ECG Recurrence Plot-Based Arrhythmia Classification Using Two-Dimensional Deep Residual CNN Features
Abstract
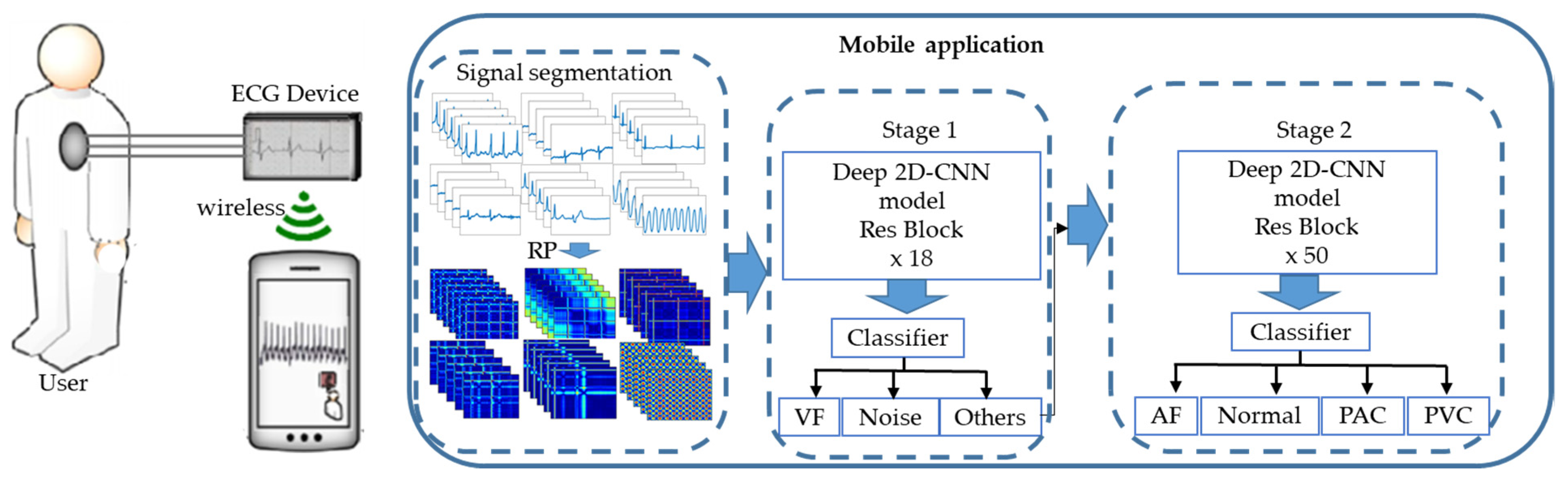
:1. Introduction
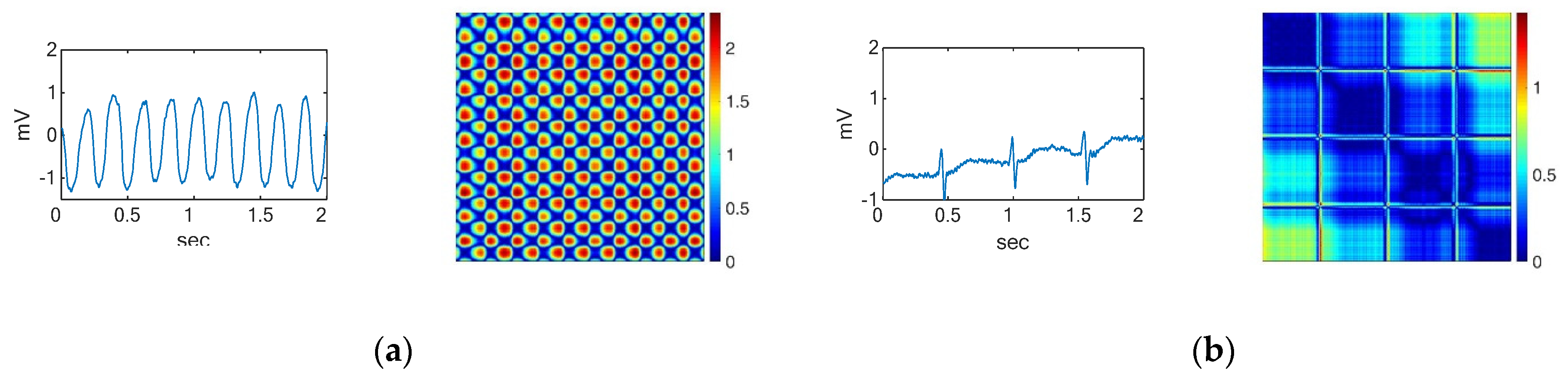
2. Time Series to Recurrent Plots
3. Materials and Methods
3.1. ECG Database
3.2. ECG Data Preprocessing
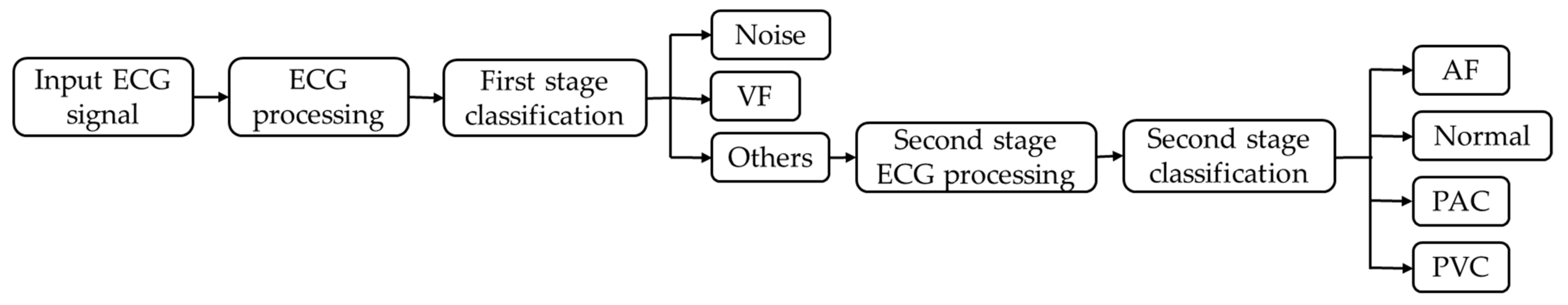
3.3. Classification
3.4. Performance Measures
- Acc: This gives a matrix that describes how the model performs across all classes.
- 2.
- Sens: This gives the percentage of the true samples that were correctly detected by the algorithm.
- 3.
- Sp: This indicates the percentage of the samples that were correctly detected as negative segments and beats.
- 4.
- PPV: This is calculated according to Bayes’ theorem.
- 5.
- F1: This gives the harmonic mean of the sensitivity and the positive predictive value.
- 6.
- Kappa:
- 7.
- True negative (TN): This represents the number of negative samples that were correctly predicted as negative by the model. TN is calculated for each of the three classes in the example in Table 1.
- 8.
- False positive (FP): FP is the number of samples predicted by the model to be positive which, in fact, turned out to be negative.
- 9.
- False negative (FN): FN is the number of positive samples that were incorrectly predicted as negative by the model. In multiclass classification, FNs are also calculated for each class.
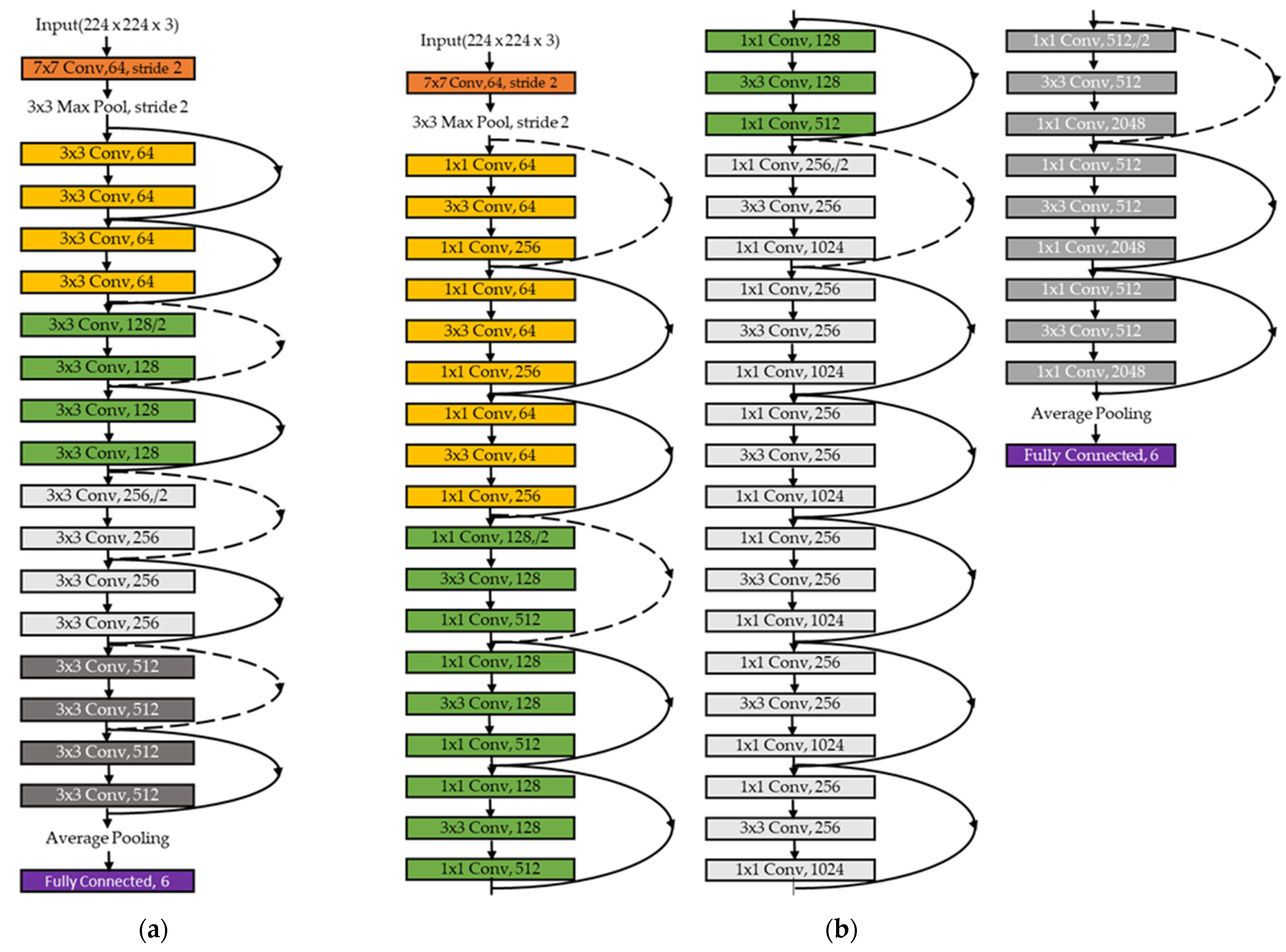
3.5. Two-Dimensional CNN Classifier
3.6. Training
4. Results
4.1. Determining the Number of Layers in ResNet
4.2. Performance Evaluation
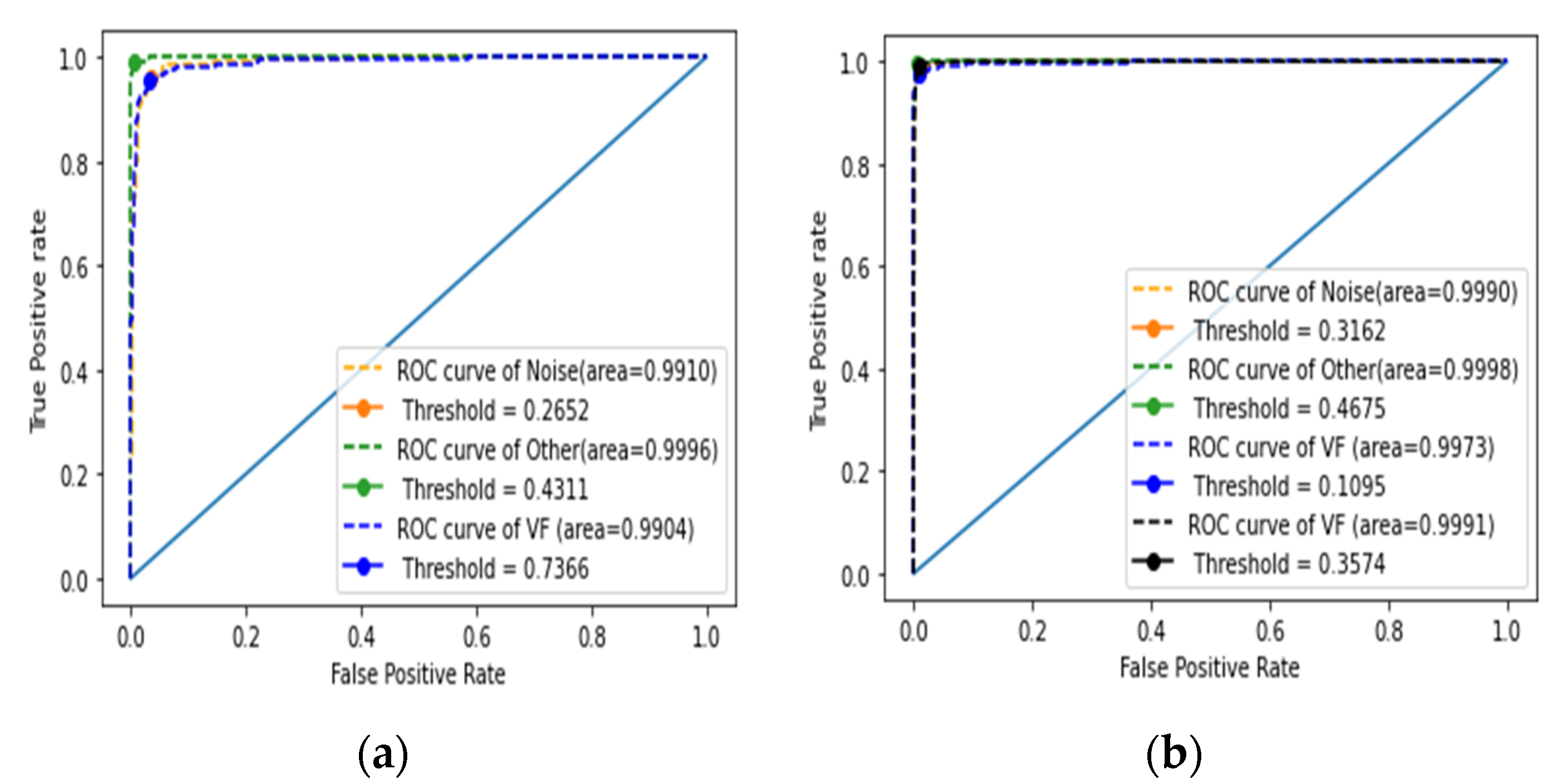
4.2.1. Assessment of the First Stage of Classification
4.2.2. Performance Evaluation of the Second Stage of Classification
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Centers for Disease Control and Prevention. Underlying Cause of Death, 1999–2018. CDC WONDER Online Database; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2018.
- Virani, S.S.; Alonso, A.; Benjamin, E.J.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Delling, F.N.; et al. Heart disease and stroke statistics—2020 update: A report from the American Heart Association. Circulation 2020, 141, e139–e596. [Google Scholar] [CrossRef] [PubMed]
- Fryar, C.D.; Chen, T.C.; Li, X. Prevalence of Uncontrolled Risk Factors for Cardiovascular Disease: United States, 1999–2010 (No. 103); US Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Health Statistics: Washington, DC, USA, 2012.
- Pławiak, P. Novel methodology of cardiac health recognition based on ECG signals and evolutionary-neural system. Expert Syst. Appl. 2018, 92, 334–349. [Google Scholar] [CrossRef]
- Luz, E.J.D.S.; Schwartz, W.R.; Cámara-Chávez, G.; Menotti, D. ECG-based heartbeat classification for arrhythmia detection: A survey. Comput. Methods Programs Biomed. 2016, 127, 144–164. [Google Scholar] [CrossRef] [PubMed]
- Padmavathi, K.; Ramakrishna, K.S. Classification of ECG signal during atrial fibrillation using autoregressive modeling. Procedia Comput. Sci. 2015, 46, 53–59. [Google Scholar] [CrossRef] [Green Version]
- Martis, R.J.; Acharya, U.R.; Min, L.C. ECG beat classification using PCA, LDA, ICA and discrete wavelet transform. Biomed. Signal Process. Control 2013, 8, 437–448. [Google Scholar] [CrossRef]
- Banerjee, S.; Mitra, M. Application of cross wavelet transform for ECG pattern analysis and classification. IEEE Trans. Instrum. Meas. 2013, 63, 326–333. [Google Scholar] [CrossRef]
- Odinaka, I.; Lai, P.H.; Kaplan, A.D.; O’Sullivan, J.A.; Sirevaag, E.J.; Kristjansson, S.D.; Sheffield, A.K.; Rohrbaugh, J.W. ECG biometrics: A robust short-time frequency analysis. In Proceedings of the 2010 IEEE International Workshop on Information Forensics and Security, Seattle, WA, USA, 12–15 December 2010; pp. 1–6. [Google Scholar]
- Raj, S.; Ray, K.C. ECG signal analysis using DCT-based DOST and PSO optimized SVM. IEEE Trans. Instrum. Meas. 2017, 66, 470–478. [Google Scholar] [CrossRef]
- Moavenian, M.; Khorrami, H. A qualitative comparison of artificial neural networks and support vector machines in ECG arrhythmias classification. Expert Syst. Appl. 2010, 37, 3088–3093. [Google Scholar] [CrossRef]
- Varatharajan, R.; Manogaran, G.; Priyan, M.K. A big data classification approach using LDA with an enhanced SVM method for ECG signals in cloud computing. Multimed. Tools Appl. 2018, 77, 10195–10215. [Google Scholar] [CrossRef]
- Celin, S.; Vasanth, K. ECG signal classification using various machine learning techniques. J. Med. Syst. 2018, 42, 241. [Google Scholar] [CrossRef]
- Kropf, M.; Hayn, D.; Schreier, G. ECG classification based on time and frequency domain features using random forests. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017; pp. 1–4. [Google Scholar]
- Acharya, U.R.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adam, M.; Gertych, A.; Tan, R.S. A deep convolutional neural network model to classify heartbeats. Comput. Biol. Med. 2017, 89, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Fujita, H.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adam, M. Application of deep convolutional neural network for automated detection of myocardial infarction using ECG signals. Inf. Sci. 2017, 415–416, 190–198. [Google Scholar] [CrossRef]
- Yildirim, O.; Tan, R.S.; Acharya, U.R. An efficient compression of ECG signals using deep convolutional autoencoders. Cogn. Syst. Res. 2018, 52, 198–211. [Google Scholar] [CrossRef]
- Al Rahhal, M.M.; Bazi, Y.; AlHichri, H.; Alajlan, N.; Melgani, F.; Yager, R.R. Deep learning approach for active classification of electrocardiogram signals. Inf. Sci. 2016, 345, 340–354. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Oh, S.L.; Raghavendra, U.; Tan, J.H.; Adam, M.; Gertych, A.; Hagiwara, Y. Automated identification of shockable and non-shockable life-threatening ventricular arrhythmias using convolutional neural network. Future Gener. Comput. Syst. 2018, 79, 952–959. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Lih, O.S.; Adam, M.; Tan, J.H.; Chua, C.K. Automated detection of coronary artery disease using different durations of ECG segments with convolutional neural network. Knowl.-Based Syst. 2017, 132, 62–71. [Google Scholar] [CrossRef]
- Wang, J.; Liu, P.; She, M.F.; Nahavandi, S.; Kouzani, A. Bag-of-words representation for biomedical time series classification. Biomed. Signal Process. Control 2013, 8, 634–644. [Google Scholar] [CrossRef] [Green Version]
- Hatami, N.; Chira, C. Classifiers with a reject option for early time-series classification. In Proceedings of the 2013 IEEE Symposium on Computational Intelligence and Ensemble Learning (CIEL), Singapore, 16–19 April 2013; pp. 9–16. [Google Scholar]
- Wang, Z.; Oates, T. Pooling sax-bop approaches with boosting to classify multivariate synchronous physiological time series data. In Proceedings of the Twenty-Eighth International Flairs Conference, Hollywood, FL, USA, 18–25 May 2015. [Google Scholar]
- Mahmoodabadi, S.Z.; Ahmadian, A.; Abolhasani, M.D. ECG feature extraction using Daubechies wavelets. In Proceedings of the Fifth IASTED International Conference on Visualization, Imaging and Image Processing, Benidorm, Spain, 7–9 September 2005; pp. 343–348. [Google Scholar]
- Mahmoodabadi, S.Z.; Ahmadian, A.; Abolhasani, M.D.; Eslami, M.; Bidgoli, J.H. ECG feature extraction based on multiresolution wavelet transform. In Proceedings of the 2005 IEEE Engineering in Medicine and Biology 27th Annual Conference, Shanghai, China, 17–18 January 2006; pp. 3902–3905. [Google Scholar]
- Karpagachelvi, S.; Arthanari, M.; Sivakumar, M. ECG feature extraction techniques-a survey approach. arXiv 2010, arXiv:1005.0957. [Google Scholar]
- Collobert, R.; Weston, J. A unified architecture for natural language processing: Deep neural networks with multitask learning. In Proceedings of the 25th International Conference on Machine Learning, New York, NY, USA, 5–9 July 2008; pp. 160–167. [Google Scholar]
- Tsantekidis, A.; Passalis, N.; Tefas, A.; Kanniainen, J.; Gabbouj, M.; Iosifidis, A. Forecasting stock prices from the limit order book using convolutional neural networks. In Proceedings of the 2017 IEEE 19th Conference on Business Informatics (CBI), Thessaloniki, Greece, 24–27 July 2017; Volume 1, pp. 7–12. [Google Scholar]
- Van Den Oord, A.; Dieleman, S.; Schrauwen, B. Deep content-based music recommendation. In Proceedings of the Neural Information Processing Systems Conference (NIPS 2013), Lake Tahoe, NV, USA, 5–10 December 2013; Volume 26. [Google Scholar]
- Avilov, O.; Rimbert, S.; Popov, A.; Bougrain, L. Deep learning techniques to improve intraoperative awareness detection from electroencephalographic signals. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Online, 20–24 July 2020; pp. 142–145. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Fradi, M.; Khriji, L.; Machhout, M. Real-time arrhythmia heart disease detection system using CNN architecture based various optimizers-networks. Multimed. Tools Appl. 2021, 1–22. [Google Scholar] [CrossRef]
- Ma, F.; Zhang, J.; Liang, W.; Xue, J. Automated classification of atrial fibrillation using artificial neural network for wearable devices. Math. Probl. Eng. 2020, 2020, 9159158. [Google Scholar] [CrossRef] [Green Version]
- Jun, T.J.; Nguyen, H.M.; Kang, D.; Kim, D.; Kim, D.; Kim, Y.H. ECG arrhythmia classification using a 2-D convolutional neural network. arXiv 2018, arXiv:1804.06812. [Google Scholar]
- Atal, D.K.; Singh, M. Arrhythmia classification with ECG signals based on the optimization-enabled deep convolutional neural network. Comput. Methods Programs Biomed. 2020, 196, 105607. [Google Scholar] [CrossRef] [PubMed]
- Essa, E.; Xie, X. An Ensemble of Deep Learning-Based Multi-Model for ECG Heartbeats Arrhythmia Classification. IEEE Access 2021, 9, 103452–103464. [Google Scholar] [CrossRef]
- Issa, Z.; Miller, J.M.; Zipes, D.P. Clinical Arrhythmology and Electrophysiology: A Companion to Braunwald’s Heart Disease; Elsevier Health Sciences: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Odutayo, A.; Wong, C.X.; Hsiao, A.J.; Hopewell, S.; Altman, D.G.; Emdin, C.A. Atrial fibrillation and risks of cardiovascular disease, renal disease, and death: Systematic review and meta-analysis. BMJ 2016, 354, i4482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, T.J.; Haarbo, J.; Pehrson, S.M.; Thomsen, B. Impact of premature atrial contractions in atrial fibrillation. Pacing Clin. Electrophysiol. 2004, 27, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Mathunjwa, B.M.; Lin, Y.T.; Lin, C.H.; Abbod, M.F.; Shieh, J.S. ECG arrhythmia classification by using a recurrence plot and convolutional neural network. Biomed. Signal Process. Control 2021, 64, 102262. [Google Scholar] [CrossRef]
- Nolle, F.M.; Badura, F.K.; Catlett, J.M.; Bowser, R.W.; Sketch, M.H. CREI-GARD, a new concept in computerized arrhythmia monitoring systems. Comput. Cardiol. 1986, 13, 515–518. [Google Scholar]
- Moody, G.B.; Muldrow, W.; Mark, R.G. A noise stress test for arrhythmia detectors. Comput. Cardiol. 1984, 11, 381–384. [Google Scholar]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [Green Version]
- Eeklnann, J.P.; Karmphorst, S.O.; Rulle, D. Recurrence plot of dynamical systems. Europhys. Lett. 1987, 4, 973–977. [Google Scholar]
- Marwan, N.; Romano, M.C.; Thiel, M.; Kurths, J. Recurrence plots for the analysis of complex systems. Phys. Rep. 2007, 438, 237–329. [Google Scholar] [CrossRef]
- Robinson, G.; Thiel, M. Recurrences determine the dynamics. Chaos Interdiscip. J. Nonlinear Sci. 2009, 19, 023104. [Google Scholar] [CrossRef] [PubMed]
- Hirata, Y.; Horai, S.; Aihara, K. Reproduction of distance matrices and original time series from recurrence plots and their applications. Eur. Phys. J. Spec. Top. 2008, 164, 13–22. [Google Scholar] [CrossRef]
- Faria, F.A.; Almeida, J.; Alberton, B.; Morellato, L.P.C.; Torres, R.D.S. Fusion of time series representations for plant recognition in phenology studies. Pattern Recognit. Lett. 2016, 83, 205–214. [Google Scholar] [CrossRef] [Green Version]
- Sipers, A.; Borm, P.; Peeters, R. Robust reconstruction of a signal from its unthresholded recurrence plot subject to disturbances. Phys. Lett. A 2017, 381, 604–615. [Google Scholar] [CrossRef]
- Chen, Y.; Su, S.; Yang, H. Convolutional neural network analysis of recurrence plots for anomaly detection. Int. J. Bifurc. Chaos 2020, 30, 2050002. [Google Scholar] [CrossRef]
- Lu, J.; Tong, K.Y. Robust single accelerometer-based activity recognition using modified recurrence plot. IEEE Sens. J. 2019, 19, 6317–6324. [Google Scholar] [CrossRef]
- Webber, C.L.; Marwan, N. Recurrence Quantification Analysis. Theory and Best Practices; Springer: Cham, Switzerland, 2015. [Google Scholar]
- Moody, G. A new method for detecting atrial fibrillation using RR intervals. Comput. Cardiol. 1983, 227–230. [Google Scholar]
- Moody, G.B.; Mark, R.G. The impact of the MIT-BIH arrhythmia database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef]
- Greenwald, S.D. The Development and Analysis of a Ventricular Fibrillation Detector. Ph.D. Thesis, Massachusetts Institute of Technology, Cambridge, MA, USA, 1986. [Google Scholar]
- Tseng, W.J.; Hung, L.W.; Shieh, J.S.; Abbod, M.F.; Lin, J. Hip fracture risk assessment: Artificial neural network outperforms conditional logistic regression in an age- and sex-matched case control study. BMC Musculoskelet. Disord. 2013, 14, 207. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving deep into rectifiers: Surpassing human-level performance on ImageNet classification. In Proceedings of the IEEE International Conference on Computer Vision 2015, Santiago, Chile, 11–18 December 2015; pp. 1026–1034. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Frid-Adar, M.; Diamant, I.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018, 321, 321–331. [Google Scholar] [CrossRef] [Green Version]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning, Lille, France, 6–11 July 2015; pp. 448–456. [Google Scholar]
- Sra, S.; Nowozin, S.; Wright, S.J. (Eds.) Optimization for Machine Learning; MIT Press: Cambridge, MA, USA, 2012. [Google Scholar]
- Lee, C.Y.; Xie, S.; Gallagher, P.; Zhang, Z.; Tu, Z. Deeply-supervised nets. In Proceedings of the Artificial Intelligence and Statistics, San Diego, CA, USA, 9–12 May 2015; pp. 562–570. [Google Scholar]
- Zhang, H.; Liu, C.; Zhang, Z.; Xing, Y.; Liu, X.; Dong, R.; He, Y.; Xia, L.; Liu, F. Recurrence Plot-Based Approach for Cardiac Arrhythmia Classification Using Inception-ResNet-v2. Front. Physiol. 2021, 12, 648950. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Tu, S.; Mehmood, R.M.; Ehatisham-ul-haq, M. A Hybrid Deep CNN Model for Abnormal Arrhythmia Detection Based on Cardiac ECG Signal. Sensors 2021, 21, 951. [Google Scholar] [CrossRef]
- Degirmenci, M.; Ozdemir, M.A.; Izci, E.; Akan, A. Arrhythmic Heartbeat Classification Using 2D Convolutional Neural Networks. IRBM 2021. [Google Scholar] [CrossRef]
- Izci, E.; Ozdemir, M.A.; Degirmenci, M.; Akan, A. Cardiac arrhythmia detection from 2D ECG images by using deep learning technique. In Proceedings of the 2019 Medical Technologies Congress (TIPTEKNO), Izmir, Turkey, 3–5 October 2019; pp. 1–4. [Google Scholar]
- Le, M.D.; Rathour, V.S.; Truong, Q.S.; Mai, Q.; Brijesh, P.; Le, N. Multi-module Recurrent Convolutional Neural Network with Transformer Encoder for ECG Arrhythmia Classification. In Proceedings of the 2021 IEEE EMBS International Conference on Biomedical and Health Informatics (BHI), Online, 27–30 July 2021; pp. 1–5. [Google Scholar]
- Li, Q.; Liu, C.; Li, Q.; Shashikumar, S.P.; Nemati, S.; Shen, Z.; Clifford, G.D. Ventricular ectopic beat detection using a wavelet transform and a convolutional neural network. Physiol. Meas. 2019, 40, 055002. [Google Scholar] [CrossRef]
- Chen, C.; Hua, Z.; Zhang, R.; Liu, G.; Wen, W. Automated arrhythmia classification based on a combination network of CNN and LSTM. Biomed. Signal Process. Control 2020, 57, 101819. [Google Scholar] [CrossRef]
- Yıldırım, Ö.; Pławiak, P.; Tan, R.S.; Acharya, U.R. Arrhythmia detection using deep convolutional neural network with long duration ECG signals. Comput. Biol. Med. 2018, 102, 411–420. [Google Scholar] [CrossRef]
- He, R.; Liu, Y.; Wang, K.; Zhao, N.; Yuan, Y.; Li, Q.; Zhang, H. Automatic cardiac arrhythmia classification using combination of deep residual network and bidirectional LSTM. IEEE Access 2019, 7, 102119–102135. [Google Scholar] [CrossRef]
- Yildirim, Ö. A novel wavelet sequence based on deep bidirectional LSTM network model for ECG signal classification. Comput. Biol. Med. 2018, 96, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Wang, R.; Fan, X.; Liu, J.; Li, Y. Multi-class arrhythmia detection from 12-lead varied-length ECG using attention-based time-incremental convolutional neural network. Inf. Fusion 2020, 53, 174–182. [Google Scholar] [CrossRef]
- Wang, Z.; Li, H.; Han, C.; Wang, S.; Shi, L. Arrhythmia Classification Based on Multiple Features Fusion and Random Forest Using ECG. J. Med. Imaging Health Inform. 2019, 9, 1645–1654. [Google Scholar] [CrossRef]
- El-Saadawy, H.; Tantawi, M.; Shedeed, H.A.; Tolba, M.F. Electrocardiogram (ECG) heart disease diagnosis using PNN, SVM and Softmax regression classifiers. In Proceedings of the 2017 Eighth International Conference on Intelligent Computing and Information Systems (ICICIS), Cairo, Egypt, 5–7 December 2017; pp. 106–110. [Google Scholar]
- Sahoo, S.; Mohanty, M.; Behera, S.; Sabut, S.K. ECG beat classification using empirical mode decomposition and mixture of features. J. Med. Eng. Technol. 2017, 41, 652–661. [Google Scholar] [CrossRef]
- Khairuddin, A.M.; Azir, K.K. Using the HAAR Wavelet Transform and K-nearest Neighbour Algorithm to Improve ECG Detection and Classification of Arrhythmia. In Proceedings of the 34th International Conference on Industrial, Engineering and Other Applications of Applied Intelligent Systems, Kuala Lumpur, Malaysia, 26–29 July 2021; pp. 310–322. [Google Scholar]
- Oh, S.L.; Eddie, Y.K.N.; Ru, S.T.; Acharya, U.R. Automated diagnosis of arrhythmia using combination of CNN and LSTM techniques with variable length heart beats. Comput. Biol. Med. 2018, 102, 278–287. [Google Scholar] [CrossRef]
- Zhu, W.; Chen, X.; Wang, Y.; Wang, L. Arrhythmia Recognition and Classification Using ECG Morphology and Segment Feature Analysis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 16, 131–138. [Google Scholar] [CrossRef]
- Aphale, S.S.; John, E.; Banerjee, T. ArrhyNet: A High Accuracy Arrhythmia Classification Convolutional Neural Network. In Proceedings of the 2021 IEEE International Midwest Symposium on Circuits and Systems (MWSCAS), Online, 9–11 August 2021; pp. 453–457. [Google Scholar]
- Sellami, A.; Hwang, H. A robust deep convolutional neural network with batch-weighted loss for heartbeat classification. Expert Syst. Appl. 2019, 122, 75–84. [Google Scholar] [CrossRef]
- Gan, Y.; Shi, J.C.; He, W.M.; Sun, F.J. Parallel classification model of arrhythmia based on DenseNet-BiLSTM. Biocybern. Biomed. Eng. 2021, 41, 1548–1560. [Google Scholar] [CrossRef]


| Predicted | ||||
|---|---|---|---|---|
| A | B | C | ||
| Actual | A | |||
| B | ||||
| C | ||||
| Layer Name | Output Size | ResNet-18 | ResNet-50 |
|---|---|---|---|
| Conv 1 | 112 × 112 | 7 × 7, 64, stride 2 | 7 × 7, 64, stride 2 |
| Conv 2_x | 56 × 56 | 3 × 3 max pool, stride 2 | 3 × 3 max pool, stride 2 |
| Conv 3_x | 28 × 28 | ||
| Conv 4_x | 14 × 14 | ||
| Conv 5_x | 7 × 7 | ||
| 1 × 1 | Average pool, 6-d fc, softmax | Average pool, 6-d fc, softmax |
| ResNet Number of Layers | Training | Validation | Testing | Size (MB) |
|---|---|---|---|---|
| ResNet-18 | 98.77% | 96.61% | 97.04% | 43 |
| ResNet-34 | 98.12% | 96.74% | 97.45% | 83 |
| ResNet-50 | 98.40% | 96.23% | 96.53% | 94 |
| ResNet-101 | 98.55% | 96.42% | 97.26% | 169 |
| ResNet-152 | 98.60% | 96.09% | 96.72% | 230 |
| ResNet Number of Layers | Training | Validation | Testing | Size (MB) |
|---|---|---|---|---|
| ResNet-18 | 99.43% | 94.06% | 94.65% | 43 |
| ResNet-34 | 99.21% | 95.59% | 95.26% | 83 |
| ResNet-50 | 98.95% | 97.22% | 98.15% | 94 |
| ResNet-101 | 98.59% | 97.91% | 98.46% | 169 |
| ResNet-152 | 98.56% | 97.62% | 98.33% | 230 |
| ResNet Number of Layers | Sens | Sp | ||||
|---|---|---|---|---|---|---|
| Noise | Other | VF | Noise | Other | VF | |
| ResNet-18 | 96.33% | 92.96% | 99.42% | 97.16% | 99.30% | 99.28% |
| ResNet-34 | 95.93% | 99.17% | 94.35% | 98.32% | 99.00% | 98.91% |
| ResNet-50 | 92.64% | 98.58% | 96.61% | 97.03% | 98.31% | 99.33% |
| ResNet-101 | 93.20% | 99.50% | 96.89% | 97.29% | 99.37% | 99.40% |
| ResNet-152 | 93.43% | 98.75% | 95.76% | 97.34% | 98.51% | 99.17% |
| ResNet Number of Layers | Sens | Sp | ||||||
|---|---|---|---|---|---|---|---|---|
| AF | Normal | PAC | PVC | AF | Normal | PAC | PVC | |
| ResNet-18 | 87.27% | 99.67% | 92.08% | 98.49% | 94.37% | 99.84% | 99.11% | 99.46% |
| ResNet-34 | 88.35% | 99.08% | 96.04% | 98.69% | 94.83% | 99.56% | 99.55% | 99.53% |
| ResNet-50 | 97.17% | 99.67% | 95.51% | 98.49% | 99.11% | 99.80% | 99.41% | 99.28% |
| ResNet-101 | 98.34% | 99.67% | 96.31% | 97.98% | 99.22% | 99.84% | 99.59% | 99.28% |
| ResNet-152 | 97.84% | 99.42% | 94.72% | 98.99% | 98.99% | 99.73% | 99.41% | 99.64% |
| CV | Training | Validation | Testing |
|---|---|---|---|
| Mean ± SD | 98.56 ± 0.16% | 96.76 ± 0.31% | 97.21 ± 0.34% |
| Noise | Other | VF | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CV | Sens | Sp | F1-Score | Sens | Sp | F1-Score | Sens | Sp | F1-Score |
| Mean | 96.44 | 97.80 | 93.01 | 93.77 | 99.22 | 95.46 | 99.27 | 98.97 | 99.42 |
| STD | 0.47% | 0.76% | 1.10% | 1.39% | 0.24% | 0.63% | 0.09% | 0.74% | 0.06% |
| CV | Noise | Other | VF | |||
|---|---|---|---|---|---|---|
| AUC | Threshold | AUC | Threshold | AUC | Threshold | |
| Mean | 0.991 | 0.273 | 0.999 | 0.536 | 0.991 | 0.626 |
| STD | 0.002 | 0.040 | 0.001 | 0.334 | 0.001 | 0.160 |
| CV | Training | Validation | Testing |
|---|---|---|---|
| Mean ± SD | 98.72 ± 0.16% | 97.71 ± 0.16% | 98.36 ± 0.16% |
| AF | N | PAC | PVC | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CV | Sens | Sp | F1-Score | Sens | Sp | F1-Score | Sens | Sp | F1-Score | Sens | Sp | F1-Score |
| Mean | 97.64 | 98.90 | 98.07 | 99.65 | 99.84 | 99.18 | 95.73 | 99.52 | 96.59 | 98.67 | 99.52 | 98.37 |
| STD | 0.42% | 0.19% | 0.08% | 0.22% | 0.10% | 0.19% | 1.11% | 0.12% | 0.46% | 0.48% | 0.17% | 0.18% |
| CV | AF | Normal | PAC | PVC | ||||
|---|---|---|---|---|---|---|---|---|
| AUC | Threshold | AUC | Threshold | AUC | Threshold | AUC | Threshold | |
| Mean | 0.998 | 0.234 | 0.999 | 0.624 | 0.998 | 0.144 | 0.999 | 0.264 |
| STD | 0.001 | 0.139 | 0.000 | 0.228 | 0.001 | 0.096 | 0.000 | 0.110 |
| Av Sens | Av Sp | PPV | Av F1-score | Kappa | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CV | 1st Stage | 2nd Stage | 1st Stage | 2nd Stage | 1st Stage | 2nd Stage | 1st Stage | 2nd Stage | 1st Stage | 2nd Stage |
| Mean | 96.49 | 97.92 | 98.66 | 99.45 | 93.29 | 95.18 | 95.28 | 97.71 | 95.28 | 97.71 |
| STD | 0.39% | 0.30% | 0.14% | 0.04% | 0.68% | 0.37% | 0.57% | 0.20% | 0.57% | 0.20% |
| Acc | Av Sens | Av Sp | Av PPV | Av F1-score | Kappa |
|---|---|---|---|---|---|
| 94.85% | 94.44 ± 2.94% | 94.96 ± 7.31% | 93.37 ± 7.31% | 94.05 ± 4.61% | 93.37% |
| Classifier | Layer Size | Model Size (MB) | Training Time (h) | Accuracy (%) First Stage | Accuracy (%) Second Stage |
|---|---|---|---|---|---|
| ResNet-18 | 18 | 43 | 6.26 | 97.21 | 94.65 |
| ResNet-34 | 34 | 83 | 6.22 | 97.45 | 95.26 |
| ResNet-50 | 50 | 94 | 6.32 | 96.53 | 98.36 |
| ResNet-101 | 101 | 169 | 6.25 | 97.26 | 98.46 |
| ResNet-152 | 152 | 230 | 6.26 | 96.72 | 98.33 |
| AlexNet | 8 | 228 | 5.37 | 96.59 | 98.53 |
| VGG16 | 16 | 525 | 7.6 | 87.35 | 86.86 |
| VGG19 | 19 | 545 | 7.75 | 81.01 | 94.09 |
| Studies | Databases | No. of Classes | Segment Length(s) | Method | Acc (%) | Sens (%) | PPV (%) | F1-Score |
|---|---|---|---|---|---|---|---|---|
| Zhang et al. [64] | CPSC | 9 | 5 | Inception-ResNet-v2 | N/A | 84.7 | 84.7 | 84.4 |
| Ullah et al. [65] | MITDB | 8 | N/A | 2D CNN | 99.02 | N/A | N/A | N/A |
| Degirmenci et al. [66] | MITDB | 5 | N/A | CNN | 99.70 | 99.70 | 99.22 | N/A |
| Izci et al. [67] | MITDB | 5 | N/A | CNN | 97.42 | N/A | N/A | N/A |
| Le et al. [68] | MITDB | 6 | N/A | RCNN | 98.29 | N/A | N/A | 99.14 |
| Li et al. [69] | MITDB | - | N/A | CNN | 97.96 | N/A | N/A | 84.94 |
| Chen et al. [70] | MITDB | 6 | 10 | CNN + LSTM | 99.32 | 97.75 | 97.66 | N/A |
| Yıldırım et al. [71] | MITDB | 17 | 10 | 1D CNN | 91.33 | 83.91 | N/A | 91.33 |
| He et al. [72] | CPSC | 9 | 30 | CNN + LSTM | N/A | N/A | N/A | 80.6 |
| Yildirim et al. [73] | MITDB | 5 | 1 | DULSTM DBLSTM | 99.25 99.39 | N/A | N/A | N/A |
| Yao et al. [74] | CPSC | 9 | 1.5 | ResNet + BLSTM-GMP | N/A | 80.1 | 82.6 | 81.2 |
| Fradi et al. [44] | MITDB, TPB | 5 | 1.496 | 1D CNN | 99.61 | N/A | N/A | 99 |
| Wang et al. [75] | MITDB | 5 | N/A | Random forest | 92.31 | 89.98 | N/A | N/A |
| El-Saadawy et al. [76] | MITDB | 5 | N/A | SVM + PNN | 88.7 | N/A | N/A | N/A |
| Sahoo et al. [77] | MITDB | 6 | N/A | PNN + RBF-NN | 99.54 99.89 | N/A | N/A | N/A |
| Khairuddin et al. [78] | MITDB | 17 | N/A | K-NN | 97.30 | N/A | N/A | N/A |
| Proposed | AFDB, MITDB, CUDB, VFDB | 3 4 6 | 2 | ResNet-18 ResNet-50 ResNet-18&50 | 97.21 98.36 94.85 | 96.49 97.92 94.44 | 95.54 98.20 93.37 | 95.96 98.05 94.05 |
| AAMI Type | No. of Beats |
|---|---|
| Normal (N) | 8980 |
| Ventricular ectopic (V) | 7202 |
| Supraventricular ectopic (S) | 2758 |
| Fusion (F) | 799 |
| Unknown (Q) | 8307 |
| Type | CV_1 | CV_2 | CV_3 | CV_4 | CV_5 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sens | SP | F1 | Sens | SP | F1 | Sens | SP | F1 | Sens | SP | F1 | Sens | SP | F1 | |
| Mean | 96.14% | 97.00% | 96.56% | 96.00% | 97.76% | 96.84% | 95.89% | 97.26% | 96.55% | 97.03% | 95.63% | 96.31% | 96.92% | 96.78% | 96.84% |
| STD | 4.35% | 3.39% | 3.79% | 4.96% | 1.90% | 3.38% | 4.83% | 1.88% | 3.33% | 2.89% | 5.42% | 4.15% | 2.39% | 3.07% | 2.64% |
| CV | Acc | Sens | Sp | PPV | F1 | Kappa |
|---|---|---|---|---|---|---|
| Mean ± STD | 98.21 ± 0.11% | 96.40 ± 0.54% | 96.89 ± 0.79% | 93.26 ± 2.61% | 96.65 ± 0.19% | 97.44 ± 0.15% |
| Studies | Classes | Method | Acc (%) | Sens | Sp | PPV | F1 |
|---|---|---|---|---|---|---|---|
| Acharya et al. [15] | 5 | CNN | 94.03 | 96.71 | 91.54 | N/A | N/A |
| Oh et al. [79] | 5 | CNN + LSTM | 98.10 | 97.50 | 98.70 | N/A | N/A |
| Izci et al. [67] | 5 | CNN | 97.42 | N/A | N/A | N/A | N/A |
| Zhu et al. [80] | 5 | SVM | 97.80 | 88.83 | 93.76 | N/A | N/A |
| Aphale et al. [81] | 5 | CNN | 92.73 | 92.00 | 91.00 | N/A | 91.00 |
| Sellami et al. [82] | 5 | CNN | 99.48 | 96.97 | 99.87 | 96.83 | N/A |
| Gan et al. [83] | 4 | DenseNet-BiLSTM | 99.44 | 95.89 | 99.32 | 96.11 | 95.89 |
| Proposed | 5 | ResNet-50 | 98.21 | 96.40 | 96.89 | 93.26 | 96.65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mathunjwa, B.M.; Lin, Y.-T.; Lin, C.-H.; Abbod, M.F.; Sadrawi, M.; Shieh, J.-S. ECG Recurrence Plot-Based Arrhythmia Classification Using Two-Dimensional Deep Residual CNN Features. Sensors 2022, 22, 1660. https://doi.org/10.3390/s22041660
Mathunjwa BM, Lin Y-T, Lin C-H, Abbod MF, Sadrawi M, Shieh J-S. ECG Recurrence Plot-Based Arrhythmia Classification Using Two-Dimensional Deep Residual CNN Features. Sensors. 2022; 22(4):1660. https://doi.org/10.3390/s22041660
Chicago/Turabian StyleMathunjwa, Bhekumuzi M., Yin-Tsong Lin, Chien-Hung Lin, Maysam F. Abbod, Muammar Sadrawi, and Jiann-Shing Shieh. 2022. "ECG Recurrence Plot-Based Arrhythmia Classification Using Two-Dimensional Deep Residual CNN Features" Sensors 22, no. 4: 1660. https://doi.org/10.3390/s22041660
APA StyleMathunjwa, B. M., Lin, Y.-T., Lin, C.-H., Abbod, M. F., Sadrawi, M., & Shieh, J.-S. (2022). ECG Recurrence Plot-Based Arrhythmia Classification Using Two-Dimensional Deep Residual CNN Features. Sensors, 22(4), 1660. https://doi.org/10.3390/s22041660








