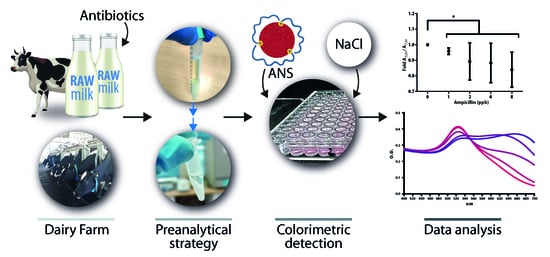
A Simple Yet Effective Preanalytical Strategy Enabling the Application of Aptamer-Conjugated Gold Nanoparticles for the Colorimetric Detection of Antibiotic Residues in Raw Milk
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Synthesis of Gold Nanoparticles (AuNPs)
2.3. AuNPs Characterization
2.4. Synthesis of NAS and the Determination of Detection Parameters
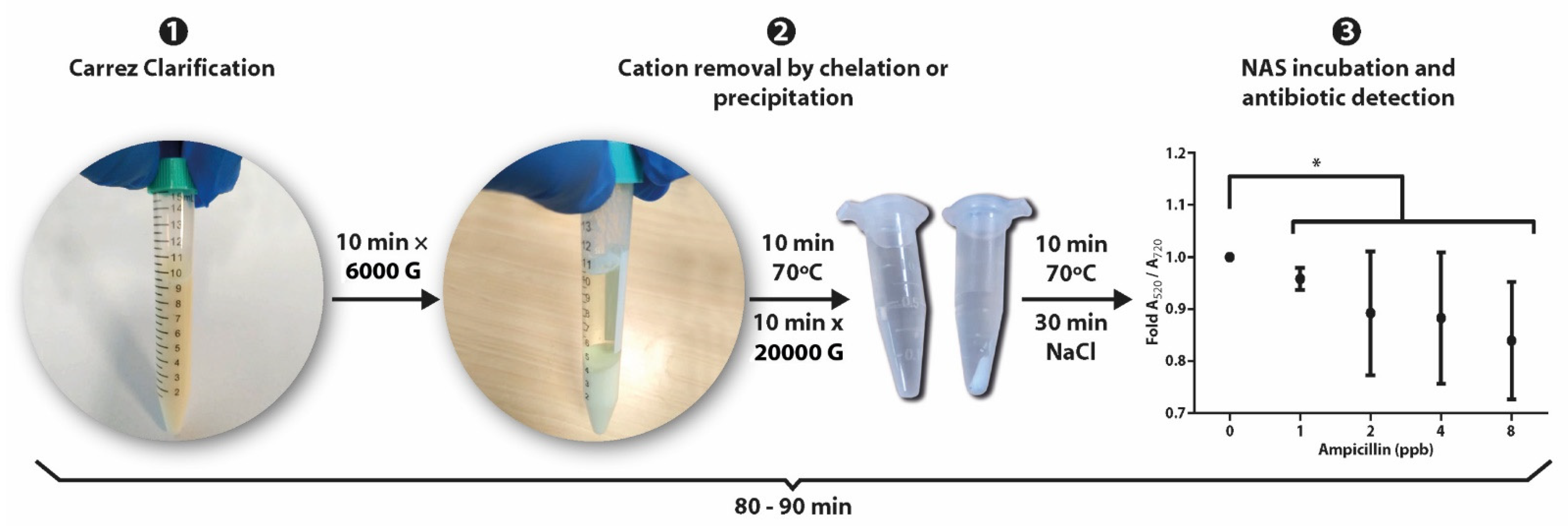
2.5. Preanalytical Processing of Raw Milk
- Carrez clarification: Five hundred milliliters of Carrez I reagent was added into 10 mL of raw milk without antibiotics (control) or with kanamycin, oxytetracycline, sulfadimethoxine or ampicillin, and vortexed for 1 min. Then, 500 mL of Carrez II reagent was added and vortexed for 1 min until the mixture was homogeneous, which was subsequently centrifuged at 1000× g for 5 min. The supernatant (milk whey) was recovered and immediately used for the detection of antibiotics with the NAS.
- Ethyl acetate clarification: Four milliliters of raw milk was centrifuged for 20 min at 1000 and 10 °C to separate the fat. Then, 2 mL of the supernatant was diluted with 2 mL of nanopure water and stirred for 10 min in a vortex. Seven milliliters of ethyl acetate was added, vortexed for 15 min and centrifuged for 15 min at 1500× g and 4 °C, which gave rise to a three-phase mixture. The bottom layer (milk whey) was recovered and centrifugated again before being stored at 4 °C.
2.6. Milk and Milk Whey Characterization
2.7. Colorimetric Detection of Antibiotics in Clarificatecd Raw Milk
2.8. Colorimetric Detection of Antibiotics in Cation-Removed Milk Whey
2.9. Statistical Analysis
3. Results and Discussions
3.1. NAS and Detection Principle
3.2. Determination of the Detection Parameters of NAS
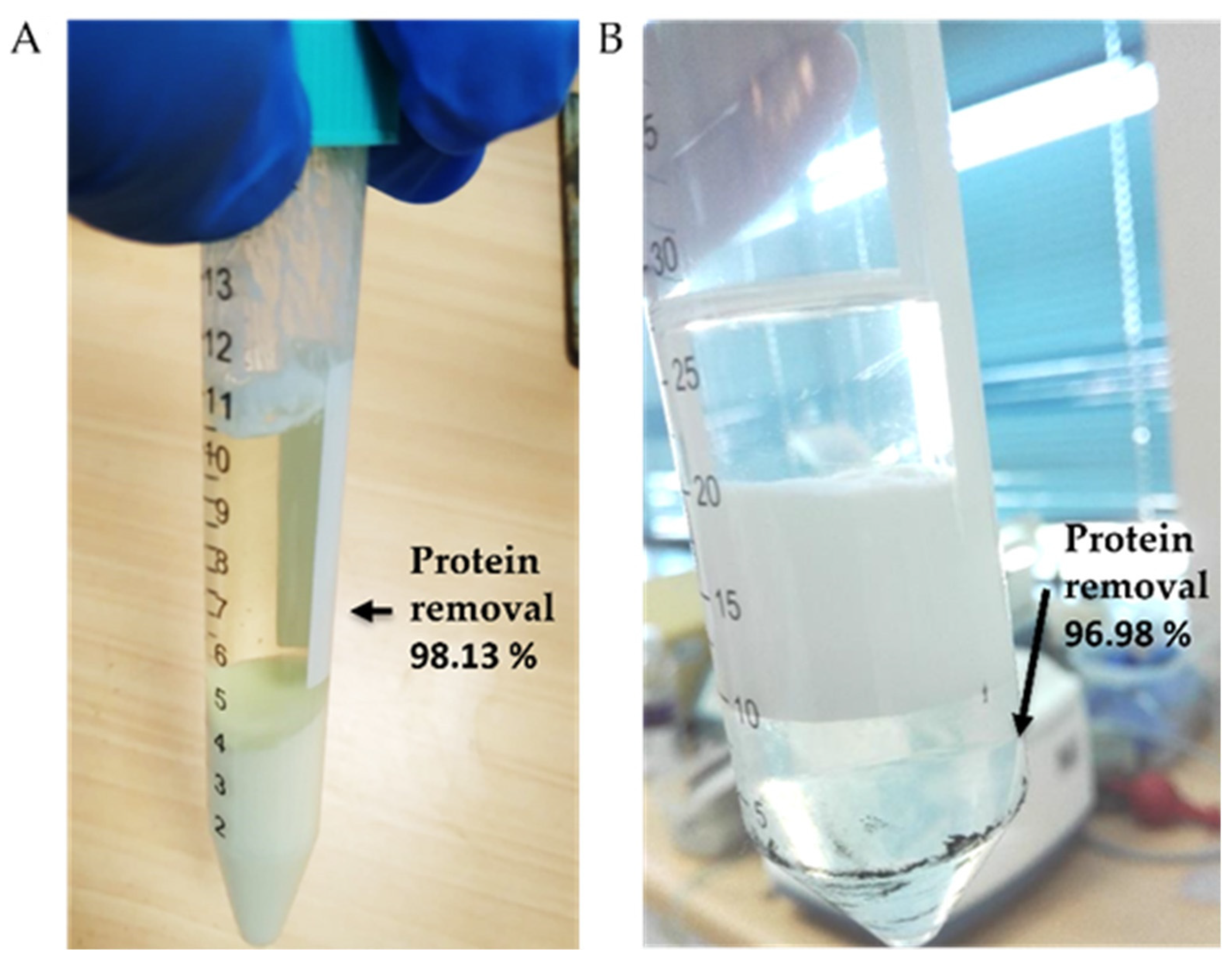
3.3. Raw Milk Clarification
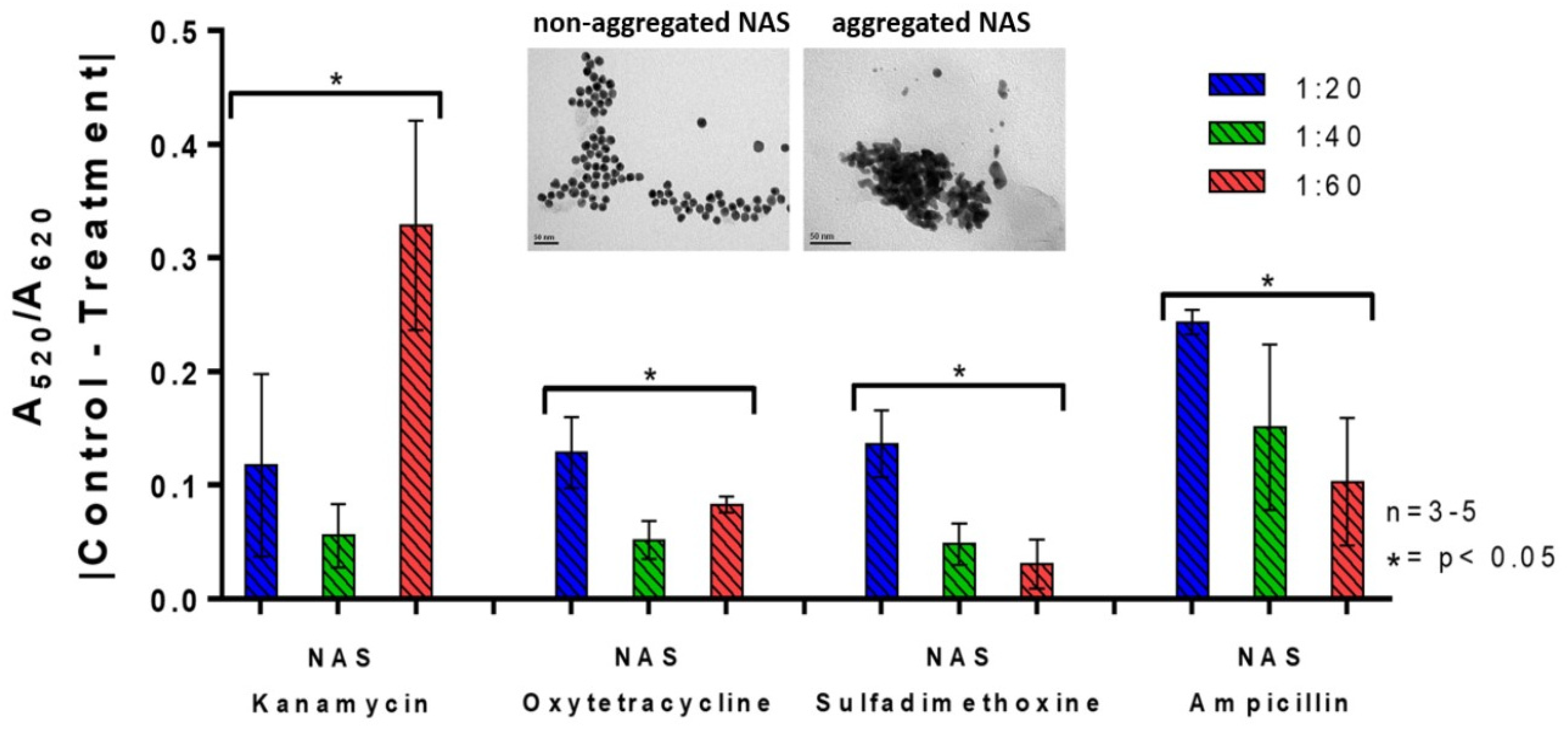
3.4. Effect of Ion and Lactose on the Aggregation of AuNPs
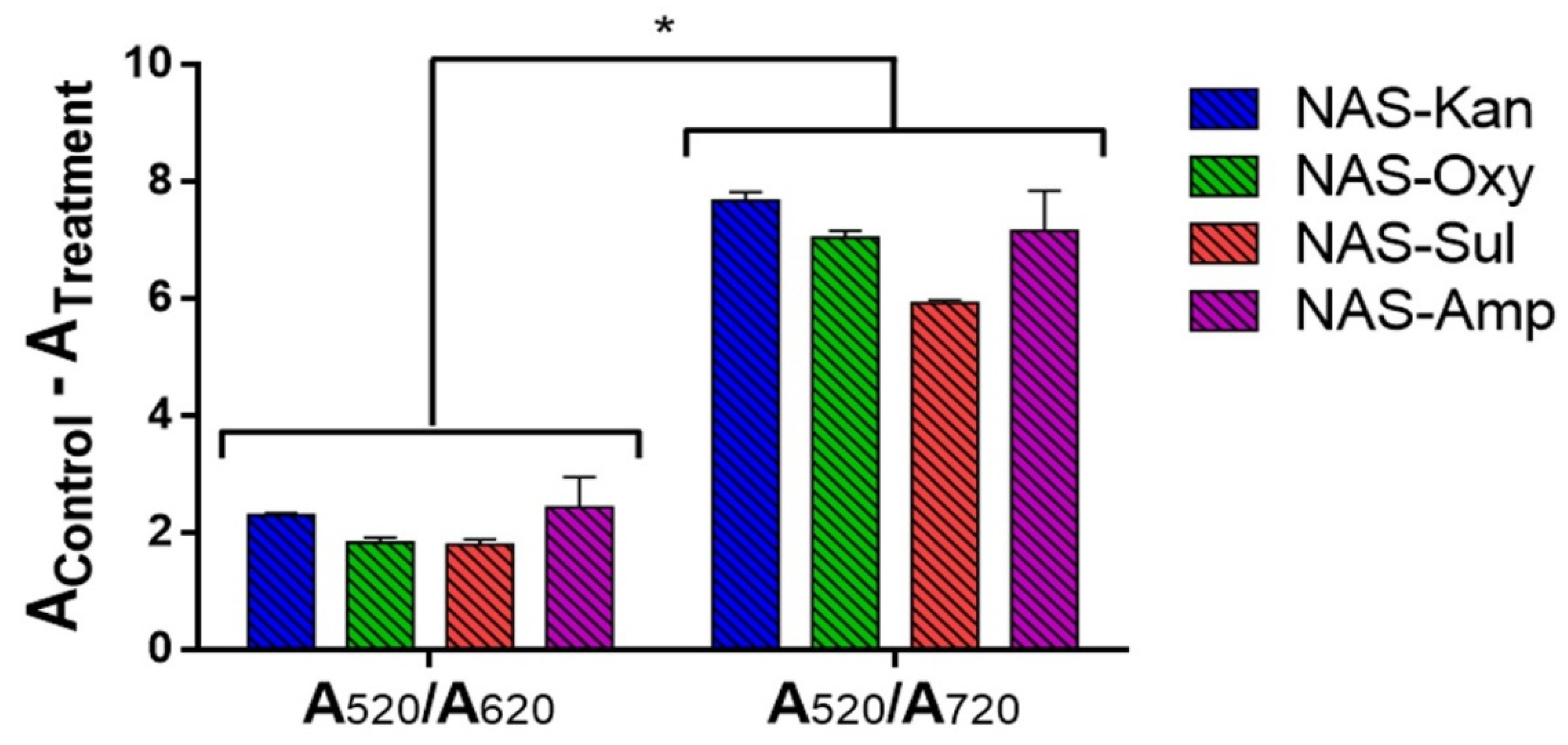
3.5. Cation Removal and Antibiotic Detection
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haug, A.; Høstmark, A.T.; Harstad, O.M. Bovine Milk in Human Nutrition—A Review. Lipids Health Dis. 2007, 6, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- USDA Foreign Agricultural Service. Dairy: World Markets and Trade. Washington, DC, USA. Available online: https://Apps.Fas.Usda.Gov/Psdonline/Circulars/Dairy.Pdf (accessed on 13 September 2021).
- Fusco, V.; Chieffi, D.; Fanelli, F.; Logrieco, A.F.; Cho, G.-S.; Kabisch, J.; Böhnlein, C.; Franz, C.M.A.P. Microbial Quality and Safety of Milk and Milk Products in the 21st Century. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2013–2049. [Google Scholar] [CrossRef] [PubMed]
- Boeckel, T.P.V.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global Trends in Antimicrobial Use in Food Animals. PNAS 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, H.; Cheng, G.; Iqbal, Z.; Ai, X.; Hussain, H.I.; Huang, L.; Dai, M.; Wang, Y.; Liu, Z.; Yuan, Z. Benefits and Risks of Antimicrobial Use in Food-Producing Animals. Front. Microbiol. 2014, 5, 288. [Google Scholar] [CrossRef] [Green Version]
- Sharma, C.; Rokana, N.; Chandra, M.; Singh, B.P.; Gulhane, R.D.; Gill, J.P.S.; Ray, P.; Puniya, A.K.; Panwar, H. Antimicrobial Resistance: Its Surveillance, Impact, and Alternative Management Strategies in Dairy Animals. Front. Vet. Sci. 2018, 4, 237. [Google Scholar] [CrossRef]
- Limmathurotsakul, D.; Dunachie, S.; Fukuda, K.; Feasey, N.A.; Okeke, I.N.; Holmes, A.H.; Moore, C.E.; Dolecek, C.; van Doorn, H.R.; Shetty, N.; et al. Improving the Estimation of the Global Burden of Antimicrobial Resistant Infections. Lancet Infect. Dis. 2019, 19, e392–e398. [Google Scholar] [CrossRef]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. HM Government and Welcome Trust: UK. 2016. Available online: https://Amr-Review.Org/Sites/Default/Files/160518_Final%20paper_with%20cover.Pdf. (accessed on 18 October 2021).
- Chiesa, L.M.; DeCastelli, L.; Nobile, M.; Martucci, F.; Mosconi, G.; Fontana, M.; Castrica, M.; Arioli, F.; Panseri, S. Analysis of Antibiotic Residues in Raw Bovine Milk and Their Impact toward Food Safety and on Milk Starter Cultures in Cheese-Making Process. LWT 2020, 131, 109783. [Google Scholar] [CrossRef]
- Commission Regulation (EU) No 37/2010 of 22 December 2009 on Pharmacologically Active Substances and Their Classification Regarding Maximum Residue Limits in Foodstuffs of Animal Origin. Official Journal of the European Union. 2010 L15: 1–72. Available online: https://Ec.Europa.Eu/Health/Sites/Default/Files/Files/Eudralex/Vol-5/Reg_2010_37/Reg_2010_37_en.Pdf (accessed on 18 October 2021).
- MINSAL Subsecretaría de Salud Pública. Resolución 1560 Exenta Fija Límites Máximos de Residuos de Medicamentos Veterinarios En Alimentos Destinados al Consumo Humano. Santiago, Chile. Available online: https://www.Bcn.Cl/Leychile/Navegar?IdNorma=1135977 (accessed on 18 October 2021).
- Gaudin, V. The Growing Interest in Development of Innovative Optical Aptasensors for the Detection of Antimicrobial Residues in Food Products. Biosensors 2020, 10, 21. [Google Scholar] [CrossRef] [Green Version]
- Somwanshi, S.; Somvanshi, S.; Kharat, P. Nanocatalyst: A Brief Review on Synthesis to Applications. J. Phys. Conf. Ser. 2020, 1644, 012046. [Google Scholar] [CrossRef]
- Jain, K.; Patel, A.S.; Pardhi, V.P.; Flora, S.J.S. Nanotechnology in Wastewater Management: A New Paradigm Towards Wastewater Treatment. Molecules 2021, 26, 1797. [Google Scholar] [CrossRef]
- McClements, D. Nanotechnology Approaches for Improving the Healthiness and Sustainability of the Modern Food Supply. ACS Omega 2020, 5, 29623–29630. [Google Scholar] [CrossRef] [PubMed]
- Zuliani, A.; Ranjan, P.; Luque, R.; Van der Eycken, E.V. Heterogeneously Catalyzed Synthesis of Imidazolones via Cycloisomerizations of Propargylic Ureas Using Ag and Au/Al SBA-15 Systems. ACS Sustain. Chem. Eng. 2019, 7, 5568–5575. [Google Scholar] [CrossRef]
- Moram, S.S.B.; Shaik, A.K.; Byram, C.; Hamad, S.; Soma, V.R. Instantaneous Trace Detection of Nitro-Explosives and Mixtures with Nanotextured Silicon Decorated with Ag–Au Alloy Nanoparticles Using the SERS Technique. Anal. Chim. Acta 2020, 1101, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Dev, A.; Karmakar, S. Nanosensors and Nanobiosensors in Food and Agriculture. Environ. Chem. Lett. 2018, 16, 161–182. [Google Scholar] [CrossRef]
- Prajapati, S.; Padhan, B.; Amulyasai, B.; Sarkar, A. Chapter 11—Nanotechnology-Based Sensors. In Biopolymer-Based Formulations; Pal, K., Banerjee, I., Sarkar, P., Kim, D., Deng, W.-P., Dubey, N.K., Majumder, K., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 237–262. ISBN 978-0-12-816897-4. [Google Scholar] [CrossRef]
- Díaz-García, V.; Retamal-Morales, G. Aptamer and Riboswitches: A Novel Tool for the Need of New Antimicrobial Active Compounds. In Antibiotic Materials in Healthcare; Kokkarachedu, V., Kanikireddy, V., Sadiku, R., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 231–247. [Google Scholar] [CrossRef]
- Hu, X.; Chang, K.; Wang, S.; Sun, X.; Hu, J.; Jiang, M. Aptamer-Functionalized AuNPs for the High-Sensitivity Colorimetric Detection of Melamine in Milk Samples. PLoS ONE 2018, 13, e0201626. [Google Scholar] [CrossRef] [Green Version]
- Bai, W.; Zhu, C.; Liu, J.; Yan, M.; Yang, S.; Chen, A. Gold Nanoparticle-Based Colorimetric Aptasensor for Rapid Detection of Six Organophosphorous Pesticides. Environ. Toxicol. Chem. 2015, 34, 2244–2249. [Google Scholar] [CrossRef]
- Yin, X.; Wang, S.; Liu, X.; He, C.; Tang, Y.; Li, Q.; Liu, J.; Su, H.; Tan, T.; Dong, Y. Aptamer-Based Colorimetric Biosensing of Ochratoxin A in Fortified White Grape Wine Sample Using Unmodified Gold Nanoparticles. Anal. Sci. 2017, 33, 659–664. [Google Scholar] [CrossRef] [Green Version]
- Priyadarshini, E.; Pradhan, N. Gold Nanoparticles as Efficient Sensors in Colorimetric Detection of Toxic Metal Ions: A Review. Sens. Actuators B Chem. 2017, 238, 888–902. [Google Scholar] [CrossRef]
- Liu, G.; Lu, M.; Huang, X.; Li, T.; Xu, D. Application of Gold-Nanoparticle Colorimetric Sensing to Rapid Food Safety Screening. Sensors 2018, 18, 4166. [Google Scholar] [CrossRef] [Green Version]
- Zahra, Q.; Luo, Z.; Ali, R.; Khan, M.I.; Li, F.; Qiu, B. Advances in Gold Nanoparticles-Based Colorimetric Aptasensors for the Detection of Antibiotics: An Overview of the Past Decade. Nanomaterials 2021, 11, 840. [Google Scholar] [CrossRef]
- Zhang, F.; Liu, J. Label-Free Colorimetric Biosensors Based on Aptamers and Gold Nanoparticles: A Critical Review. Anal. Sens. 2021, 1, 30–43. [Google Scholar] [CrossRef]
- Amendola, V.; Pilot, R.; Frasconi, M.; Maragò, O.M.; Iatì, M.A. Surface Plasmon Resonance in Gold Nanoparticles: A Review. J. Phys. Condens. Matter 2017, 29, 203002. [Google Scholar] [CrossRef]
- Yan, S.; Foroughi, M.; Safaei, M.; Jahani, S.; Ebrahimpour, N.; Borhani, F.; Zade, N.; Aramesh-Boroujeni, Z.; Foong, L. A review: Recent advances in ultrasensitive and highly specific recognition aptasensors with various detection strategies. Int. J. Biol. Macrom. 2020, 155, 184–207. [Google Scholar] [CrossRef]
- Andrew, S.M. Effect of Fat and Protein Content of Milk from Individual Cows on the Specificity Rates of Antibiotic Residue Screening Tests. J. Dairy Sci. 2000, 83, 2992–2997. [Google Scholar] [CrossRef]
- Zhou, N.; Zhang, J.; Tian, Y. Aptamer-Based Spectrophotometric Detection of Kanamycin in Milk. Anal. Methods 2014, 6, 1569–1574. [Google Scholar] [CrossRef]
- He, L.; Luo, Y.; Zhi, W.; Wu, Y.; Zhou, P.; He, L.; Luo, Y.; Zhi, W.; Wu, Y.; Zhou, P. A Colorimetric Aptamer Biosensor Based on Gold Nanoparticles for the Ultrasensitive and Specific Detection of Tetracycline in Milk. Aust. J. Chem. 2013, 66, 485–490. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.-H.; Lee, L.-P.; Min, J.-R.; Lim, M.-W.; Jeong, S.-H. An Indirect Competitive Assay-Based Aptasensor for Detection of Oxytetracycline in Milk. Biosens. Bioelectron. 2014, 51, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Emrani, A.S.; Danesh, N.M.; Lavaee, P.; Ramezani, M.; Abnous, K.; Taghdisi, S.M. Colorimetric and Fluorescence Quenching Aptasensors for Detection of Streptomycin in Blood Serum and Milk Based on Double-Stranded DNA and Gold Nanoparticles. Food Chem. 2016, 190, 115–121. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, Y.; Tao, H.; Chen, H.; Yang, W.; Qiu, S. Colorimetric Detection of Streptomycin in Milk Based on Peroxidase-Mimicking Catalytic Activity of Gold Nanoparticles. RSC Adv. 2017, 7, 38471–38478. [Google Scholar] [CrossRef] [Green Version]
- Adami, A.; Mortari, A.; Morganti, E.; Lorenzelli, L. Microfluidic Sample Preparation Methods for the Analysis of Milk Contaminants. J. Sens. 2015, 2016, e2385267. [Google Scholar] [CrossRef] [Green Version]
- Contreras-Trigo, B.; Díaz-García, V.; Guzmán-Gutierrez, E.; Sanhueza, I.; Coelho, P.; Godoy, S.E.; Torres, S.; Oyarzún, P. Slight pH Fluctuations in the Gold Nanoparticle Synthesis Process Influence the Performance of the Citrate Reduction Method. Sensors 2018, 18, 2246. [Google Scholar] [CrossRef] [Green Version]
- Maye, M.M.; Han, L.; Kariuki, N.N.; Ly, N.K.; Chan, W.-B.; Luo, J.; Zhong, C.-J. Gold and Alloy Nanoparticles in Solution and Thin Film Assembly: Spectrophotometric Determination of Molar Absorptivity. Anal. Chim. Acta 2003, 496, 17–27. [Google Scholar] [CrossRef]
- Hill, H.D.; Mirkin, C.A. The Bio-Barcode Assay for the Detection of Protein and Nucleic Acid Targets Using DTT-Induced Ligand Exchange. Nat. Protoc. 2006, 1, 324–336. [Google Scholar] [CrossRef] [PubMed]
- Song, K.-M.; Cho, M.; Jo, H.; Min, K.; Jeon, S.H.; Kim, T.; Han, M.S.; Ku, J.K.; Ban, C. Gold Nanoparticle-Based Colorimetric Detection of Kanamycin Using a DNA Aptamer. Anal. Biochem. 2011, 415, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Kim, J.H.; Kim, I.A.; Lee, S.J.; Jurng, J.; Gu, M.B. A Novel Colorimetric Aptasensor Using Gold Nanoparticle for a Highly Sensitive and Specific Detection of Oxytetracycline. Biosens. Bioelectron. 2010, 26, 1644–1649. [Google Scholar] [CrossRef]
- Chen, A.; Jiang, X.; Zhang, W.; Chen, G.; Zhao, Y.; Tunio, T.M.; Liu, J.; Lv, Z.; Li, C.; Yang, S. High Sensitive Rapid Visual Detection of Sulfadimethoxine by Label-Free Aptasensor. Biosens. Bioelectron. 2013, 42, 419–425. [Google Scholar] [CrossRef]
- Song, K.-M.; Jeong, E.; Jeon, W.; Cho, M.; Ban, C. Aptasensor for Ampicillin Using Gold Nanoparticle Based Dual Fluorescence-Colorimetric Methods. Anal. Bioanal. Chem. 2012, 402, 2153–2161. [Google Scholar] [CrossRef]
- Horwitz, W.; Latimer, G.W. Official Methods of Analysis of AOAC International; AOAC International: Gaithersburg, MA, USA, 2005. [Google Scholar]
- Xue, Y.; Li, X.; Li, H.; Zhang, W. Quantifying Thiol–Gold Interactions towards the Efficient Strength Control. Nat. Commun. 2014, 5, 4348. [Google Scholar] [CrossRef] [Green Version]
- Adeel, M.; Rahman, M.; Lee, J.-J. Label-Free Aptasensor for the Detection of Cardiac Biomarker Myoglobin Based on Gold Nanoparticles Decorated Boron Nitride Nanosheets. Biosens. Bioelectron. 2019, 126, 143–150. [Google Scholar] [CrossRef]
- Peng, Y.; Li, L.; Mu, X.; Guo, L. Aptamer-Gold Nanoparticle-Based Colorimetric Assay for the Sensitive Detection of Thrombin. Sens. Actuators B Chem. 2013, 177, 818–825. [Google Scholar] [CrossRef]
- Azzam EM, S.; El-Farargy AF, M.; Hegazy, M.A.; Abd El-Aal, A.A. Detection of Heavy Metal Ions Using Synthesized Amino Thiol Surfactants Assembled on Gold Nanoparticles. J. Disp. Sci. Technol. 2014, 35, 175–184. [Google Scholar] [CrossRef]
- Li, H.; Rothberg, L. Colorimetric Detection of DNA Sequences Based on Electrostatic Interactions with Unmodified Gold Nanoparticles. PNAS 2004, 101, 14036–14039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Q.; Wang, Y.; Jia, J.; Xiang, Y. Colorimetric aptasensors for determination of tobramycin in milk and chicken eggs based on DNA and gold nanoparticles. Food Chem. 2018, 30, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, P.; Godoy, S.E.; Torres, S.; Oyarzún, P.; Sanhueza, I.; Díaz-García, V.; Contreras-Trigo, B.; Coelho, P. Improved Antibiotic Detection in Raw Milk Using Machine Learning Tools over the Absorption Spectra of a Problem-Specific Nanobiosensor. Sensors 2020, 20, 4552. [Google Scholar] [CrossRef]
- Holt, C. The Milk Salts and Their Interaction with Casein. In Advanced Dairy Chemistry: Lactose, Water, Salts and Vitamins; Fox, P.F., Ed.; Springer: Boston, MA, USA, 1997; Volume 3, pp. 233–256. [Google Scholar] [CrossRef]
- Bachran, K.; Bernhard, R.A. Interaction of Iron(II) with Lactose. J. Agric. Food Chem. 1980, 28, 536–540. [Google Scholar] [CrossRef]
- Reynolds, A.J.; Haines, A.H.; Russell, D.A. Gold Glyconanoparticles for Mimics and Measurement of Metal Ion-Mediated Carbohydrate−Carbohydrate Interactions. Langmuir 2006, 22, 1156–1163. [Google Scholar] [CrossRef]
- Ho, T.T.-T.; Dang, C.-H.; Huynh, T.K.-C.; Hoang, T.K.-D.; Nguyen, T.-D. In Situ Synthesis of Gold Nanoparticles on Novel Nanocomposite Lactose/Alginate: Recyclable Catalysis and Colorimetric Detection of Fe(III). Carbohydr. Polym. 2021, 251, 116998. [Google Scholar] [CrossRef]
- He, L.; Luo, Y.; Zhi, W.; Zhou, P. Colorimetric Sensing of Tetracyclines in Milk Based on the Assembly of Cationic Conjugated Polymer-Aggregated Gold Nanoparticles. Food Anal. Methods 2013, 6, 1704–1711. [Google Scholar] [CrossRef]
- Chen, M.; Gan, N.; Zhou, Y.; Li, T.; Xu, Q.; Cao, Y.; Chen, Y. A novel aptamer- metal ions- nanoscale MOF based electrochemical biocodes for multiple antibiotics detection and signal amplification. Sens. Actuators B Chem. 2017, 242, 1201–1209. [Google Scholar] [CrossRef]
- Giannone, G.; Santi, M.; Ermini, M.L.; Cassano, D.; Voliani, V. A Cost-Effective Approach for Non-Persistent Gold Nano-Architectures Production. Nanomaterials 2020, 10, 1600. [Google Scholar] [CrossRef]
- Gan, P.P.; Ng, S.H.; Huang, Y.; Li, S.F. Green synthesis of gold nanoparticles using palm oil mill effluent (POME): A low-cost and eco-friendly viable approach. Bioresour. Technol. 2012, 113, 132–135. [Google Scholar] [CrossRef] [PubMed]
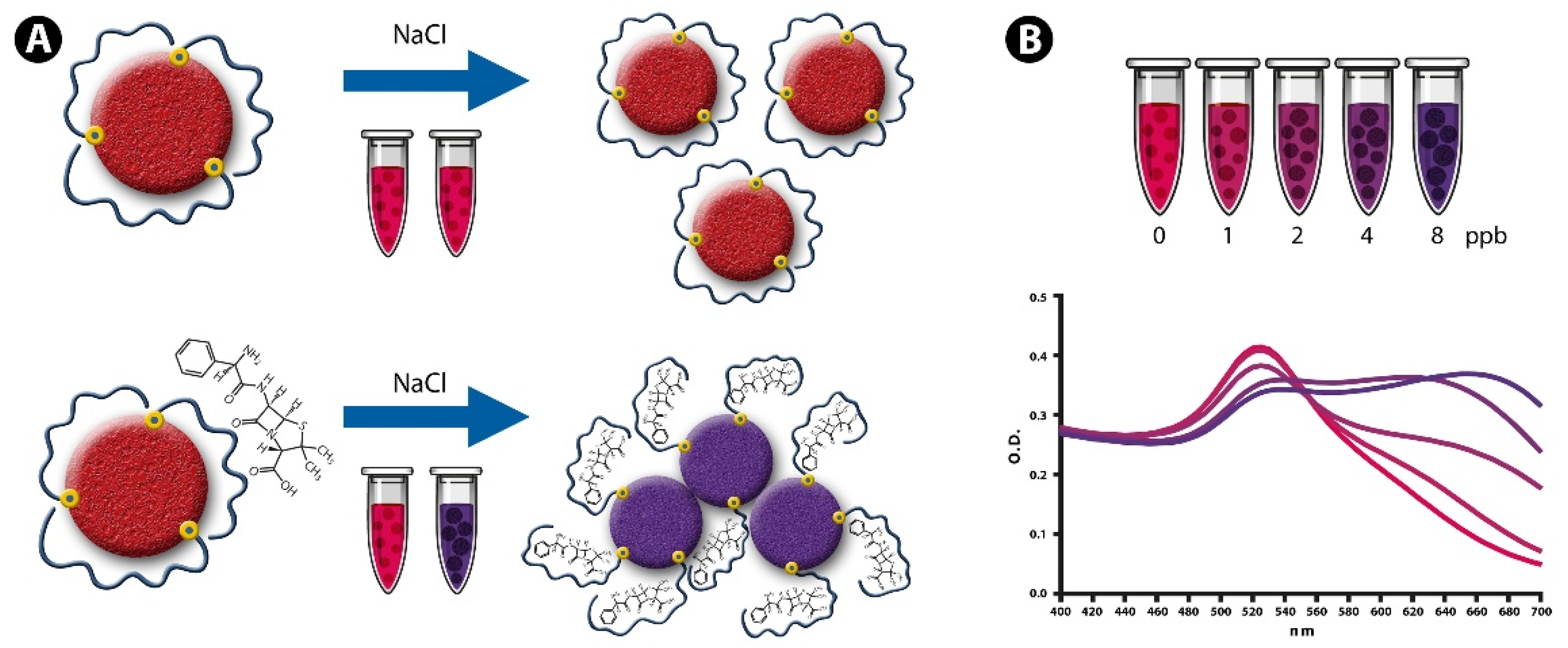




| Antibiotic | Maximum Residual Limit (MRL) | Aptamer Sequence (ssDNA) | Aptamer Ref. |
|---|---|---|---|
| Kanamycin | 150 μg/L * | 5′-TGGGGGTTGAGGCTAAGCCGA-3′ (21b) | [40] |
| Oxytetracycline | 100 μg/L * | 5′-CGTACGGAATTCGCTAGCGGGCGGGGGTGCTGGGGGAATGGAGTGCTGCGTGCTGCGGGGATCCGAGCTCCACGTG-3′ (76b) | [41] |
| Sulfadimethoxine | 25 μg/L † 100 μg/L * | 5°′-GAGGGCAACGAGTGTTTATAGA-3′ (22b) | [42] |
| Ampicillin | 4 μg/L *,† | 5′-GCGGGCGGTTGTATAGCGG-3′ (19b) | [43] |
| Element | Concentration | Synthetic Milk Whey | Lactose Solution | Ionic Solution |
|---|---|---|---|---|
| Lactose | 5% w/v | X | X | |
| Ca2+ | 30.120 mM (60.232 mEq/L) | X | X | |
| Mg2+ | 4.750 mM (9.506 mEq/L) | X | X | |
| Fe3+ | 0.011 mM (0.032 mEq/L) | X | X | |
| PO43− | 30.582 mM (91.745 mEq/L) | X | X | |
| Na+ | 23.587 mM (23.587 mEq/L) | X | X | |
| K+ | 38,930 mM (38.930 mEq/L) | X | X | |
| Zn2+ | 0.066 mM (0.131 mEq/L) | X | X | |
| Citrate3− | 9.292 mM (18.584 mEq/L) | X | X | |
| Cu2+ | 0.002 mM (0.005 mEq/L) | X | X |
| Components | Raw Milk | Milk Whey |
|---|---|---|
| Lipids | 5.3% | 0.0% |
| Proteins | 3.2% | 0.2% |
| Raw fiber | 0.4% | 0.6% |
| Total carbohydrates | 4.7% | 4.5% |
| Lactose | 5.0% | 5.3% |
| Ashes | 0.7% | 1.2% |
| pH | 5.94 | 6.05 |
| Nanoaptasensor | Best Method for Interference Elimination | Detection Limit |
|---|---|---|
| Kanamycin | EDTA | 37.50 μg/L (0.25 MRL) |
| Oxytetracycline | Dilution | 25.00 μg/L (0.25 MRL) |
| Sulfadimethoxine | NaHCO3 | 6.25 μg/L (0.25 MRL) |
| Ampicillin | EDTA | 1.00 μg/L (0.25 MRL) |
| Milk Sample | Antibiotic Residue | Preanalytical Time | Assay Time | LOD | Range | Pretreatment/Antibiotic Addition | Ref. |
|---|---|---|---|---|---|---|---|
| Supermarket milk | Ampicillin (AMP) | 1.5 h | 80 min | 10 ng/mL (28.6 nM) | 1–100 ng/mL (2.86–286 nM) | Ethyl acetate addition/centrifugation at 5000 rpm to obtain supernatant/nitrogen blow-down at 40 °C/pellet resuspension in water (total dilution: 2-fold)/AMP addition (after pretreatment). | [43] |
| N.S. | Tetracycline (TET) | N.S. | 25 min | 45.8 nM | 10–400 nM | Milk dilution with water (1:5)/acetic acid addition/centrifugation (total dilution: 5-fold)/TET addition (protocol details unspecified). | [56] |
| Milk powder | Tetracycline (TET) | N.S. | 60 min | 122 nM | 10–500 nM | Acetic acid addition/centrifugation/TET addition (protocol details unspecified). | [32] |
| N.S. | Kanamycin (KAN) | 35 min | 100 min | 1 nM | 1–8 nM 100–500 nM | KAN addition (before pretreatment)/acetic acid addition/incubation at 45 °C/centrifugation at 10,000 rpm/filtration 0.22 μm/pH adjustment (total dilution unspecified). | [31] |
| N.S. | Streptomycin (STR) | N.A. | 70 min | 73.1 nM | 30–1030 nM | Milk dilution with water (total dilution: 50-fold)/STR addition (after pretreatment). | [34] |
| Supermarket milk | Streptomycin (STR) | 30 min | 60 min | 86 nM | 100–500 nM | Milk dilution with water (total dilution: 5-fold)/STR addition (after dilution)/EDTA and trifluoracetic acid addition/centrifugation at 6000 rpm/supernatant collection. | [35] |
| Raw milk from a dairy farm | Ampicillin Oxytetracycline Sulfadimethoxine Kanamycin | 40 min | 30 min | 1 μg/L (2.9 nM) 25 μg/L (54.3 nM) 6.25 μg/L (20.9 nM) 37.5 μg/L (77.4 nM) | 1–16 μg/L (2.9–46.4 nM) 25–200 μg/L (54.3–434.4 nM) 6.25–100 μg/L (20.9–334.4 nM) 37.5–600 μg/L (77.4–1238.4 nM) | Antibiotic addition (before pretreatment)/Carrez reagents/centrifugation at 1000 rpm/dilution, NaHCO3 or EDTA treatments/centrifugation at 1000 rpm/supernatant collection (total dilution: 1.16-fold). | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz-García, V.; Contreras-Trigo, B.; Rodríguez, C.; Coelho, P.; Oyarzún, P. A Simple Yet Effective Preanalytical Strategy Enabling the Application of Aptamer-Conjugated Gold Nanoparticles for the Colorimetric Detection of Antibiotic Residues in Raw Milk. Sensors 2022, 22, 1281. https://doi.org/10.3390/s22031281
Díaz-García V, Contreras-Trigo B, Rodríguez C, Coelho P, Oyarzún P. A Simple Yet Effective Preanalytical Strategy Enabling the Application of Aptamer-Conjugated Gold Nanoparticles for the Colorimetric Detection of Antibiotic Residues in Raw Milk. Sensors. 2022; 22(3):1281. https://doi.org/10.3390/s22031281
Chicago/Turabian StyleDíaz-García, Víctor, Braulio Contreras-Trigo, Camila Rodríguez, Pablo Coelho, and Patricio Oyarzún. 2022. "A Simple Yet Effective Preanalytical Strategy Enabling the Application of Aptamer-Conjugated Gold Nanoparticles for the Colorimetric Detection of Antibiotic Residues in Raw Milk" Sensors 22, no. 3: 1281. https://doi.org/10.3390/s22031281
APA StyleDíaz-García, V., Contreras-Trigo, B., Rodríguez, C., Coelho, P., & Oyarzún, P. (2022). A Simple Yet Effective Preanalytical Strategy Enabling the Application of Aptamer-Conjugated Gold Nanoparticles for the Colorimetric Detection of Antibiotic Residues in Raw Milk. Sensors, 22(3), 1281. https://doi.org/10.3390/s22031281