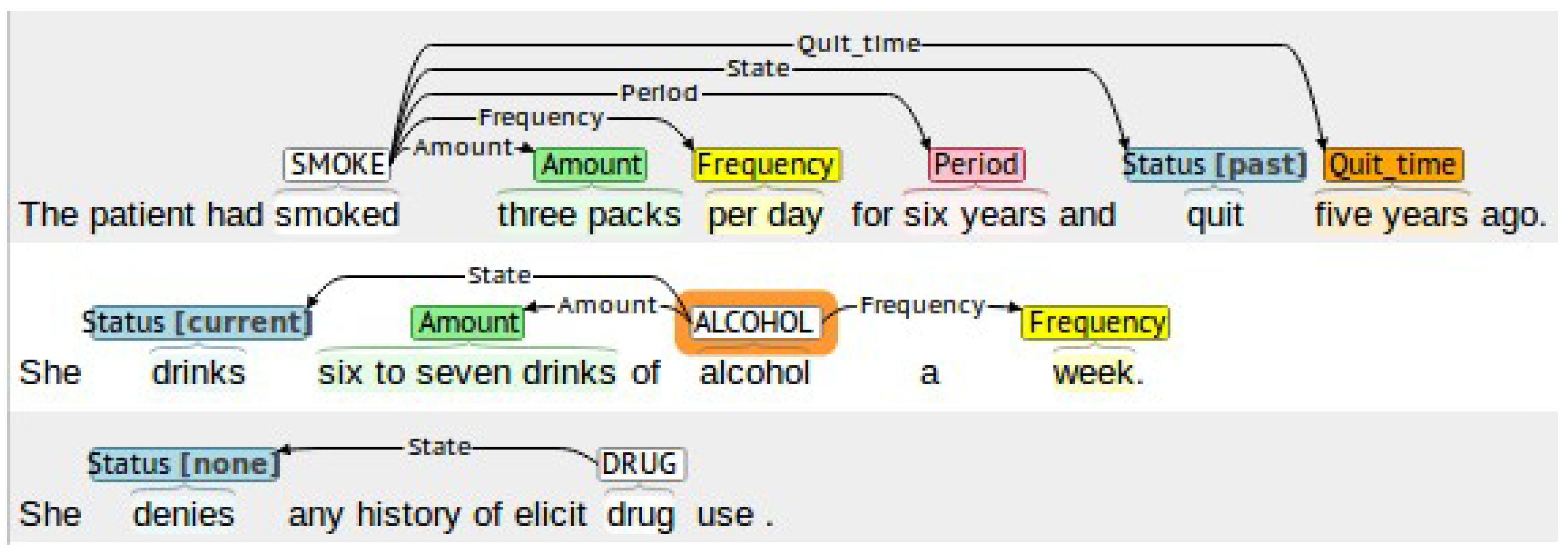
1. Introduction
Substance use (smoking, alcohol, and drug use) is a significant part of a patient’s history and can be used for clinical care and clinical research purposes. In order to improve patients’ health care, it is essential for clinicians to have a precise picture of each patient’s substance consumption. For instance, the value of the result of a common lab test (leukocyte differential count), which is used in the diagnosis of a variety of medical conditions, can cause confusion for clinicians if the patient’s status was not previously identified as a chronic alcoholic [
1]. Moreover, substance use causes a wide range of medical conditions and can exacerbate existing ones. Various published clinical research works identify substance use as a risk factor that could lead to morbidity and mortality [
2,
3]. Smoking is associated with various diseases such as cardiovascular and gastrointestinal diseases, infections, and some types of cancer [
4]. Consuming a large amount of alcohol has been shown to increase the risk of chronic pancreatitis, malnutrition, and cancer [
5,
6], whilst the long-term use of alcohol can seriously damage most organs and systems in the body [
7]. Drug use effects include addiction, heart and/or lung disease, cancer, mental illness, hepatitis, and other serious health issues [
8].
Substance use can be considered a disorder on its own if the frequent use of drugs and/or alcohol causes considerable clinical and/or functional impairment, such as health problems and disability [
9]. According to the UK’s National Health Service (NHS), in England, there were 506,100 hospital admissions and 74,600 deaths attributable to smoking for 2019/20 [
10]. Furthermore, according to the UK’s Office for National Statistics, in 2019, approximately 14.1% of the UK population aged 18 and above have smoked cigarettes [
11]. For alcohol use, in 2017 in the UK, 7327 people died as a direct result of their drinking, but when every death in which alcohol was a factor is included, the figure is closer to 24,000 [
12]. The misuse of alcohol is the biggest risk factor for death, ill-health, and disability among 15–49 year-olds in the UK and the fifth biggest risk factor across all ages [
13]. In addition, alcohol-related illness is estimated to cost the NHS around GBP 3.5 billion annually in England alone [
12]. For drug use, an estimated 1 in 11 adults aged 16 to 59 years had taken a drug in 2019 in the UK, equating to around 3.2 million people [
14]. In the same year, there were 4393 registered deaths related to drug misuse in England and Wales [
15]. Substance use can have a vast range of short- and long-term and direct and indirect effects. These effects vary based on the specific kind of substance used, how much is consumed, how it is consumed, and the overall health status of the user.
Electronic Health Records (EHRs) contain rich information about patients’ substance-use factors. This information can be either in a structured format or hidden within free text. EHRs are a source of valuable information about a vast range of diseases, health risk factors, and health outcomes [
16]. This information enables researchers to conduct high-resolution interventional and observational clinical research [
17,
18,
19]. However, most of the information is buried in unstructured narrative text [
20]. Extracting information from clinical narrative text is a challenging and arduous task due to misspellings, unconventional abbreviations, and ungrammatical text [
21]. In the last decade, Natural Language Processing (NLP) methods have received considerable attention as a way of analysing EHR narrative text and extracting the hidden information [
16,
18,
22,
23].
Recent developments in NLP techniques have demonstrated increasingly promising performance in the recognition and extraction of meaningful pieces of information from clinical narrative text [
24,
25]. NLP automates the processes required to access the vast embedded information in EHRs and to consolidate it into a coherent structure [
26]. NLP techniques have been widely adopted in different fields, including medical applications such as cohort identification [
27], genome-wide association studies [
28], sentiment analysis [
29], medical status extraction [
30], text summarisation [
31], and diagnosis code assignment [
32]. A large number of tools and frameworks are currently available for clinical information extraction purposes, such as the
Clinical Text Analysis Knowledge Extraction System (cTAKES) [
33],
MetaMap [
34], and
Medical Language Extraction and Encoding (MedLEE) [
35]. Most frameworks employ dictionaries and rule-based techniques to identify clinical entities. The cTAKES system, which was developed by the Mayo Clinic, is one of the most popular open-source NLP systems in the literature. This system has been developed based on the
Unstructured Information Management Architecture (UIMA) framework and the
OpenNLP NLP toolkit; cTAKES is able to parse the clinical free text in order to identify the types of relevant clinical concepts in addition to qualifying elements such as “negated” or “non-negated” and “current” or “history”. The concept code is provided by mapping each class to a specific terminology domain, which is responsible for handling language variations.
A range of clinical NLP challenges have been organised in order to identify clinical entities from clinical text. The Centre of Informatics for Integrating Biology and the Bedside (i2b2) has organised many challenges. For example, in 2006, they presented a challenge for discovering patient smoking status [
36]. In 2008, a challenge was conducted for recognising obesity from clinical text [
37]. In 2009, the challenge focused on medication recognition [
38]. In 2010, the challenge focused on recognising medical concept identification, classification, and relation extraction [
39]. Other challenges have been shared by i2b2, such as the co-reference challenge [
40], temporal relations challenge [
29], and de-identification and heart disease risk factors challenge [
41].
In this work, we propose a system to automatically detect smoking, alcohol, and drug-use status and the related information, such as amount, frequency, type, quit-time, and period, in medical records. Our system relies on several NLP techniques, such as chunking and keyword search, combined with rule-based methods to cope with complex clinical contexts. The developed system accurately extracts the information using four stages. In the first stage, the system excludes all records that do not contain any substance-use information using a keyword search method. In the second stage, an extension of the negation detection NegEx [
42] algorithm is proposed. Temporal status (current or past) identification is conducted in the third stage, whereas in the last stage, the system extracts related substance-use attributes. Furthermore, the proposed system can work successfully at both the document-level and sentence-level. Our experimental evaluation on a dataset created and annotated for designing and testing our system, demonstrates the efficiency of the proposed system, achieving an F1-score of up to 0.99 for the detection of substance-use status and up to 0.98 for the detection of substance-use-related attributes. In addition, experimental evaluation on an “unseen” dataset demonstrates the generalisation ability of the proposed approach, achieving an F1-score of up to 0.97 for the detection of substance-use status and up to 0.99 or the detection of substance-use-related attributes. The contribution of our work can be summarised as follows: (a) We propose an automated substance-use detection system for clinical text and evaluate it for smoking, alcohol, and drug use. (b) We propose an extension of the NegEx negation detection algorithm that improves negation detection compared to the original algorithm. (c) We evaluate our proposed system on a dataset created and annotated for designing and testing our system as well as on an “unseen” publicly available dataset. (d) We evaluate our proposed system against deep learning approaches that use trainable or pre-trained word embeddings.
The rest of this paper is organised into four sections. The background information and related works are presented in
Section 2. The proposed system is described in
Section 3.
Section 4 presents and discusses the obtained experimental results, whilst conclusions are drawn in
Section 5.
2. Related Work
The use of EHRs has become critical in medicine, healthcare delivery, operations, and medical research [
43]. However, the unstructured nature of the majority (80%) of information in EHRs makes it very difficult to process for secondary use [
44]. The importance of being able to automatically process and extract information from unstructured clinical text has led to significant amounts of research work on the topic of text classification in the context of medical text and clinical notes. Available works rely both on traditional classification approaches and on more-recent neural-network-based classification and other advances in NLP [
25]. Older works relied on classification approaches such as Support Vector Machines (SVMs) [
45] and k-Nearest Neighbours (kNN) [
46], while in recent years, deep-learning models such as Convolutional Neural Networks (CNNs) have attracted significant attention and have provided competitive results [
47]. The use of pre-trained word embeddings such as word2vec [
48] and GloVe [
49], or attention-based models such as BERT [
50,
51], ClinicalBERT [
52], EhrBERT [
53], and BioBERT [
54] have also recently been shown as viable alternatives [
55].
Substance-use status identification has been widely investigated in the literature due to the associated health risks. Some systems detect substance use as a disease risk factor for heart disease [
56,
57,
58,
59], type 2 diabetes mellitus [
60,
61], and epilepsy [
62], among others. Feller et al. [
63] attempted to infer the presence and status of social and behavioural determinants of health (SBDH) information in patient records, including alcohol and substance use. In their work, clinical documents were represented as a bag-of-words using the term frequency–inverse document frequency (TF-IDF) representation. The L2-penalised logistic regression, SVMs, Random Forests, CaRT, and AdaBoost classifiers were then tested, achieving an F1-score of 91.3% for alcohol use detection and 92.5% for substance-use detection.
Aiming to identify smoking status, in 2006, i2b2 shared a dataset containing patient discharge summaries. The challenge set up was to classify patients’ records into five categories (“current smoker”, “past smoker”, “smoker”, “non-smoker” and “unknown”). Many systems were proposed to address this challenge, with Uzuner et al. [
64] providing a summary of the reported results of these systems. Other later attempts were conducted on the same dataset to participate in the same challenge. Multiple classifications using Error Correcting Output Codes (ECOC) were used by Cohen [
65] in an attempt to detect smoking status, achieving a micro-F1 score of 90%. His proposed system used a range of different techniques, including hot-spot identification, zero-vector filtering, inverse class frequency weighting, error-correcting output codes, and post-processing rules, with results showing that hot-spot identification had the largest positive effect. Wicentowski et al. [
66] proposed a system to detect smoking status when smoking terms were present and when those terms were removed. Their system under-performed when the smoking terms were not available but performed similarly to expert human annotators. Heinze et al. [
67] applied a medical coding expert system (LifeCode
®) to the i2b2 smoking challenge. The authors concluded that smoking category temporal differentiation is a significant challenge. McCormick et al. [
68] compared a supervised classifier using lexical features to one relying on semantic features generated by an unmodified version of MedLEE, a clinical NLP engine. Results showed that their supervised classifier trained with semantic MedLEE features was competitive with the top-performing smoking classifier in the i2b2 challenge, achieving an F1-score of 89%.
Sohn et al. [
69] developed the Mayo smoking status-detection module by remodelling negation detection for non-smoker patients. Their system employed a rule-based model to classify document-level and patient-level smoking status. Their system works by employing a rule-based layer to detect the existence of smoking-related keywords. For any sentence with smoking information, a second layer is then used to detect any negation terms related to smoking terms using the NegEx algorithm. The SVM classifier is used to determine the sentence temporal status as past or current smoking. The portability of the Mayo module has been examined by Liu et al. [
70] using Vanderbilt University Hospital’s EHR data; they concluded that modifications are necessary to improve the detection performance. Khor et al. [
71] enhanced the Mayo smoking status-detection module to distinguish between current and past smokers. They retrained the SVM classifier by grouping keywords into lexically similar categories to improve efficiency and concluded that a keyword-grouping approach can increase the learning ability and enhance overall accuracy. Other approaches have compared the performance of systems that retrieve smoking status through different structured and unstructured sources [
72,
73]. SVMs were also used by Lix et al. [
74] in combination with unigrams and mixed-grams to classify patient alcohol use from unstructured text in primary care EHRs. Results showed that a higher F1-score was achieved for classification of current drinkers using unigrams (89%) than mixed-grams (83%), but the difference for the unknown category was minimal (98% vs. 97%).
Wang et al. [
75] developed an NLP system for detecting and extracting related information from substance-use sentences. The authors used the constituent parser and dependency parser along with rule-based methods to build a two-stage system. In the first stage, they detected three main subcategories of substance-use sentences (alcohol, drugs, and nicotine). In the second stage, they extracted attributes from the detected sentences, such as amount, frequency, status, method, and temporal information. They evaluated their system on two different datasets, MTSamples and notes from the University of Pittsburgh Medical Center (UPMC) de-identified clinical notes repository, achieving F1-scores of 89.8%, 84.6%, and 89.4% for alcohol-, drug-, and nicotine-use detection, respectively, as well as a maximum average F1-score of 96.6% for related attribute extraction. Yetisgen et al. [
76] used a combination of machine learning and NLP approaches to identify sentences related to tobacco, alcohol, and drug use, as well as to their related attributes. Their system involved three steps: The first step detected the sentences that contained substance-use events. In the second step, they extracted seven entities to describe the events, and finally, in the third stage, they presented the extracted entities in a structured format. They evaluated their system on the MTSamples dataset, achieving F1-scores between 35.29% and 97.61%. One limitation of the aforementioned works is that they were designed for detecting sentence status rather than document status.
4. Results and Discussion
A thorough experimental evaluation was conducted in order to examine the performance of the proposed system on the tasks of detecting unknown, none, past, or current substance use (smoking, alcohol, and drugs) from discharge records as well as on the task of detecting substance-use attributes (amount, type, frequency, quit-time, and period). To ensure a fair experimental evaluation and to avoid overfitting, the dataset was divided into training (65%) and test (35%) sets. To this end, 1128 records were used to design and fine-tune our system, and 611 records were used to test the system’s performance. For smoking, the number of related records was 503 records for training and 297 records for testing. For alcohol, there were 389 records for training and 238 testing records. Finally, for drugs, there were 128 records for training and 89 for testing. The performance of our system was measured using the following information retrieval metrics: sensitivity (Sen), precision (Pre), and F1-score (F1).
4.1. System Performance Evaluation
The performance of our proposed system on the test set is reported in s
Table 6 and
Table 7. Our system was able to accurately detect related substance-use records (Stage 1) with an F1-score between 0.9785 and 0.9974. For smoking and alcohol use, the proposed system detected all related records correctly with a sensitivity of 1.0, while sensitivity reached 0.9785 for drug use detection. Precision exceeded 0.97 for all substance-use detection types, resulting in very few unrelated records being retrieved.
In order to identify the negated records (Stage 2), we first tested using the cTAKES NegEx method. However, NegEx failed to identify the negated records. For instance, when identifying negated smoking records, 11 records were identified out of 113. Results were similar for identifying negated alcohol records, where only 9 out of 135 were detected correctly. In addition, none of the negated drug records were identified. We then tested our proposed negation-detection method. As shown in
Table 6, our negation-detection module (Stage 2) was successful in detecting negation, achieving an F1-score between 0.9710 and 0.9821. Sensitivity reached 0.9735 for detecting negation for smoking, 0.9630 for alcohol use, and 0.9571 for drug use. Furthermore, our proposed extended negation-detection module reduced false positives significantly, achieving a precision value of more than 0.98.
Temporal status classification into current or past (Stage 3) was the most challenging stage for our system. The proposed system was able to detect the smoking temporal status with an F1-score of 0.9463 for “Past” and 0.9294 for “Current” statuses, thus achieving an average F1-score of 0.9379. For the detection of alcohol-use temporal status, the average F1-score was 0.8731, with an F1-score of 0.80 for “Past” and 0.9461 for “Current” statuses. Finally, for the detection of the drug-use temporal status, the average F1-score was 0.8286, with an F1-score of 0.8571 for “Past” and 0.80 for “Current” statuses. From these results, it is evident that the proposed system is more efficient in detecting the temporal status for smoking compared to alcohol and drug use.
The performance of the proposed system for the extraction of substance-use attributes (Stage 4) is presented in
Table 7. Attributes related to smoking were accurately identified, with an F1-score between 0.9244 and 1. For alcohol use, the proposed system successfully identified the amount, type, and frequency attributes with an F1-score between 0.9333 and 0.9831, while performance suffered for the quit-time attribute, reaching an F1-score of 0.80. This decrease in performance can be attributed to the various syntactic patterns appearing in alcohol-related text. Finally, the proposed system was successful in identifying the amount, type, and period attributes for drug use, achieving an F1-score between 0.8780 and 1, with performance dropping for the quit-time attribute, reaching an F1-score of 0.75. It must also be noted that the achieved precision was very high in most cases, indicating that our system returned a very small number of incorrect results.
4.2. Performance Comparison to Deep Learning Approaches
To further evaluate the suitability of the proposed system, its performance was compared to that of six deep learning approaches for the task of substance-use status classification. To this end, the task was modelled as a four-class classification problem with the classes being “Unknown”, “None”, “Past”, and “Current”. The following six models were then trained for each one of the three substances in the dataset following the approaches of [
80,
81,
82]: (i) a Multilayer Perceptron (MLP) model using a trainable embedding layer, (ii) a Bidirectional Long Short-Term Memory (Bi-LSTM) [
83] model using a trainable embedding layer, (iii) an MLP model using the GloVe-50 [
49] pre-trained word embeddings, (iv) a Bi-LSTM model using the GloVe-50 pre-trained word embeddings, (v) the pre-trained Bidirectional Encoder Representations from Transformers (BERT) [
50] base uncased model, and (vi) a fine-tuned BERT base uncased model. The trainable embedding layer used in the first two models had 128 dimensions. In addition, a sequence length of 512 tokens was used for BERT, as it was the maximum supported by the BERT base uncased model, whereas the sequence length for the MLP and Bi-LSTM models was set to 2500 tokens. Cross-entropy loss and the Adam optimiser were used for training the models using the Keras API, with the loss function being weighted according to the class ratios in order to account for the class imbalance. It must be noted that in order to achieve a fair comparison to the proposed approach, no additional preprocessing was applied to the text. Each text was first tokenised, and the list of tokens was used as an input to the models. Results for each substance and for each class for all the examined deep learning models are presented in
Table 8.
From
Table 8, it is evident that the examined deep learning approaches performed significantly worse that the proposed method. For detecting “Unknown” use status, the fine-tuned BERT model performed the best for smoking and alcohol use, with F1-scores of 0.7611 and 0.80, respectively, and the MLP with trainable embedding performed the best for drug use, with an F1-score of 0.9442, compared to 0.9969, 0.9974, and 0.9785, respectively, for the proposed method. For the detection of negated records (“None”), the fine-tuned BERT model performed the best for smoking, with an F1-score of 0.4768, and the MLP with trainable embedding performed the best for alcohol and drug use, with F1-scores of 0.3866 and 0.2424, respectively, compared to 0.9821, 0.9738, and 0.9710, respectively, for the proposed method. For the detection of “Past” use status, the MLP with trainable embedding performed the best for smoking and drug use, with F1-scores of 0.2264 and 0.50, respectively, and the MLP with GloVe-50 embeddings performed the best for alcohol use, with an F1-score of 0.0769, compared to 0.9463, 0.8571, and 0.80, respectively, for the proposed method. It must be noted that for the detection of “Past” status for drug use, the Bi-LSTM with trainable embeddings and the MLP with GloVe-50 embeddings also achieved an F1-score of 0.50. Finally, for the detection of “Current” use status, the BERT model performed the best for smoking, with an F1-score of 0.2072, the fine-tuned BERT performed the best for alcohol use, with an F1-score of 0.2873, and the MLP with GloVe-50 or trainable embeddings performed the best for drug use, with an F1-score of 0.1818, compared to 0.9294, 0.9461, and 0.80, respectively, for the proposed method. It is evident that, apart from the detection of records with “Unknown” substance-use status, the examined deep learning approaches struggled to distinguish between the other classes, something that can be partially attributed to the lack of sufficient training samples (as shown in
Table 8) combined with the high dimensional feature space due to the length of the medical discharge records.
4.3. Evaluation on an Unseen Dataset
In order to demonstrate our system’s generalisability and superiority, we tested our system using the MTSamples dataset [
78] that has also been used by Yetisgen et al. [
76]. Yetisgen et al. employed machine learning for the detection of the substance-use status, as previously mentioned in the related works (
Section 2).
Table 9 provides a comparison between the results of our proposed system on the MTSamples dataset and the results published by Yetisgen et al. for the same dataset. Since the compared work detects the status at the sentence level, we also configured our proposed system to detect the status from sentences instead of records in order to conduct a fair comparison. It is evident from
Table 9 that the proposed system outperforms Yetisgen et al.’s approach, achieving a higher F1-score in all cases except for alcohol use with a status of “Current” and drug use with a status of “None”. It must also be noted that our system led to significant improvements in F1-scores in most cases compared to Yetisgen et al.’s and to more stable performance across the three different statuses for each substance.
In addition to detecting the substance-use status from the MTSamples dataset, we also extracted the related substance-use attributes. Results are provided in
Table 10 and are consistent with the results achieved for the dataset used to create and evaluate our system (
Table 7). The generalisability of systems and models across different datasets is one of the greatest challenges in this field. The results of our proposed system on an unseen independent dataset demonstrate its generalisation ability, indicating its potential for real-world use.
4.4. Error Analysis
By analysing the performance of each stage of our system, we identified the sources of errors as follows:
In the first stage, the proposed system fails to exclude the records when the substance-use keywords that appear in the text belong to one of the patient’s family members, such as father, mother, or wife. The cTAKES assertion module detects the main subject of the statement. However, we avoid using this module to identify the family member in our system because it dramatically increases the false-positive rate. Another source of error is misspelled keywords. For example, when identifying drug-related records, the system fails to include records that contain misspelled keywords such as “barbituate abuse”. Moreover, while the system sets extra restrictions over the patient record sections in detecting drug-related keywords, these keywords could appear in one of the restricted sections. For example, the existence of “remote drug abuse” could appear in the section “HISTORY OF PRESENT ILLNESS”.
In the remaining stages, the main error source that increases false positives and false negatives is the case when sentences include keywords of different substance use types. For example, in negation detection, the system fails to detect the following negation correctly: “Smoking, no alcohol”. While the previous sentence means that the patient is a smoker and does not drink alcohol, the system assigns negative status for both smoking and alcohol keywords. In some cases where the negation word is too far from the substance-use-related keywords, the system did not detect the negation. For example in the sentence “Abuse of drugs or alcohol were denied”, the word “drugs” is too far from “denied” to be negated, as our system uses a two-word window after and nine-word window before the substance-use keyword to detect negation. However, expanding the window after the keyword is likely to increase the false-positive rate.
Finally, some error sources come from patterns that did not appear in the training set. Since drug abuse is not as widespread as smoking and alcohol use, it is normal to have fewer samples in the dataset. In our case of 1739 records, only 45 were identified as patient drug-use records. Moreover, out of these 45 records, the frequency of drug use was mentioned only in 4 records. Consequently, our system was not able to detect the drug-use frequency value due to the absence of any cases in the testing set. The same situation was also faced when identifying alcohol-use period values.
4.5. Further Discussion
From
Table 8, it is evident that the examined deep learning models underperfomed significantly compared to the proposed rule-based system. We hypothesise that this is a result of the combination of an insufficient number of samples, especially for some pairs of substance and class, and the large size of the patient discharge summaries. The maximum length of the discharge summaries in the dataset was 3821 words and the median length was 948 words. For the MLP and Bi-LSTM models, the number of words led to a significantly large feature space, especially after their conversion to their respective word embeddings. The large feature space combined with the small number of samples for some pairs of substance and class plus the overall class imbalance hindered the performance of the MLP and Bi-LSTM models and led to overfitting. In addition, despite BERT not being affected by the size of the word representation, the limitation of BERT’s base uncased pre-trained model to inputs of a maximum of 512 tokens resulted in the model ignoring a significant amount of text in the case of larger discharge summaries, resulting in decreased performance. Furthermore, the use of pre-trained word embeddings, such as GloVe, may lead to discarding some valuable words, such as abbreviations, names of substances, and medical terms, as these words may not exist in the corpus used for training the embeddings. Indeed, results in this work showed that the two models with trainable embeddings performed better that the same models using the GloVe-50 embeddings.
5. Conclusions
In this work, we built a system to automatically detect substance use (smoking, alcohol, and drug use) and to extract related attributes from clinical text, more specifically, from patient discharge records. A suitable dataset was created and annotated and was finally divided into a training set that was used for designing and fine-tuning the proposed system and a test set that was only used for the final performance evaluation. The proposed system was built using NLP and rule-based techniques and consists of four stages: In the first stage, the system excludes unrelated records using a keyword search technique, achieving an F1-score of up to 0.99. In the second stage, an extension of the NegEx negation detection algorithm detects the negation status of patient records, achieving an F1-score of up to 0.98. In the third stage, the system extracts the temporal status of the substance use and classifies the records into current or past status, achieving a maximum F1-score of 0.94. Finally, the substance-use-related attributes (amount, frequency, type, quit-time, and period) were extracted from each patient record using rule-based techniques, achieving satisfactory performance.
The experimental evaluation showed that the proposed system is able to successfully detect the substance-use status and related attributes from clinical notes at both document-and sentence-level. Furthermore, in order to demonstrate its generalisability, the proposed system was also evaluated on the independent, unseen MTSamples dataset, and its performance was compared with a state-of-the-art approach. Results showed that the proposed system outperformed the compared approach on an unseen dataset, thus demonstrating its generalisation ability. To conclude, the results suggest that using NLP and rule-based techniques is an efficient way of extracting embedded information from clinical text.
For future work, we hypothesise that working with a larger dataset could potentially improve the ability to generate more rules and thus increase the system’s overall performance. Furthermore, apart from the rule-based approach, we plan to work on deep learning approaches for the task at hand. The mediocre performance achieved by the examined deep learning approaches in this work demonstrated various shortcomings that must be addressed. The large size of the clinical discharge records, in combination with the limited number of samples for some pairs of substance and use status, hindered the ability of state-of-the-art deep learning models to perform efficiently. To this end, we plan to increase the size of the available training datasets by gathering and curating additional clinical discharge records; we also plan to develop deep learning approaches tailored to the examined task. Finally, we plan to examine whether a hybrid approach that combines rules-based and deep learning approaches for different stages of the processing pipeline may be more suited for the examined task by training separate deep learning models for each stage of the pipeline and combining them with rule-based methods. In addition, the issue of the large text length could be potentially addressed by training the models at sentence-level, thus significantly reducing the number of input tokens, and then using a rule-based strategy to combine the statuses of all sentences in a text.