, 10 and 30 patients were used, respectively (Table 1).
4.3. Evaluation Metrics and Baselines
Both the MRBrainS and ACDC datasets provided manual annotations (ground truths). To evaluate our method, we used the Dice metric to measure the overlap of the moving and fixed annotations. Increased Dice scores represent high registration performance and vice versa.
Although there are recent developments in image registration (described in the Introduction and detailed in Related Work), the mutual information (MI) and Symmetric Normalization (SyN) techniques through using the Advanced Normalization Toolbox (ANTs) (http://stnava.github.io/ANTs/, last accessed on 1 February 2022) [37] are still considered the current reference standard techniques for affine and non-rigid registration, respectively [4,9,10,11,12,13,14,15,16,17,18,30,31,32,33,34,35,36,37,38,39,40,41].
Hence, the proposed model is evaluated against the standard MI and SyN techniques to assess affine and non-rigid registration performance in multi-slice multi-modality brain and multi-phase/multi-slice single-modality cardiac MRI data, respectively.
5. Results
5.1. Inter-Modality Registration
Initially, 2D and 3D T1 to T2-FLAIR image registration was examined. The proposed model was consistent in achieving higher scores against the SyN method for all brain anatomical areas investigated (Table 2). As presented in Table 2, the proposed model outperformed the SyN method across all 2D and 3D T1 to T2-FLAIR image registration experiments examined. These results were maintained when both affine and deformable image registration were evaluated.

Table 2.
Results obtained from the 2D and 3D T1 to T2-FLAIR registration, on the MRBrainS data. Dice scores were measured on the brain stem (BS), cerebellum (CE) and white matter (WHM). Standard deviations are included within the parenthesis. Bold shows our FIRE model-derived results.
A visual representation of the T1 to T2-FLAIR registration is illustrated in Figure 5. In this representation, the FIRE model shows better alignment between the outer contour (in the extracerebral space) outlining the cerebrospinal fluid (shown with blue) and the actual brain tissue delineation, versus the Syn technique.
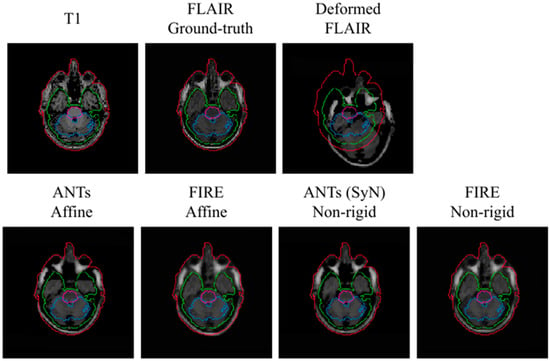
Figure 5.
Visual representation of the T1 to T2-FLAIR registration using the MRBrainS data. Brain stem, cerebellum and white matter used to assess registration performance are illustrated with purple, blue and green, respectively. Red illustrates the outer contour outlining the entire head.
Subsequently, IR to T2-FLAIR registration was evaluated. Our proposed model consistently reached improved registration performance against the Syn method: a mean Dice score of 0.68 (0.3), 0.69 (0.2) and 0.70 (0.3) for the BS, Ce and WHM brain structures was reached, respectively. A visual representation of the IR to T2-FLAIR registration is illustrated in Figure 6.
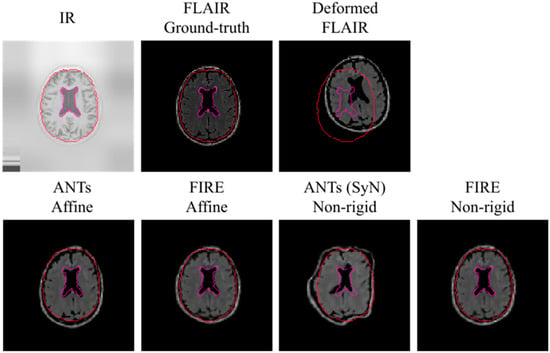
Figure 6.
Visual representation of the IR to the T2-FLAIR registration using the MRBrainS data. To increase clarity because of having less anatomical information in IR and T2-FLAIR data, we use the outer brain region and ventricles (with red and purple respectively) to visually demonstrate registration results. Note that the upper left image shows an IR image before pixel normalisation occurred during model fitting.
It is important to note that the average Dice score obtained across all brain anatomical areas was below 0.45 when the SyN technique was evaluated: there was a mean Dice score of 0.43 (0.2), 0.42 (0.3) and 0.44 (0.3) for the BS, Ce and WHM brain structures, respectively. These results were repeated when a grid search for the Syn method within ANTs was carefully examined, making it impossible to derive a visual alignment of the IR to T2-FLAIR registration.
5.2. Intra-Modality Registration
Intra-modality registration was then investigated using 4D cardiac cine-MRI data. On the left ventricle anatomical areas, the FIRE model and the Syn method showed high and comparable Dice scores (Table 3, Figure 7). On the myocardial anatomical areas, the FIRE model marginally outperformed the Syn method (Table 3).

Table 3.
Results on ACDC data. Dice scores computed on left ventricular endocardium (LVe) and myocardium (Myo). Standard deviations are included within the parenthesis. LVe: left ventricle, Myo: myocardial tissue. Bold shows higher model performance, versus the other model.
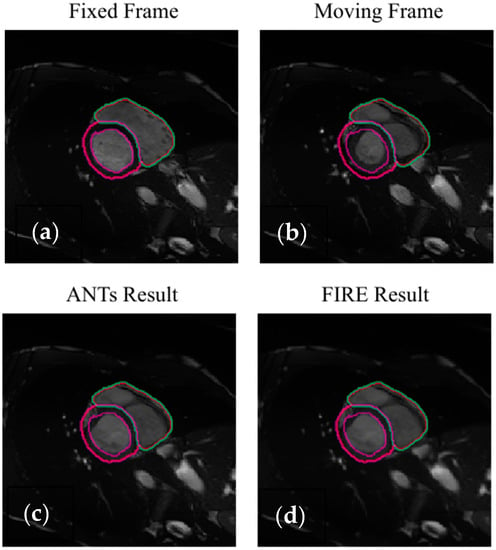
Figure 7.
Illustration of the intra-modality image registration using ACDC data (the purple and magenda contours demonstrate the endocardial and epicardial left ventricle delineations, respectively). The red and green contours illustrate the right ventricle delineations, respectively (not used in the analysis due to showing small changes across the cardiac cycle). Full diastole (a,c) and systole (b,d) are illustrated.
5.3. Computational Times
We methodologically assessed the computational costs required for each image registration process using the FIRE and the baseline SyN method implemented in ANTs. For both the FIRE model and SyN methods, the running times at the testing phase were calculated on a CPU system (Intel(R) Xeon(R) Silver 4112 CPU @ 2.60 GHz, RAM: 256 GB). Furthermore, the GPU-accelerated performance of our FIRE model was also measured using a Tesla P40 GPU with 24 G memory. Note that GPU acceleration is not available for the SyN method within ANTs.
Average running times for registering 3D and 2D data were obtained from 30 volumes, each volume containing 48 2D slices. For the 4D cardiac MR data provided in the ACDC dataset, we computed the average time for registering 10 3D volumes (of 10 slices each) at full systole to their first cardiac cine MR phase representing full diastole, therefore using 100 image pairs in total. All results are shown in Table 4.

Table 4.
Mean computational times of the FIRE and the Syn method in the testing phase. The parentheses show standard deviations.
Under the CPU mode, our FIRE model is consistently faster compared to the baseline Syn method, for both affine and non-rigid registration of the 4D cardiac data. The FIRE model shows comparable speed with the baseline in non-rigid registration of 3D and 2D brain data, whilst it was only slower for affine registration of the 3D brain data (Table 4).
When accelerated by a GPU system, our FIRE model considerably reduced the computational cost (by at least 30 times), when examined on 2D and 3D brain data. The FIRE model saved over 99.7% running time compared to the SyN method, while achieving higher accuracy in non-rigid registration of the 4D ACDC data. Bold shows improved computational time, versus the other technique.
6. Discussion
We demonstrate that the FIRE method is an efficient DL model that performs accurate and fast inter- and intra-modality affine and is capable of both affine and non-rigid image registration. The FIRE model efficiently learns inverse-consistent topology-preserving deformations when evaluated on 2D and 3D multi-modal brain MR and 4D single-modal cardiac MR data, showing higher accuracy and robustness against the reference standard, the Syn method.
6.1. Inter-Modality Registration on Brain MRI
We showed that when both 2D and 3D image registration were examined on T1 to T2-FLAIR registration, the proposed model was consistent in showing higher performance against the Syn technique across all brain structures assessed (Table 2). This finding indicates that the proposed model can robustly model different anatomical and semantic features in these multi-tissue/multi-modal MR data; thus behaving as a brain tissue and MR modality-agnostic model (for T1 to T2-FLAIR registration).
The IR to T2-FLAIR image registration was compromised by the lack of anatomical information in either input modality. Implementing the Syn technique reference standard, it was not possible to reach accurate image registration results on quantitative assessments (all DICE scores were lower than 0.45), whilst we did not reach optimal alignments on visual assessments (Figure 6). It is important to note that the Syn method is the reference standard baseline technique for model evaluation in image registration [3,4,9,10,11,12,13,14,15,16,17,18,30,31,32,33,34,35,36,37,38,39,40,41]. Despite this, our FIRE model outperformed the Syn method and was able to achieve moderate mean Dice scores across all brain structures (Section 5.1).
Moreover, when examined in the testing phase, the FIRE model showed comparable speed with the baseline in non-rigid registration of 3D and 2D brain data (Table 4). The only case for which the FIRE model was slower was when affine registration of 3D brain data was measured. Nevertheless, all FIRE model-derived affine and non-rigid registration processes for all 3D brain volumes were performed in less than 40 s.
The FIRE model can therefore perform multi-modal affine and non-rigid registration on a memory-saving mode, without requiring supercomputers or GPU systems in the testing phase.
6.2. Intra-Modality Registration on Cardiac MRI
We demonstrated that the FIRE model can optimally model deformable transformations in cardiac cine-MR data (Table 3). Although there were only two images segmented across each cine-MR dataset (used in the testing phase), these represent the maximum geometric difference within each cardiac data-set (one segmented image is always on full diastole and the other on full systole within ACDC data). Moreover, despite the local displacements between diastole and systole being relatively small (in the 3D space), the tissue deformation is the largest within cardiac cine-MR data. In other words, this means that the cardiac tissue is non-uniformly non-rigidly deformed across phases, with the maximum deformation occurring between diastole and systole (Figure 7). Hence, while using all cardiac phases (in between diastole and systole) in the training phase (representing 4D information), our results in the testing phase show that the FIRE model can optimally learn to perform accurate registration between largely deformed geometries (different cardiac shapes between systole and diastole).
In addition, the computational cost for the FIRE model was substantially lower compared to the Syn method, when non-rigid 4D registration of cardiac MR data was examined. Thus, the FIRE model demonstrated overall improved registration speed when dealing with 4D data, which can be particularly important across numerous cardiac MR applications involving time series [41,42,43,44]. To our knowledge, the only other technique that can effectively perform inverse-consistent registration directly in the training phase is the CycleMorph model [33]. In our work, we define the output deformation fields with simple inverse-consistency constraints, devising a model that can inherently learn topology-preserving image registration in a consistent mode (via the synthesis encoder G and a single transformation network).
6.3. FIRE Model Explainability
Despite recent developments in DL, inter-modality image registration is not yet widely investigated using deep networks [30,31,33,34]. A major reason is the absence of explainability regarding why a DL model learns and/or where it fails to learn anatomical image information and semantics. This becomes more evident in image registration, as the lack of standardised annotations (ground truths) may have diversified methods for model evaluation and thus, discouraged model explainability [5,6,7,26,27,28,29,30,31,32,33,34,35,38,41]. Our main consideration here is that DL explainability can help to interpret model-data interrelationships before (or beyond) deriving class activation maps [45,46], which can encourage thorough (and explainable) DL investigations in inter-modality image registration. We showed that the FIRE model can learn anatomical and semantic representations across modalities, and demonstrated improved performance for inter-modality image registration, versus the SyN method.
It is important to note that T1, T2-FLAIR and IR are routinely used MR sequences, but there are fundamental disparities in terms of the MR physics involved and, thus, their imaging content [47]. These MR modalities are optimised to provide different imaging information in clinical MRI: T1 is designed to provide detailed anatomical information, whereas IR and T2-FLAIR are primarily implemented to extract functional information [47]. In T1 to T2-FLAIR registration, our cross-domain synthesis component (encoder/decoders) can effectively leverage anatomical information from the T1 data and can subsequently map this information to complementary functional T2-FLAIR information. This complementary anatomical-functional information is compromised in the IR to T2-FLAIR data registration during cross-domain synthesis. Hence, not the anatomical information per se, but the presence of (heterogeneous) complementary anatomical-functional information makes the multi-modal transformations invertible in relation to each other, which in turn encourages our model to produce inverse-consistent topology-preserving image registrations. Although not in the scope of the current study, further work is underway to quantify this complementary information in our model, through gradient-weighted class activation mapping (Grad-CAM) [48], across each of the model main components (enconder, STN, decoders).
We should also highlight that there can be multiple methodologies by which it can become possible to enrich “poor” anatomical information within a dataset, towards enhancing inverse-consistency through complementary information. For example, an accurate semi-supervised inter-modality learning model could be explored to derive anatomical annotations and guide the registration process [49,50], hence, potentially adapting our technique for functional (non-anatomical) MR modalities [49,50]. Other active contour-based segmentation algorithms for fast image segmentation have shown excellent segmentation accuracies even for images with large noise interference and intensity inhomogeneities [51,52,53], and therefore may be applicable to enrich anatomical information in multi-modal registration of imaging modalities with decreased anatomical information.
7. Conclusions
We have demonstrated a robust method that efficiently models inter- and intra-modality image registration through bi-directional cross-domain synthesis and factorised spatial transformation. To our knowledge, parallelising cross-domain synthesis while modelling spatial transformation to dynamically learn anatomical and latent representations simultaneously across modalities has not been previously investigated.
Our work showed the efficiency of our methodology as we improve intra- and inter-modality registration by maintaining fast computational times in the testing phase, across all registration tasks.
In broader terms, the main significance of our work is the simultaneous bi-directional cross-domain image synthesis and spatial transformation per synthesis channel, which enables us to diversify our technique across numerous multi-modality medical image analysis scenarios. Through providing a model explainability framework, we can suggest that it is possible to adapt our technique to learn anatomical-semantic information when we have at least one dataset with enriched anatomical information. This means that we can customise our methodology to address image registration on additional anatomically enriched imaging data, such as with computed tomography (CT) or ultrasound data, next to MRI.
To date, the main limitation of our work is that we have only assessed multi-modal MRI data. However, we examined two organ areas (brain and heart), intra- and inter-modality registration, and investigated 2D, 3D and 4D registrations frames. This inspired our future work as we are currently investigating model generalisation frameworks to allow further multi-organ and multi-modal applications. In conclusion, we show here that our methodology demonstrated improved performance and efficiency against the current standard Syn method, thus presenting a versatile image registration technique that may have merit in the clinical setting.
Author Contributions
C.W. conceptualised the work. C.W., G.Y. and G.P. worked on data analysis, curation, engineering and investigation. Writing, review and editing were performed by all authors. G.P. edited the final draft and performed project management. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported in part by the British Heart Foundation (TG/18/5/34111, PG/16/78/32402), the European Research Council Innovative Medicines Initiative (101005122), the European Commission H2020 (952172), the Medical Research Council (MC/PC/21013) and the UKRI Future Leaders Fellowship (MR/V023799/1).
Informed Consent Statement
We confirm that we used public data for which all subjects gave informed consent to use their data for research purposes.
Data Availability Statement
For our data analysis, we used publicly available datasets and we explicitly describe how to access these data. Code will be made available at GitHub.
Conflicts of Interest
G.P. is currently an employee of Pfizer. The other authors have no conflict of interest to disclose.
References
- Rueckert, D.; Schnabel, J.A. Medical image registration. In Biomedical Image Processing; Springer: Berlin, Germany, 2010; pp. 131–154. [Google Scholar]
- Zhou, S.K.; Greenspan, H.; Davatzikos, C.; Duncan, J.S.; Van Ginneken, B.; Madabhushi, A.; Prince, J.L.; Rueckert, D.; Summers, R.M. A Review of Deep Learning in Medical Imaging: Imaging Traits, Technology Trends, Case Studies with Progress Highlights, and Future Promises. Proc. IEEE 2021, 109, 820–838. [Google Scholar] [CrossRef]
- Fu, Y.; Lei, Y.; Wang, T.; Curran, W.J.; Liu, T.; Yang, X. Deep learning in medical image registration: A review. Phys. Med. Biol. 2020, 65, 20TR01. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haskins, G.; Kruger, U.; Yan, P. Deep learning in medical image registration: A survey. Mach. Vis. Appl. 2020, 31, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.; Yang, J.; Zhang, J.; Nie, D.; Kim, M.; Wang, Q.; Shen, D. Deformable image registration based on similarity-steered cnn regression. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec City, QC, Canada, 10–14 September 2017; Springer: Cham, Switzerland, 2017; pp. 300–308. [Google Scholar]
- Krebs, J.; Mansi, T.; Delingette, H.; Zhang, L.; Ghesu, F.C.; Miao, S.; Maier, A.K.; Ayache, N.; Liao, R.; Kamen, A. Robust non-rigid regis-tration through agent based action learning. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec City, QC, Canada, 10–14 September 2017; Springer: Cham, Switzerland, 2017; pp. 344–352. [Google Scholar]
- Rohé, M.M.; Datar, M.; Heimann, T.; Sermesant, M.; Pennec, X. Svf-net: Learning deformable image registration using shape matching. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec City, QC, Canada, 10–14 September 2017; Springer: Cham, Switzerland, 2017; pp. 266–274. [Google Scholar]
- Jaderberg, M.; Simonyan, K.; Zisserman, A. Spatial transformer networks. Adv. Neural Inf. Process. Syst. 2015, 28, 2017–2025. [Google Scholar]
- Fan, J.; Cao, X.; Wang, Q.; Yap, P.-T.; Shen, D. Adversarial learning for mono- or multi-modal registration. Med. Image Anal. 2019, 58, 101545. [Google Scholar] [CrossRef]
- Fan, J.F.; Cao, X.H.; Yap, E.A.; Shen, D.G. BIRNet: Brain image registration using dual-supervised fully convolutional networks. Med. Image Anal. 2019, 54, 193–206. [Google Scholar] [CrossRef]
- De Vos, B.D.; Berendsen, F.F.; Viergever, M.A.; Sokooti, H.; Staring, M.; Isgum, I. A deep learning framework for unsupervised affine and deformable image registration. Med. Image Anal. 2019, 52, 128–143. [Google Scholar] [CrossRef] [Green Version]
- Elmahdy, M.S.; Jagt, T.; Zinkstok, R.T.; Qiao, Y.; Shahzad, R.; Sokooti, H.; Yousefi, S.; Incrocci, L.; Marijnen, C.A.M.; Hoogeman, M.; et al. Robust contour propagation using DL and image registration for online adaptive proton therapy of prostate cancer. Med. Phys. 2019, 46, 3329–3343. [Google Scholar] [CrossRef] [Green Version]
- Lei, Y.; Fu, Y.; Wang, T.; Liu, Y.; Patel, P.; Curran, W.J.; Liu, T.; Yang, X. 4D-CT deformable image registration using multiscale unsu-pervised DL. Phys. Med. Biol. 2020, 65, 085003. [Google Scholar] [CrossRef]
- Vos, B.; Berendsen, F.F.; Viergever, M.A.; Staring, M.; Išgum, I. End-to-end unsupervised deformable image registration with a convolutional neural network. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; DLMIA 2017, ML-CDS; Lecture Notes in Computer Science; Cardoso, M., Ed.; Springer: Berlin, Germany, 2017; Volume 10553, pp. 204–212. [Google Scholar]
- Ferrante, E.; Oktay, O.; Glocker, B.; Milone, D.H. On the Adaptability of Unsupervised CNN-Based Deformable Image Registration to Unseen Image Domains. In Proceedings of the International Workshop on Machine Learning in Medical Imaging, Granada, Spain, 16 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 294–302. [Google Scholar]
- Sheikhjafari, A.; Noga, M.; Punithakumar, K.; Ray, N. Unsupervised deformable image registration with fully connected gener-ative neural network. In Proceedings of the 1st Conference on Medical Imaging with Deep Learning (MIDL 2018), Amsterdam, The Netherlands, 4–6 July 2018. [Google Scholar]
- Jiang, Z.; Yin, F.-F.; Ge, Y.; Ren, L. A multi-scale framework with unsupervised joint training of convolutional neural networks for pulmonary deformable image registration. Phys. Med. Biol. 2019, 65, 015011. [Google Scholar] [CrossRef]
- Nguyen-Duc, T.; Yoo, I.; Thomas, L.; Kuan, A.; Lee, W.C.; Jeong, W.K. Weakly supervised learning in deformable EM image registration using slice interpolation. In Proceedings of the IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 670–673. [Google Scholar]
- Christensen, G.E.; Johnson, H. Consistent image registration. IEEE Trans. Med. Imaging 2001, 20, 568–582. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Shi, B.; Liao, R.; Mansi, T.; Rueckert, D.; Kamen, A. Unsupervised Deformable Registration for Multi-modal Images via Disentangled Representations. In Proceedings of the International Conference on Information Processing in Medical Imaging, Hong Kong, China, 2–7 June 2019; Springer: Cham, Switzerland, 2019; pp. 249–261. [Google Scholar]
- Zhang, J. Inverse-consistent deep networks for unsupervised deformable image registration. arXiv 2018, arXiv:1809.03443. [Google Scholar]
- Wang, C.; Yang, G.; Papanastasiou, G. FIRE: Unsupervised bi-directional inter- and intramodality registration using deep networks. In Proceedings of the International Symposium on Computer-Based Medical Systems (CBMS), Aveiro, Portugal, 7–9 July 2021; pp. 510–514. [Google Scholar]
- Wu, G.; Kim, M.; Wang, Q.; Gao, Y.; Liao, S.; Shen, D. Unsupervised deep feature learning for deformable registration of mr brain images. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MIC-CAI), Nagoya, Japan, 22–26 September 2013; Springer: Cham, Switzerland, 2013; pp. 649–656. [Google Scholar]
- Blendowski, M.; Heinrich, M.P. Combining MRF-based deformable registration and deep binary 3D-CNN descriptors for large lung motion estimation in COPD patients. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Simonovsky, M.; Gutierrez-Becker, B.; Mateus, D.; Navab, N.; Komodakis, N. A deep metric for multimodal registration. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Athens, Greece, 17–21 October 2016; Springer: Cham, Switzerland, 2016; pp. 10–18. [Google Scholar]
- Liao, R.; Miao, S.; de Tournemire, P.; Grbic, S.; Kamen, A.; Mansi, T.; Comaniciu, D. An artificial agent for robust image registration. In Proceedings of the 31 AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Ma, K.; Wang, J.; Singh, V.; Tamersoy, B.; Chang, Y.J.; Wimmer, A.; Chen, T. Multimodal image registration with deep context rein-forcement learning. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Quebec City, QC, Canada, 11–13 September 2017; Springer: Cham, Switzerland, 2017; pp. 240–248. [Google Scholar]
- Hu, J.; Luo, Z.; Wang, X.; Sun, S.; Yin, Y.; Cao, K.; Song, Q.; Lyu, S.; Wu, X. End-to-end multimodal image registration via reinforcement learning. Med. Image Anal. 2020, 68, 101878. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Yang, J.; Wang, L.; Xue, Z.; Wang, Q.; Shen, D. Deep learning based inter-modality image registration supervised by intra-modality similarity. In Proceedings of the International Workshop on Machine Learning in Medical Imaging, Granada, Spain, 16 September 2018; Springer: Cham, Switzerland, 2018; pp. 55–63. [Google Scholar]
- Balakrishnan, G.; Zhao, A.; Sabuncu, M.R.; Guttag, J.; Dalca, A.V. An unsupervised learning model for deformable medical image registration. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 9252–9260. [Google Scholar]
- Balakrishnan, G.; Zhao, A.; Sabuncu, M.R.; Guttag, J.; Dalca, A.V. VoxelMorph: A Learning Framework for Deformable Medical Image Registration. IEEE Trans. Med. Imaging 2019, 38, 1788–1800. [Google Scholar] [CrossRef] [Green Version]
- Krebs, J.; Mansi, T.; Mailhé, B.; Ayache, N.; Delingette, H. Learning structured deformations using diffeomorphic registration. arXiv 2018, arXiv:1804.07172. [Google Scholar]
- Kim, B.; Kim, D.H.; Park, S.H.; Kim, J.; Lee, J.-G.; Ye, J.C. CycleMorph: Cycle consistent unsupervised deformable image registration. Med. Image Anal. 2021, 71, 102036. [Google Scholar] [CrossRef]
- Dalca, A.V.; Balakrishnan, G.; Guttag, J.; Sabuncu, M.R. Unsupervised learning for fast probabilistic diffeomorphic registration. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Granada, Spain, 16–20 September 2018; Springer: Cham, Switzerland, 2018; pp. 729–738. [Google Scholar]
- Dalca, A.V.; Guttag, J.; Sabuncu, M.R. Anatomical Priors in Convolutional Networks for Unsupervised Biomedical Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 9290–9299. [Google Scholar]
- Sokooti, H.; de Vos, B.; Berendsen, F.; Lelieveldt, B.P.; Isgum, I.; Staring, M. Nonrigid image registration using multi-scale 3D convolutional neural networks. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Quebec City, QC, Canada, 11–13 September 2017; Springer: Cham, Switzerland, 2017; pp. 232–239. [Google Scholar]
- Avants, B.; Epstein, C.; Grossman, M.; Gee, J. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 2008, 12, 26–41. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Kwitt, R.; Styner, M.; Niethammer, M. Quicksilver: Fast predictive image registration—A deep learning approach. NeuroImage 2017, 158, 378–396. [Google Scholar] [CrossRef]
- Nielsen, R.K.; Darkner, S.; Feragen, A. Topaware: Topology-aware registration. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Shenzhen, China, 13–17 October 2019; Springer: Cham, Switzerland, 2019; pp. 364–372. [Google Scholar]
- Zhu, J.-Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Wei, D.; Ahmad, S.; Huo, J.; Peng, W.; Ge, Y.; Xue, Z.; Yap, P.T.; Li, W.; Shen, D.; Wang, Q. Synthesis and inpainting-based MR-CT regis-tration for image-guided thermal ablation of liver tumors. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Shenzhen, China, 13–17 October 2019; Springer: Cham, Switzerland, 2019; pp. 512–552. [Google Scholar]
- Papanastasiou, G.; Williams, M.C.; Kershaw, L.E.; Dweck, M.R.; Alam, S.; Mirsadraee, S.; Connell, M.; Gray, C.; MacGillivray, T.; Newby, D.E.; et al. Measurement of myocardial blood flow by cardiovascular magnetic resonance perfusion: Comparison of distributed parameter and Fermi models with single and dual bolus. J. Cardiovasc. Magn. Reson. 2015, 17, 17. [Google Scholar] [CrossRef] [Green Version]
- Papanastasiou, G.; Williams, M.C.; Dweck, M.R.; Alam, S.; Cooper, A.; Mirsadraee, S.; Newby, D.E.; Semple, S.I. Quantitative assessment of myocardial blood flow in coronary artery disease by cardiovascular magnetic resonance: Comparison of Fermi and distributed parameter modeling against invasive methods. J. Cardiovasc. Magn. Reson. 2016, 18, 57. [Google Scholar] [CrossRef] [Green Version]
- Papanastasiou, G.; Williams, M.C.; Dweck, M.; Mirsadraee, S.; Weir, N.; Fletcher, A.; Lucatelli, C.; Patel, D.; Van Beek, E.J.R.; Newby, D.E.; et al. Multimodality Quantitative Assessments of Myocardial Perfusion Using Dynamic Contrast Enhanced Magnetic Resonance and 15O-Labeled Water Positron Emission Tomography Imaging. IEEE Trans. Radiat. Plasma Med. Sci. 2018, 2, 259–271. [Google Scholar] [CrossRef]
- Linardatos, P.; Papastefanopoulos, V.; Kotsiantis, S. Explainable AI: A Review of Machine Learning Interpretability Methods. Entropy 2021, 23, 18. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2921–2929. [Google Scholar]
- Biglands, J.D.; Radjenovic, A.; Ridgway, J.P. Cardiovascular magnetic resonance physics for clinicians: Part II. J. Cardiovasc. Magn. Reson. 2012, 14, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar] [CrossRef] [Green Version]
- Chartsias, A.; Joyce, T.; Papanastasiou, G.; Semple, S.; Williams, M.; Newby, D.E.; Dharmakumar, R.; Tsaftaris, S.A. Disentangled representation learning in cardiac image analysis. Med. Image Anal. 2019, 58, 101535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chartsias, A.; Papanastasiou, G.; Wang, C.; Semple, S.; Newby, D.E.; Dharmakumar, R.; Tsaftaris, S.A. Disentangle, Align and Fuse for Multimodal and Semi-Supervised Image Segmentation. IEEE Trans. Med. Imaging 2020, 40, 781–792. [Google Scholar] [CrossRef]
- Ding, K.; Xiao, L.; Weng, G. Active contours driven by region-scalable fitting and optimized Laplacian of Gaussian energy for image segmentation. Signal Process. 2017, 134, 224–233. [Google Scholar] [CrossRef]
- Jin, R.; Weng, G. Active contours driven by adaptive functions and fuzzy c-means energy for fast image segmentation. Signal Process. 2019, 163, 1–10. [Google Scholar] [CrossRef]
- Weng, G.; Dong, B.; Lei, Y. A level set method based on additive bias correction for image segmentation. Expert Syst. Appl. 2021, 185, 115633. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).