Experiments and Evaluation
The proposed automated image-matching process based on the fuzzy image concept, which was combined with the use of similarity measures in order to find the most similar MR scans in the inter-object matching (scans containing bone structures of the knee joint), was tested on 107 clinical T1-weighted MR studies of the knee joint on the sagittal plane. The obtained results were verified by two independent experts, and correct results (for the inter-object matching of the whole scans containing the bone structures of the knee joint) were yielded in 84 cases (the accuracy of the method is equal to 78.5%).
Figure 6 shows the results of the matching process for two selected MR scans containing the bone structures of the knee joint. In both cases (scan no.5 and scan no.8 of the first group), the highest value of the similarity measure indicates the most similar scan (scan no.6 and scan no.10) from the second group.
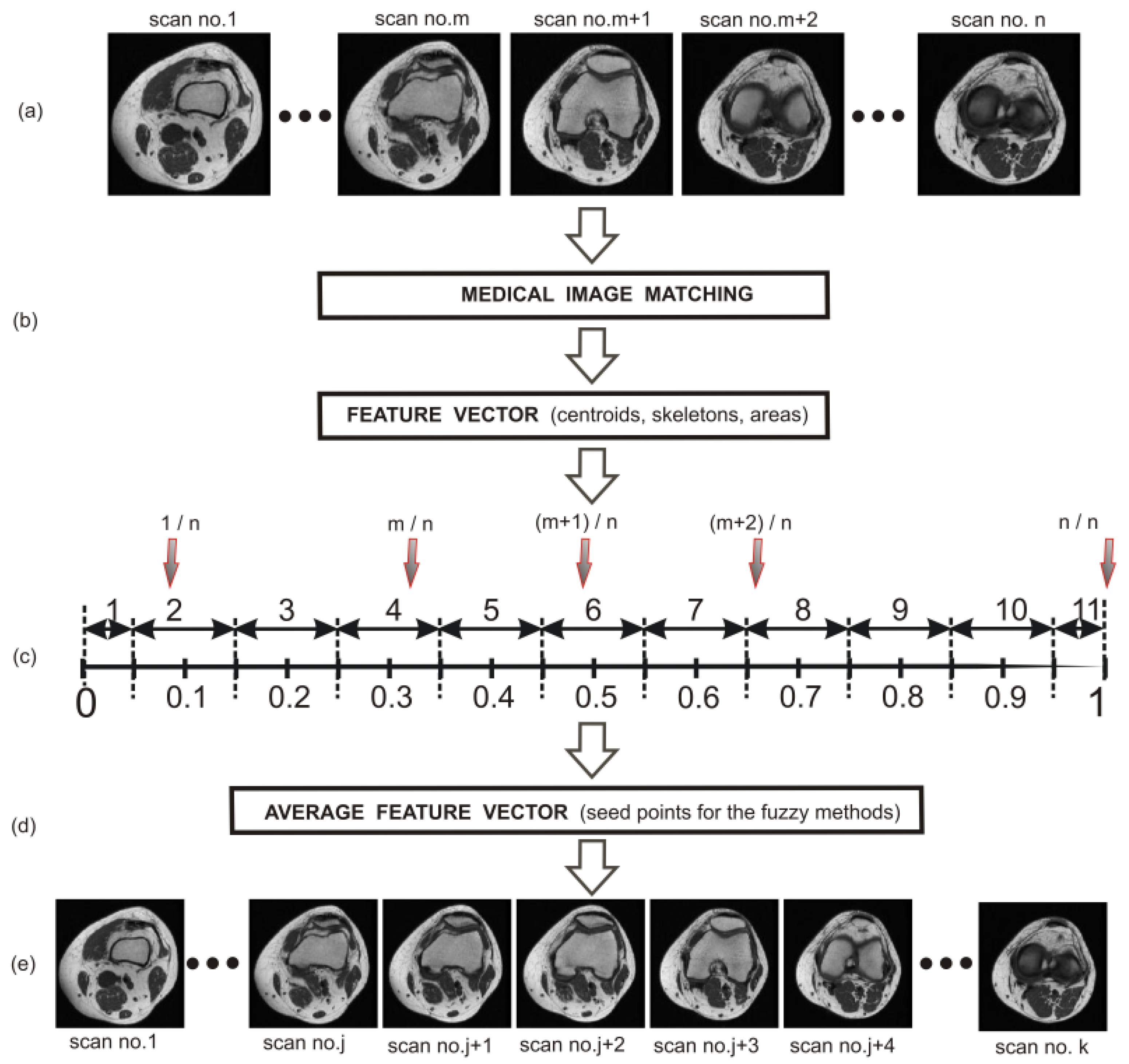
In the next step, the obtained dataset of clinical scans was divided into two groups. These were the teaching and the testing groups. The teaching group consisted of 50 arbitrarily selected clinical T1-weighted MRI studies of the knee joint and 30 clinical CT studies of the lower limb. Based on this group, a set of centroids, edges, areas, and perimeters from the extracted bone structures (corresponding to a given scan) for the patella, femur, and tibia as well as a set of areas of the entire knee joint (with respect to a given scan) were determined. The rest of the obtained clinical dataset constituted the testing group.
Based on the images of the teaching group (50 clinical T1-weighted MRI studies of the knee joint and 30 clinical CT studies of the lower limb), the normalized values of the x and y coordinates of the centroid (patella, femur, and tibia) were obtained in view of the size of the scans on the transverse plane (
Table 1) and on the sagittal plane (
Table 2). In the next step, the x and y coordinates were used as starting points (seed points) in both implemented methods (FCM and FC).
In the case of the transverse plane, the determined centroids proved to be excellent starting points for the bone structures of the knee joint. The situation was much worse in the case of the sagittal plane, especially for the bone structures of the patella (
Figure 5). From the practical point of view, the centroids of the patellar structures determined on the basis of the teaching group for 11 sets had to be modified before using them as starting points. This modification consisted in choosing such a point that belonged to the skeleton as the starting point, for which the distance between the average centroid and this point was the smallest. The choosing operation was carried out until the area of the obtained patellar extraction conformed with the area given in (
Table 3) for each set. In the case of the femur and tibia on the sagittal plane, such a modification was not necessary.
In
Table 3 and
Table 4, the normalized (to the size of the knee) areas of the patella, femur, and tibia structures on the transverse and sagittal planes, respectively, are given. These areas were used in the extraction of the bone structures from the T1-weighted MRI studies of the knee joint and the CT studies of the lower limb as a protection against over-segmentation and in the process of shifting the designated centroids of the patella on the sagittal plane (
Table 4).
The final stage in the automatic extraction of bone structures of the knee joint from MRI and CT scans based on the atlas segmentation was accomplished by means of post-processing. The post-processing operation consisted in performing consecutive steps, the aim of which was to obtain separate patellar, femoral, and tibial structures with smoothed edges. In order to achieve this, the operation was based on averaging filtration combined with the implementation of morphological operations. This approach, in the most typical cases, resulted in the correct extraction of bone structures of the knee joint.
Atlas-based segmentation combined with the fuzzy connectedness method applied to bone structures of the knee joint gave the following Dice index results (average values for the testing group): 89.48% for the patella, 86.59% for the femur, and 87.94% for the tibia, with regard to the clinical CT studies of the lower limb. For atlas-based segmentation combined with the FCM method, similar values were obtained. For the same clinical CT studies, atlas-based segmentation combined with the FCM method applied to the same anatomical structures of the knee joint gave Dice index results (average value for the testing group) at the following levels: 88.23% for the patella, 85.75% for the femur, and 87.14% for the tibia.
Atlas-based segmentation combined with the same fuzzy methods (FC and FCM), but applied to bone structures for T1-weighted MRI studies of the knee joint on the sagittal plane, gave the following Dice index results (average value for the testing group): 87.59% (FC) and 86.65% (FCM) for the patella, 88.66% (FC) and 87.49% (FCM) for the femur, and 86.59% (FC) and 85.52% (FCM) for the tibia.
The obtained Dice index values for the analyzed CT scans (testing group) are presented in
Table 5 (atlas-based segmentation combined with the FC and FCM methods), and those for the MRI scans are shown in
Table 6.
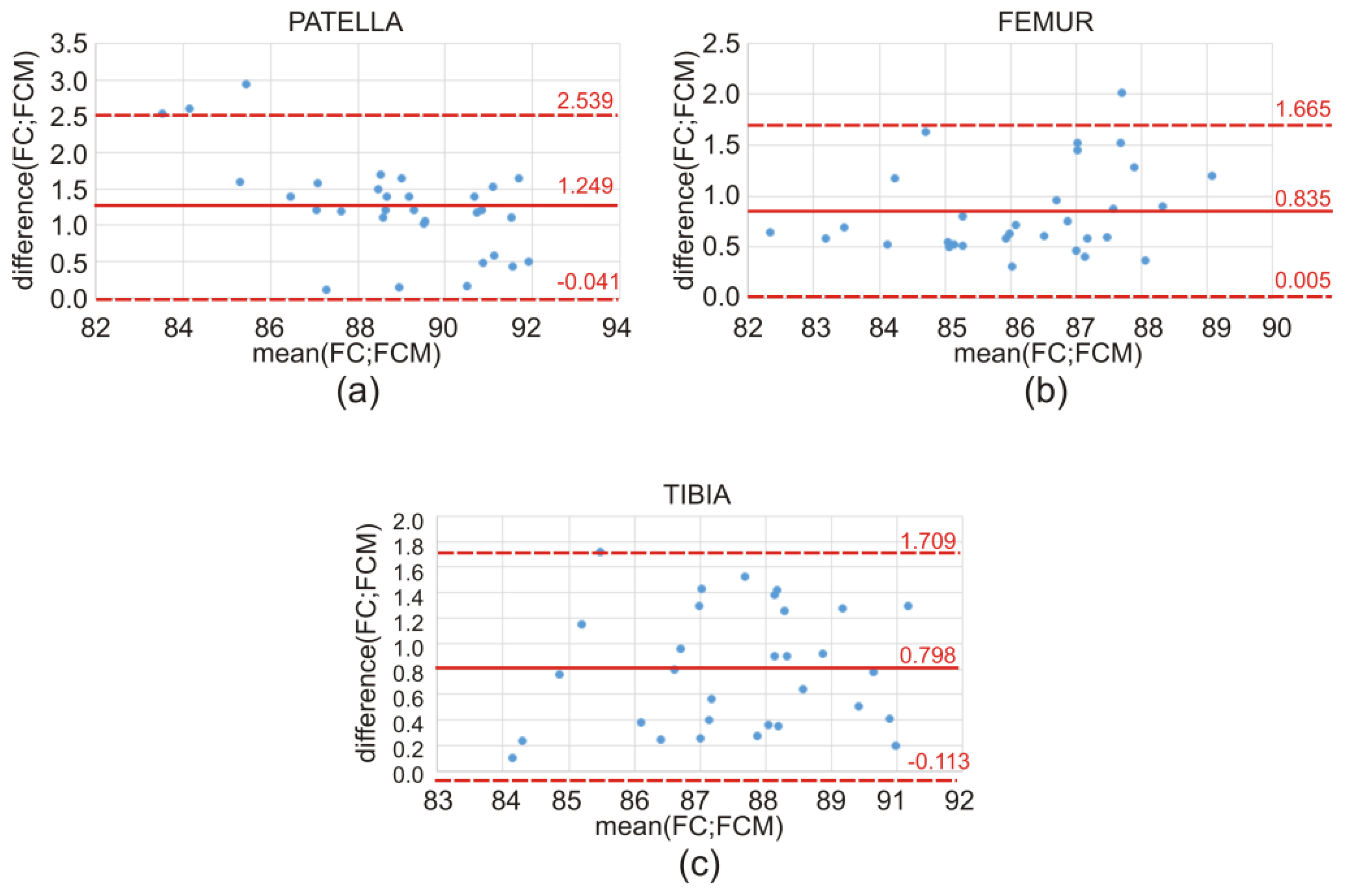
The discrepancy in the obtained values of the Dice index between the atlas-based segmentation combined with the FC method and the atlas-based segmentation of the same bone structure combined with the FCM method for CT clinical data is illustrated by means of a box-and-whisker plot (
Figure 7) and Bland–Altman plots (
Figure 8). Further on, the box-and-whisker plot (
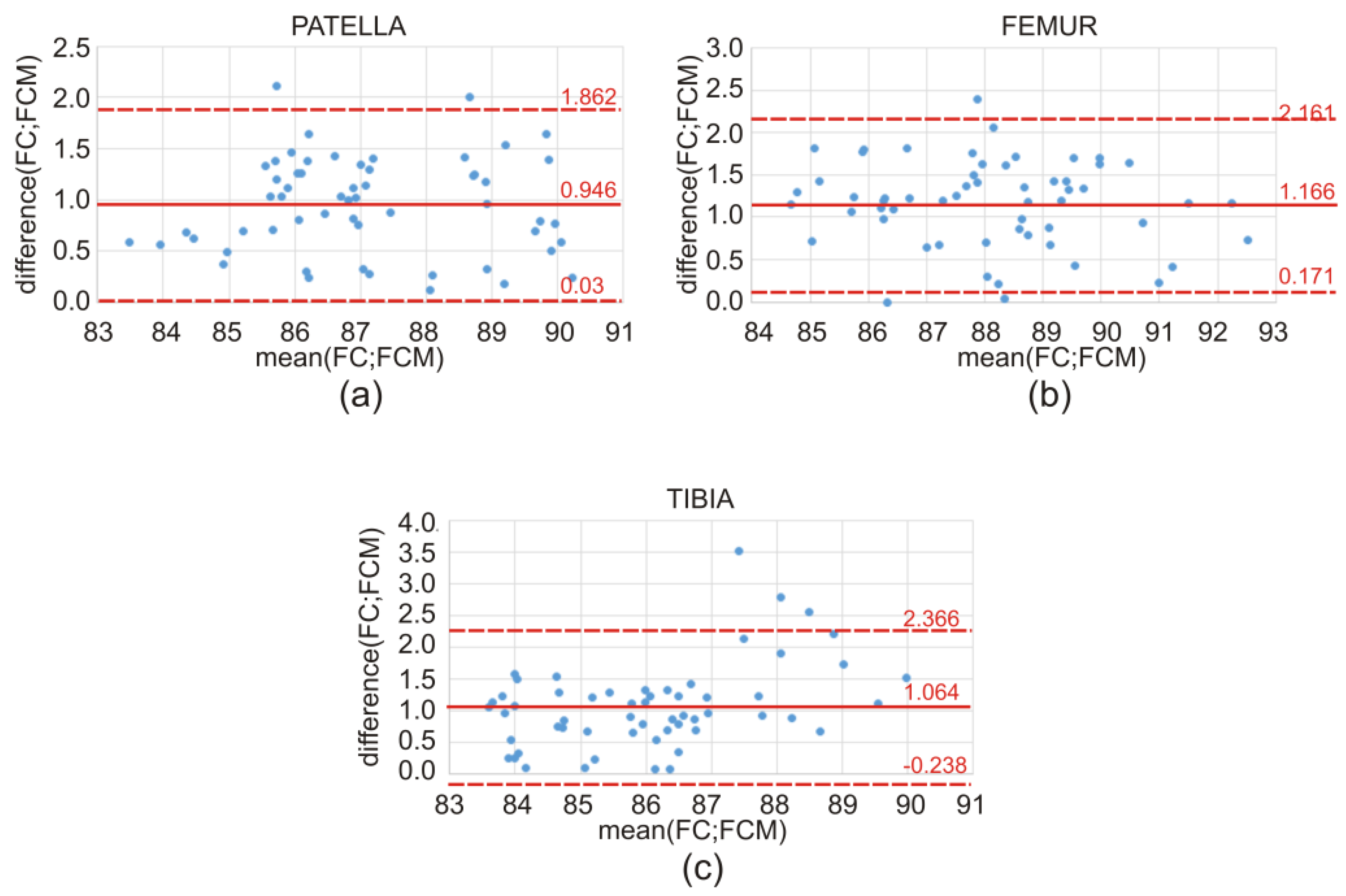
Figure 9) and the Bland–Altman plots (
Figure 10) show the discrepancy of the obtained Dice index values between the atlas-based segmentation in combination with the FC method and then with the FCM method; both methods were tested here for clinical T1-weighted MRI studies of the knee joint.
After the tests described above, the phantom tests were carried out. In these studies, atlas-based segmentation was used, leading to the creation of femoral imprints [
11]. Firstly, a CT study of the artificial femur was performed. Then, atlas-based segmentation combined with the FC method was used in order to perform femoral head extraction, after which a 3D model of this structure was created (
Figure 11c). Finally, an imprint of this artificial structure was made. The imprint thus obtained was applied to the artificial femur, and two independent experts (radiologist and orthopedist) assessed the degree of fit. In their opinion, the degree of fit achieved by the imprint was sufficient. There was no slack, rotation, nor sideslip, and there was no lateral movement either. The central and side surfaces of the artificial femur closely adhered to the imprint. The artificial femur was stable and remained motionless.
The following tests were performed in the described studies: the Wilcoxon test and the
t-test. The reason for this step was the lack of distribution normality for the compared variables. The obtained results from the performed statistical tests clearly show the small difference between the results obtained from the two implemented fuzzy approaches, FC and FCM. The analysis of the calculated
p-values for the Wilcoxon test and the
t-test (
Table 7) led to the following conclusions: The difference between both methods (FC and FCM) is not statistically significant (for all cases, the calculated
p-values were lower than 0.05). The described method has acceptable performance.
Table 8 shows the following: mean, standard deviation, and
p-values, calculated by using the Bland–Altman plots, followed by a one-sided
t-test for the patella, femur, and tibia. Furthermore, in this situation, for all cases, the calculated p-values were much lower than 0.05, indicating that the difference between the FC and FCM measurements is not statistically significant, and that both proposed fuzzy methods performed quite well.
The data (10 MRI T1-weighted series on the sagittal plane) used to verify the results of this study were obtained from the NYU fastMRI Initiative database (
fastmri.med.nyu.edu) [
25]. As starting points for both fuzzy methods (FC and FCM), those determined in this study (
Table 2) were used. The obtained values of the Dice index for the analyzed MRI series are presented in
Table 9. It can be noted that the atlas-based segmentation combined with the same fuzzy methods (FC and FCM) but applied to bone structures for the fastMRI Dataset (10 T1-weighted MRI studies of the knee joint on the sagittal plane) gave the following Dice index results (average value for the testing group): 86.18% (FC) and 85.33% (FCM) for the patella, 88.19% (FC) and 87.55% (FCM) for the femur, and 87.12% (FC) and 86.57% (FCM) for the tibia. These values do not differ significantly from those obtained in the study, therefore the results can be considered as convergent and reliable.