Sensing of Soil Organic Matter Using Laser-Induced Breakdown Spectroscopy Coupled with Optimized Self-Adaptive Calibration Strategy
Abstract
:1. Introduction
2. Materials and Methods
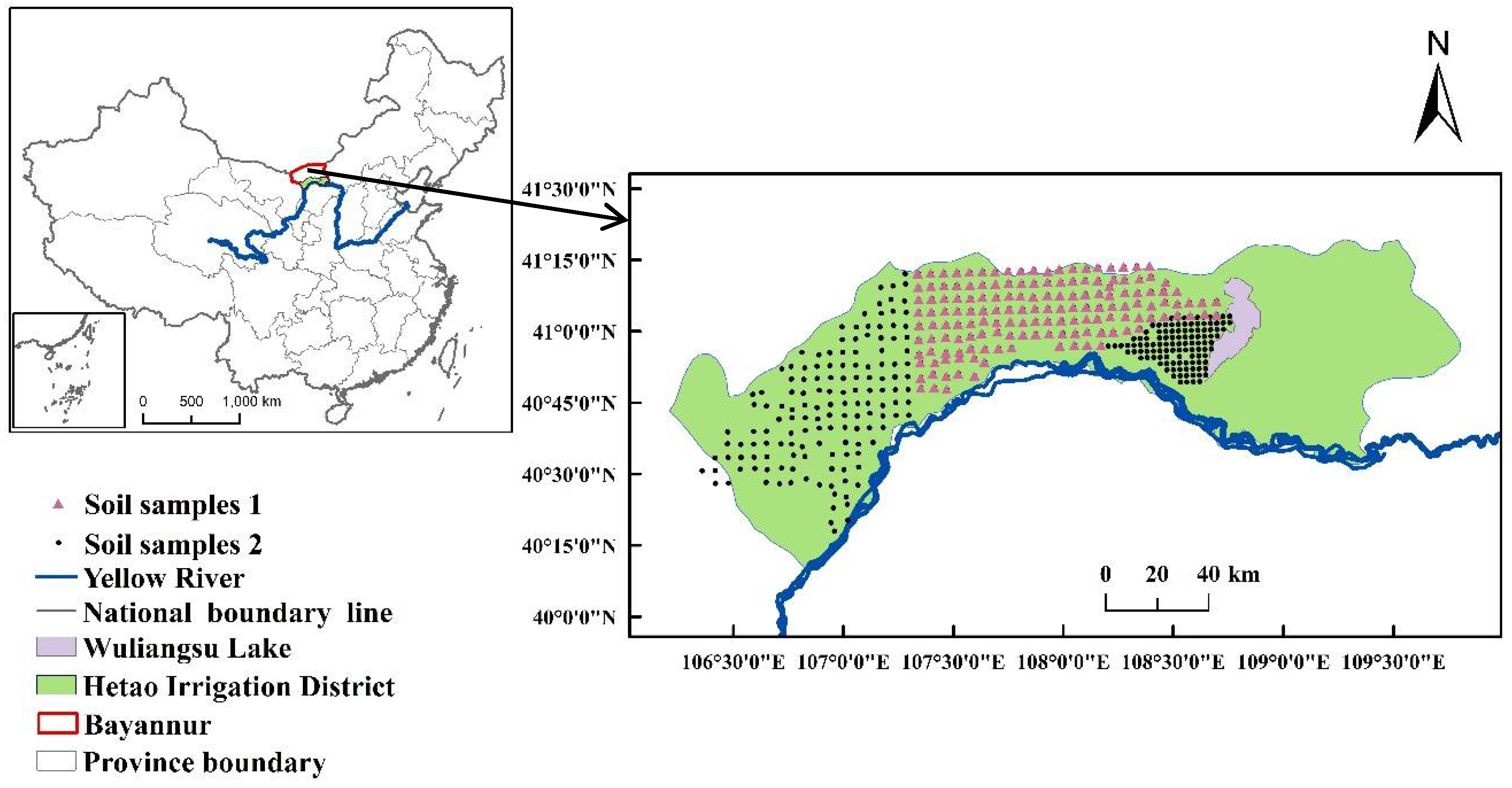
2.1. Study Area and Soil Sample Collection
2.2. Soil Property Measurement
2.3. LIBS Spectra Measurement
2.4. Self-Adaptive Model Partial Least Squares Regression (SAM–PLSR)
2.5. Model Evaluation Standard
2.6. Software and Statistical Analysis
3. Results
3.1. The Distribution of SOM
3.2. The LIBS Spectra of Soil Samples
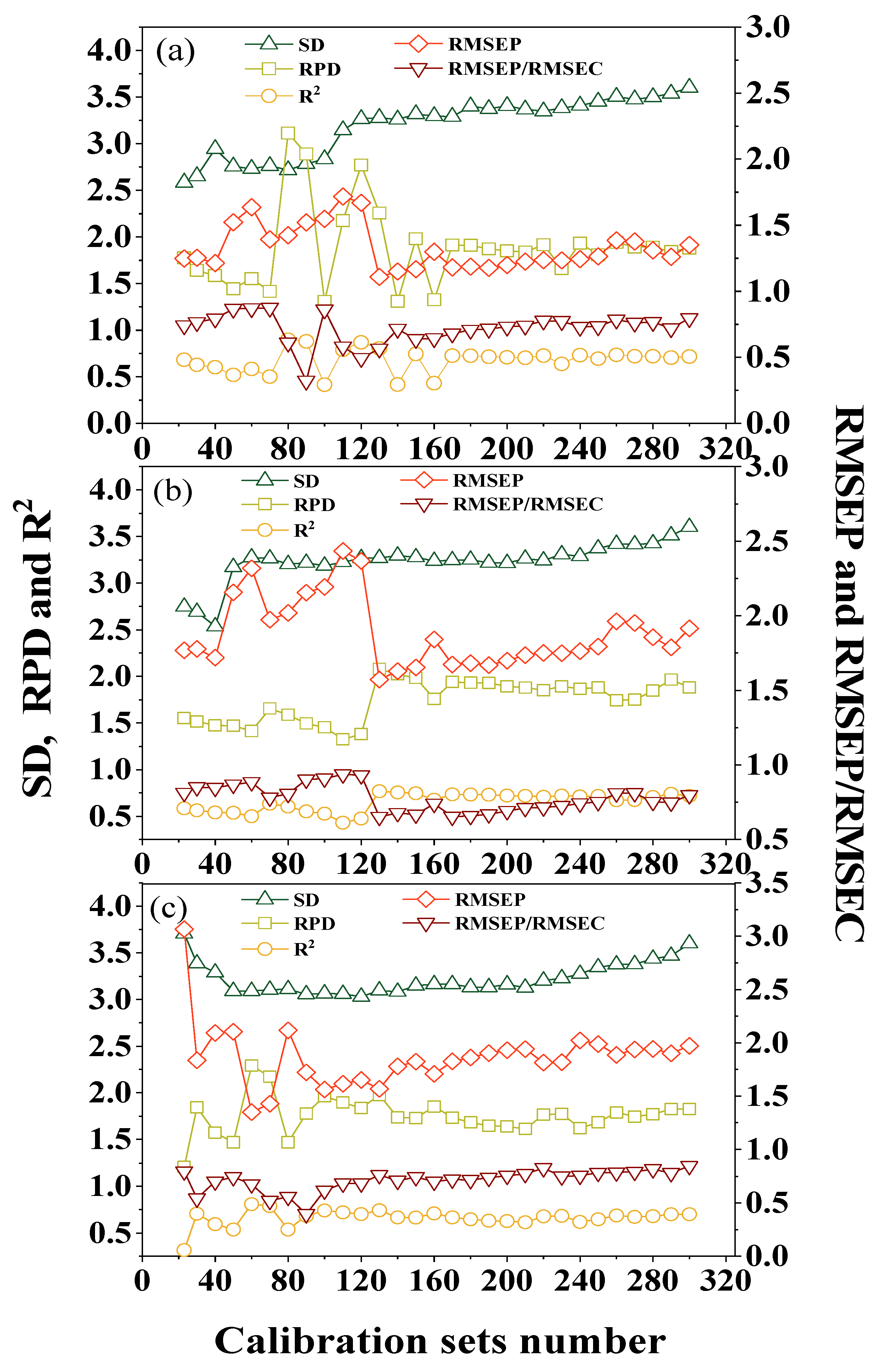
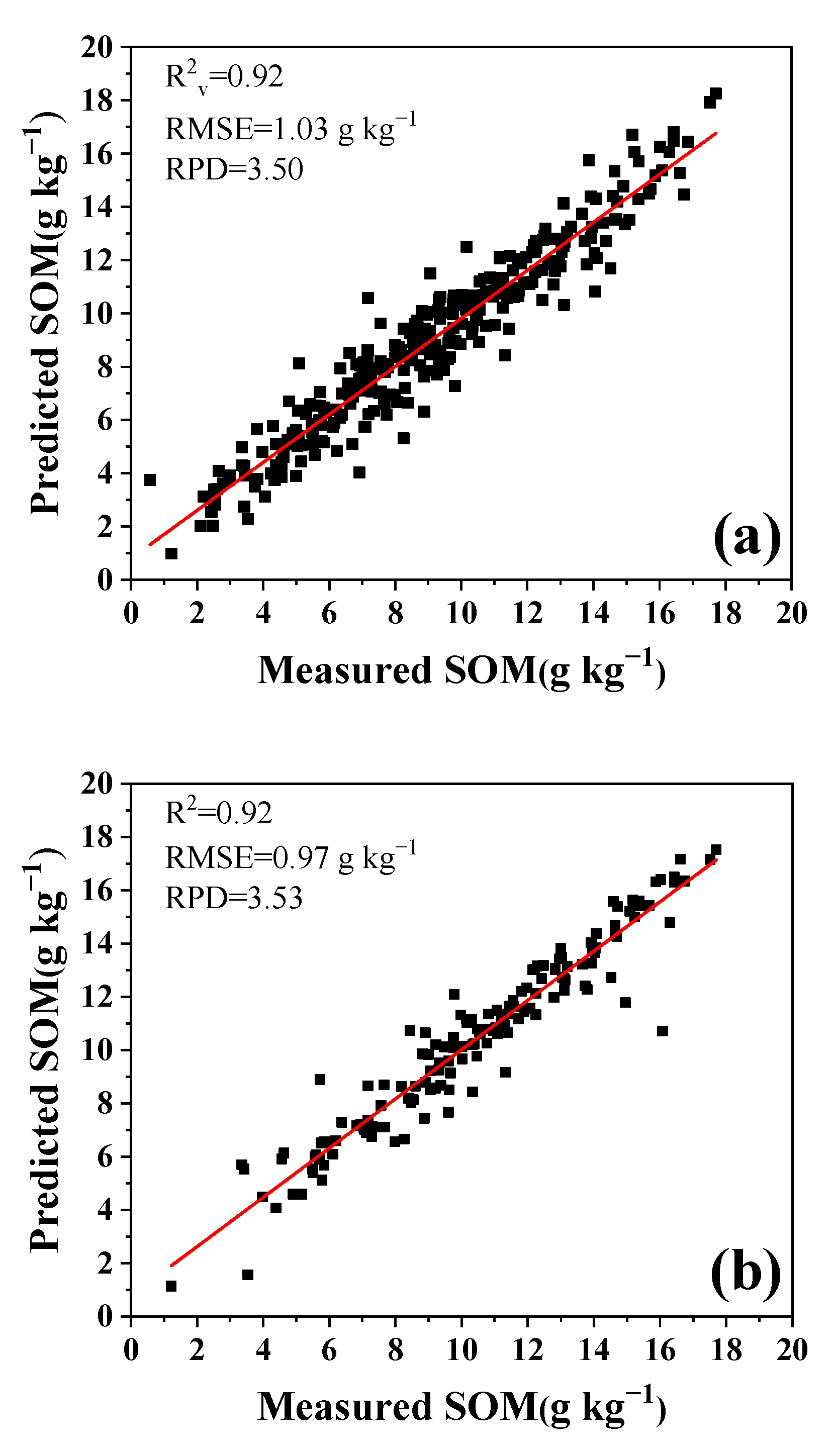
3.3. Optimization for Calibration Dataset
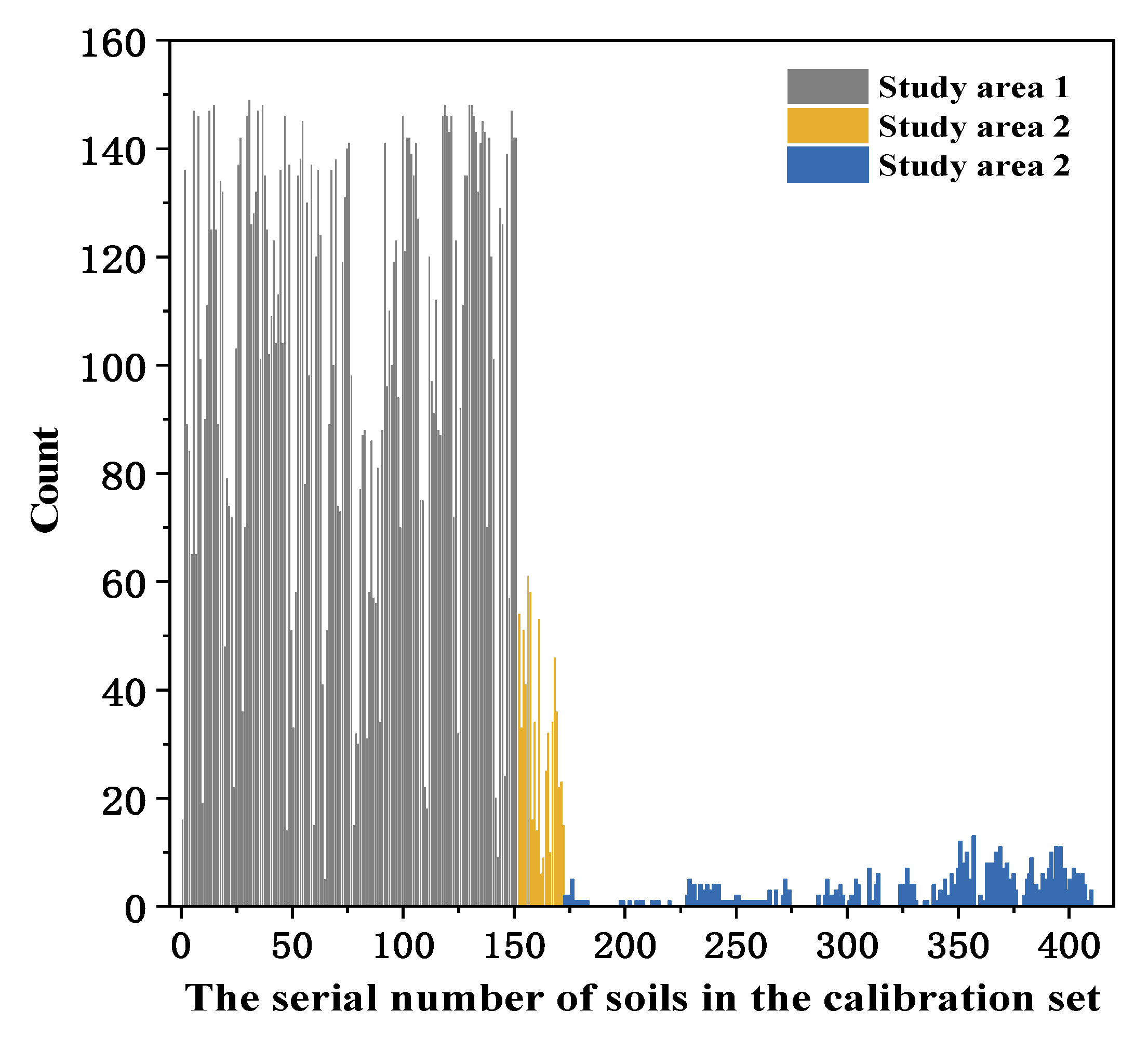
3.3.1. Re-Ordered Spectra Database
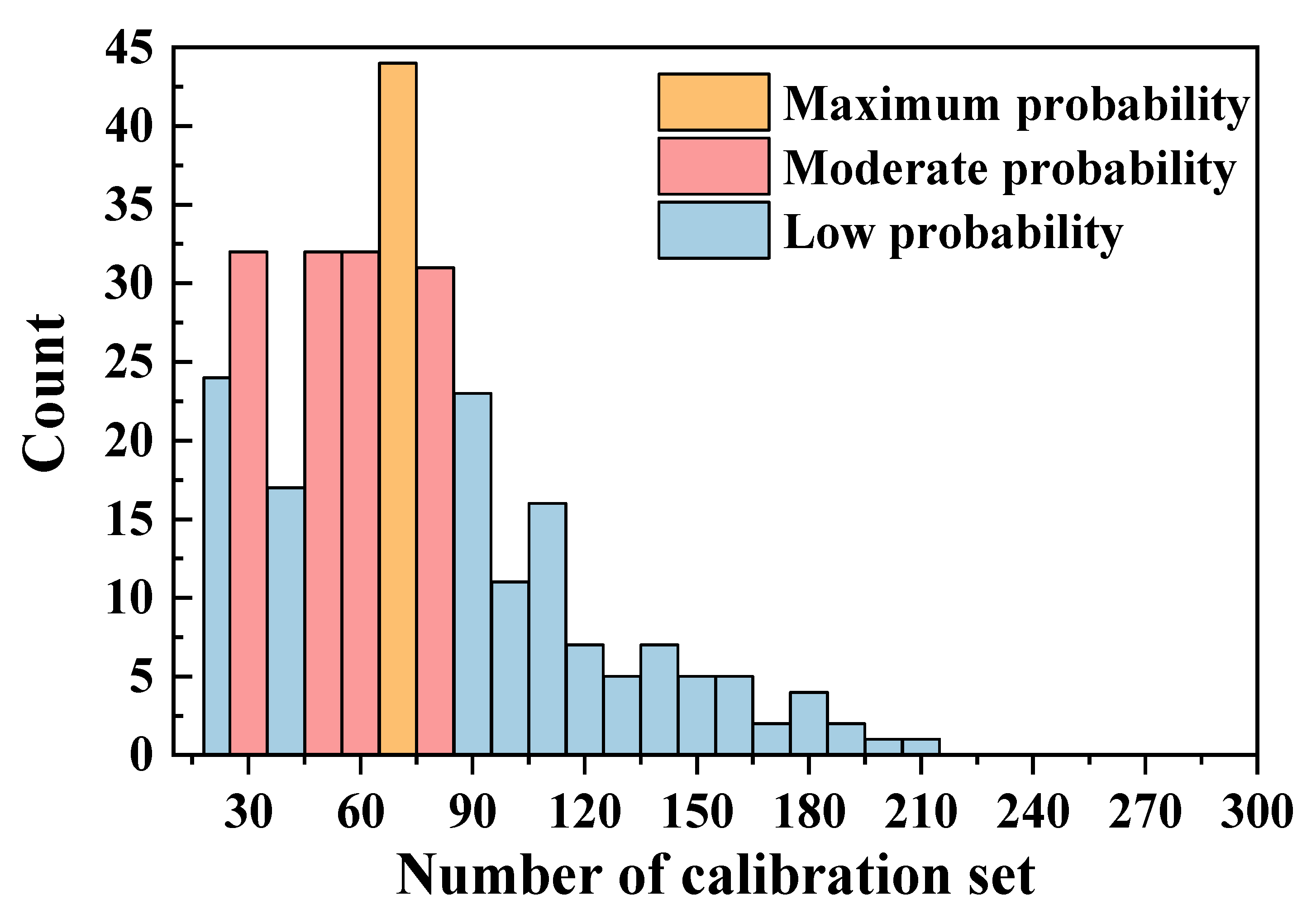
3.3.2. Optimal Sample Numbers for Calibration
3.3.3. Effects of Sample Depth on Calibration Dataset
3.3.4. Effect of Sample Sites on Calibration Dataset
4. Discussion
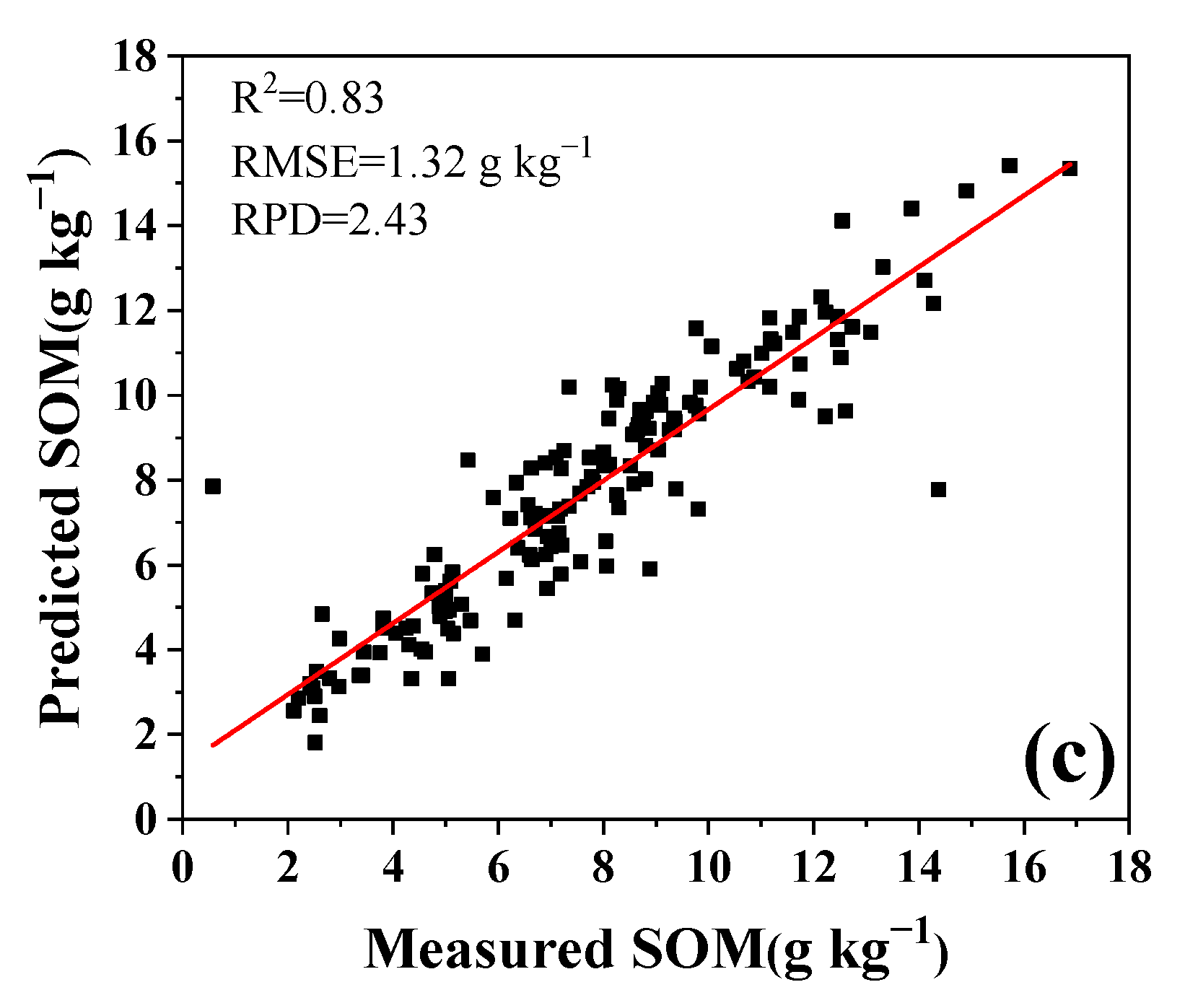
4.1. LIBS Sensor for Soil Measurement
4.2. Influences on SAM–PLSR Modeling Performance
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, C.H.; Xu, X.; Ju, C.H.; Chen, Y.H.; Wilsey, B.J.; Luo, Y.Q.; Fan, W. Long-term, amplified responses of soil organic carbon to nitrogen addition worldwide. Glob. Chang. Biol. 2021, 27, 1170–1180. [Google Scholar] [CrossRef] [PubMed]
- Lal, R. Soil carbon sequestration impacts on global climate change and food security. Science 2004, 304, 1623–1627. [Google Scholar] [CrossRef] [Green Version]
- Lal, R. Sequestering carbon in soils of agro-ecosystems. Food Policy 2011, 36, S33–S39. [Google Scholar] [CrossRef]
- Sun, X.D.; Wang, G.; Ma, Q.X.; Liao, J.H.; Wang, D.; Guan, Q.W.; Jones, D.L. Organic mulching promotes soil organic carbon accumulation to deep soil layer in an urban plantation forest. For. Ecosyst. 2021, 8, 2. [Google Scholar] [CrossRef]
- Emran, M.; Doni, S.; Macci, C.; Masciandaro, G.; Rashad, M.; Gispert, M. Susceptible soil organic matter, SOM, fractions to agricultural management practices in salt-affected soils. Geoderma 2020, 366, 114257. [Google Scholar] [CrossRef]
- McBratney, A.B.; Minasny, B.; Viscarra-Rossel, R. Spectral soil analysis and inference systems: A powerful combination for solving the soil data crisis. Geoderma 2006, 136, 272–278. [Google Scholar] [CrossRef]
- Du, C.W.; Zhou, J.M. Evaluation of soil fertility using infrared spectroscopy: A review. Environ. Chem. Lett. 2008, 7, 97–113. [Google Scholar] [CrossRef]
- Knadel, M.; Viscarra-Rossel, R.A.; Deng, F.; Thomsen, A.; Greve, M.H. Visible-near infrared spectra as a proxy for topsoil texture and glacial boundaries. Soil Sci. Soc. Am. J. 2013, 77, 568–579. [Google Scholar] [CrossRef]
- Burakov, V.S.; Tarasenko, N.V.; Nedelko, M.I.; Kononov, V.A.; Vasilev, N.N.; Isakov, S.N. Analysis of lead and sulfur in environmental samples by double pulse laser induced breakdown spectroscopy. Spectroc. Acta Pt. B Atom. Spectr. 2009, 64, 141–146. [Google Scholar] [CrossRef]
- Capitelli, F.; Colao, F.; Provenzano, M.R.; Fantoni, R.; Brunetti, G.; Senesi, N. Determination of heavy metals in soils by laser induced breakdown spectroscopy. Geoderma 2002, 106, 45–62. [Google Scholar] [CrossRef]
- Bilge, G.; Boyaci, I.H.; Eseller, K.E.; Tamer, U.; Cakir, S. Analysis of bakery products by laser-induced breakdown spectroscopy. Food Chem. 2015, 181, 186–190. [Google Scholar] [CrossRef]
- Abdel-Salam, Z.; Al Sharnoubi, J.; Harith, M.A. Qualitative evaluation of maternal milk and commercial infant formulas via LIBS. Talanta 2013, 115, 422–426. [Google Scholar] [CrossRef]
- Wei, K.; Cui, X.; Teng, G.; Khan, M.N.; Wang, Q.Q. Distinguish Fritillaria cirrhosa and non-Fritillaria cirrhosa using laser-induced breakdown spectroscopy. Plasma Sci. Technol. 2021, 23, 085507. [Google Scholar] [CrossRef]
- Wang, Q.Q.; Xiangli, W.T.; Teng, G.E.; Cui, X.T.; Wei, K. A brief review of laser-induced breakdown spectroscopy for human and animal soft tissues: Pathological diagnosis and physiological detection. Appl. Spectrosc. Rev. 2020, 56, 221–241. [Google Scholar] [CrossRef]
- Peng, J.; Liu, F.; Zhou, F.; Song, K.; Zhang, C.; Ye, L.; He, Y. Challenging applications for multi-element analysis by laser-induced breakdown spectroscopy in agriculture: A review. Trac-Trends Anal. Chem. 2016, 85, 260–272. [Google Scholar] [CrossRef]
- Bricklemyer, R.S.; Brown, D.J.; Turk, P.J.; Clegg, S. Comparing vis–NIRS, LIBS, and combined vis–NIRS-LIBS for intact soil core soil carbon measurement. Soil Sci. Soc. Am. J. 2018, 82, 1482–1496. [Google Scholar] [CrossRef]
- Hernández-García, R.; Villanueva-Tagle, M.E.; Calderón-Piñar, F.; Durruthy-Rodríguez, M.D.; Aquino, F.W.B.; Pereira-Filho, E.R.; Pomares-Alfonso, M.S. Quantitative analysis of lead zirconate titanate (PZT) ceramics by laser-induced breakdown spectroscopy (LIBS) in combination with multivariate calibration. Microchem. J. 2017, 130, 21–26. [Google Scholar] [CrossRef]
- Riebe, D.; Erler, A.; Brinkmann, P.; Beitz, T.; Lohmannsroben, H.G.; Gebbers, R. Comparison of calibration approaches in laser-induced breakdown spectroscopy for proximal soil sensing in precision agriculture. Sensors 2019, 19, 5244. [Google Scholar] [CrossRef] [Green Version]
- Kim, G.; Yoon, Y.J.; Kim, H.A.; Cho, H.J.; Park, K. Elemental composition of Arctic soils and aerosols in Ny-Alesund measured using laser-induced breakdown spectroscopy. Spectroc. Acta Pt. B Atom. Spectr. 2017, 134, 17–24. [Google Scholar] [CrossRef]
- Ferreira, E.C.; Ferreira, E.J.; Villas-Boas, P.R.; Senesi, G.S.; Carvalho, C.M.; Romano, R.A.; Martin Neto, L.; Milori, D.M.B.P. Novel estimation of the humification degree of soil organic matter by laser-induced breakdown spectroscopy. Spectroc. Acta Pt. B Atom. Spectr. 2014, 99, 76–81. [Google Scholar] [CrossRef]
- Ma, F.; Du, C.; Zhou, J. A self-adaptive model for the prediction of soil organic matter using mid-infrared photoacoustic spectroscopy. Soil Sci. Soc. Am. J. 2016, 80, 238–246. [Google Scholar] [CrossRef]
- Xu, X.B.; Du, C.W.; Ma, F.; Shen, Y.Z.; Zhang, Y.Q. Modified self-adaptive model for improving the prediction accuracy of soil organic matter by laser-induced breakdown spectroscopy. Soil Sci. Soc. Am. J. 2020, 84, 1995–2009. [Google Scholar] [CrossRef]
- Dacqui, L.P.; Pucci, A.; Janik, L.J. Soil properties prediction of western Mediterranean islands with similar climatic environments by means of mid-infrared diffuse reflectance spectroscopy. Eur. J. Soil Sci. 2010, 61, 865–876. [Google Scholar] [CrossRef]
- Asa, E.; Saafi, M.; Membah, J.; Billa, A. Comparison of linear and nonlinear kriging methods for characterization and interpolation of soil data. J. Comput. Civi. Eng. 2012, 26, 11–18. [Google Scholar] [CrossRef]
- Xue, J.; Ren, L. Assessing water productivity in the Hetao Irrigation District in Inner Mongolia by an agro-hydrological model. Irrig. Sci. 2017, 35, 357–382. [Google Scholar] [CrossRef]
- Kimble, J.M.; Knox, E.G.; Holzhey, C.S. Soil Survey Laboratory Methods for Characterizing Physical and Chemical-Properties and Mineralogy of Soils; Symp on Application of Agricultural Analysis in Environmental Studies: Atlantic City, NJ, USA, 1991; pp. 23–31. [Google Scholar]
- Soltanpour, P.N.; Jones, J.B., Jr.; Workman, S.M. Optical emission spectrometry. In Methods of Soil Analysis, Part 2: Chemicla and Microbiological Properties; ASA and SSSA: Madison, WI, USA, 1982; pp. 29–65. [Google Scholar]
- Bellon-Maurel, V.; Fernandez-Ahumada, E.; Palagos, B.; Roger, J.M.; McBratney, A. Critical review of chemometric indicators commonly used for assessing the quality of the prediction of soil attributes by NIR spectroscopy. Trac-Trends Anal. Chem. 2010, 29, 1073–1081. [Google Scholar] [CrossRef]
- Viscarra-Rossel, R.A.; McGlynn, R.N.; McBratney, A.B. Determining the composition of mineral-organic mixes using UV–vis–NIR diffuse reflectance spectroscopy. Geoderma 2006, 137, 70–82. [Google Scholar] [CrossRef]
- Couteaux, M.; Sarmiento, L.; Herve, D.; Acevedo, D. Determination of water-soluble and total extractable polyphenolics in biomass, necromass and decomposing plant material using near-infrared reflectance spectroscopy (NIRS). Soil Biol. Biochem. 2005, 37, 795–799. [Google Scholar] [CrossRef] [Green Version]
- Ciais, P.; Bombelli, A.; Williams, M.; Piao, S.L.; Chave, J.; Ryan, C.M.; Henry, M.; Brender, P.; Valentini, R. The carbon balance of Africa: Synthesis of recent research studies. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2011, 369, 2038–2057. [Google Scholar] [CrossRef]



| SOM | Ca2+ | Mg2+ | K++Na+ | ||
|---|---|---|---|---|---|
| n | (g kg−1) | (mg kg−1) | (mg kg−1) | (mg kg−1) | |
| Study Area 1 | 302 | 9.11 ± 3.61 | 120.70 ± 70.67 | 64.48 ± 53.11 | 350.98 ± 194.25 |
| 0–20 topsoil | 151 | 10.46 ± 3.45 | 126.70 ± 73.76 | 66.09 ± 55.57 | 360.81 ± 243.83 |
| 20–40 subsoil | 151 | 7.74 ± 3.23 | 109.12 ± 46.54 | 59.56 ± 41.83 | 353.3 ± 189.45 |
| Study Area 2 soil | 261 | 12.81 ± 6.08 | 116.92 ± 66.98 | 62.46 ± 47.03 | 284.08 ± 193.72 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, M.; Ma, F.; Li, Z.; Xu, X.; Du, C. Sensing of Soil Organic Matter Using Laser-Induced Breakdown Spectroscopy Coupled with Optimized Self-Adaptive Calibration Strategy. Sensors 2022, 22, 1488. https://doi.org/10.3390/s22041488
Hu M, Ma F, Li Z, Xu X, Du C. Sensing of Soil Organic Matter Using Laser-Induced Breakdown Spectroscopy Coupled with Optimized Self-Adaptive Calibration Strategy. Sensors. 2022; 22(4):1488. https://doi.org/10.3390/s22041488
Chicago/Turabian StyleHu, Mengjin, Fei Ma, Zhenwang Li, Xuebin Xu, and Changwen Du. 2022. "Sensing of Soil Organic Matter Using Laser-Induced Breakdown Spectroscopy Coupled with Optimized Self-Adaptive Calibration Strategy" Sensors 22, no. 4: 1488. https://doi.org/10.3390/s22041488
APA StyleHu, M., Ma, F., Li, Z., Xu, X., & Du, C. (2022). Sensing of Soil Organic Matter Using Laser-Induced Breakdown Spectroscopy Coupled with Optimized Self-Adaptive Calibration Strategy. Sensors, 22(4), 1488. https://doi.org/10.3390/s22041488







