Evaluation of Preprocessing Methods on Independent Medical Hyperspectral Databases to Improve Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.1.1. Colon and Esophagogastric Cancer Databases
2.1.2. Brain Cancer Database
2.1.3. Summary Databases
2.2. Processing Frameworks
2.2.1. HS Data Calibration
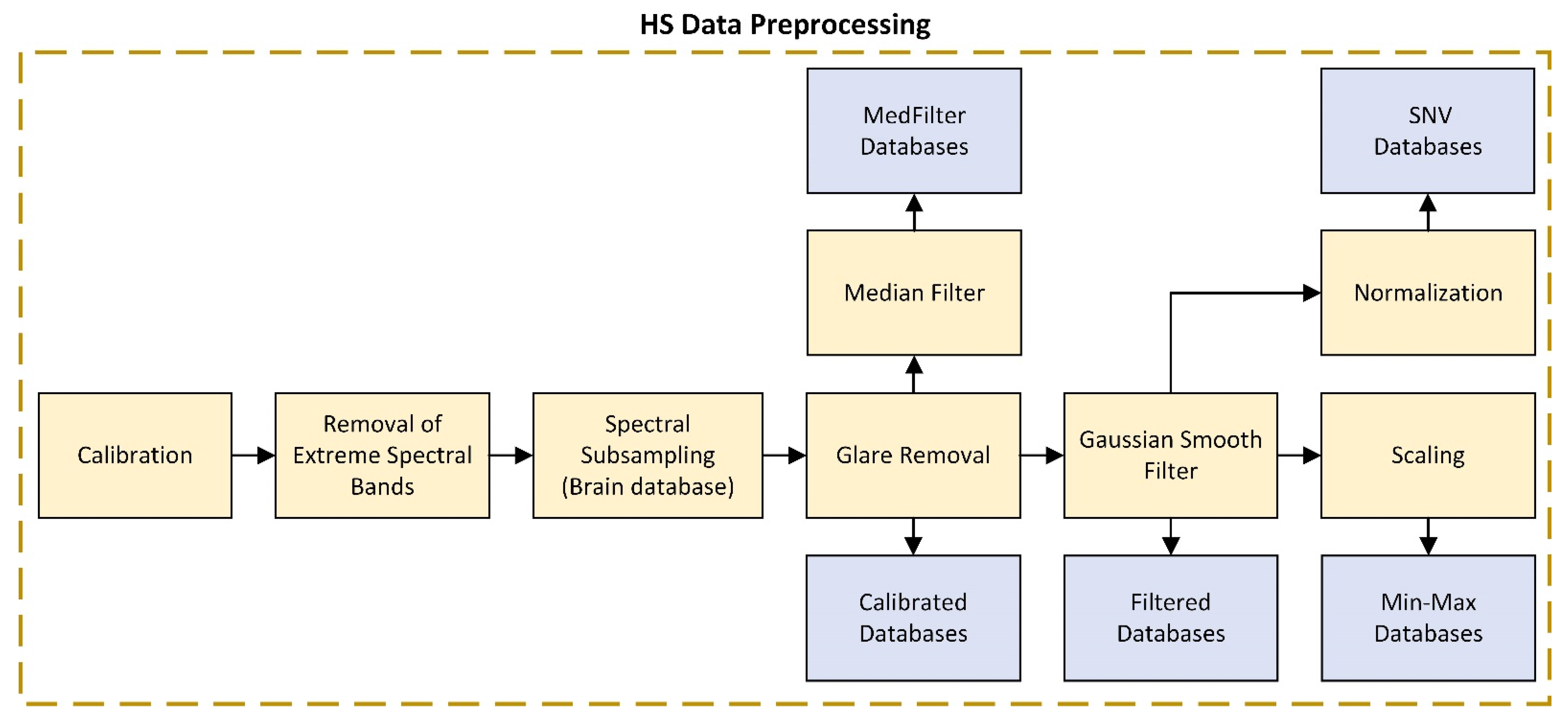
2.2.2. HS Data Preprocessing
2.2.3. Summary Data Preprocessing
2.2.4. Machine Learning (ML) Model
2.2.5. Deep Learning (DL) Model
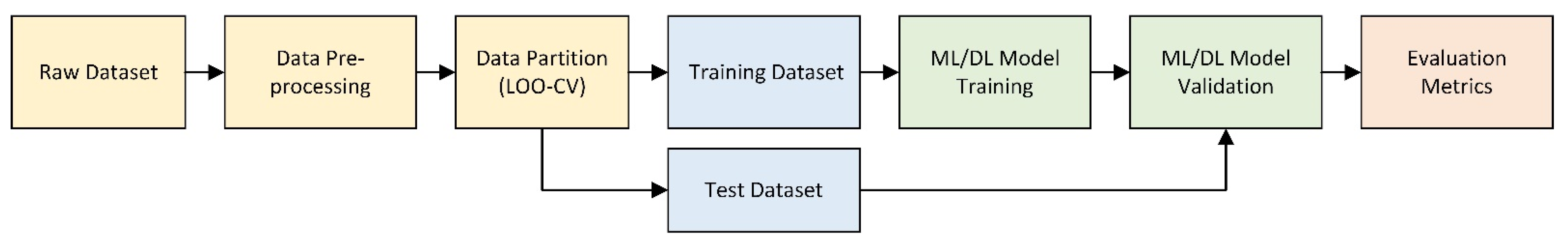
2.2.6. Data Partition
2.2.7. Evaluation Metrics
3. Results
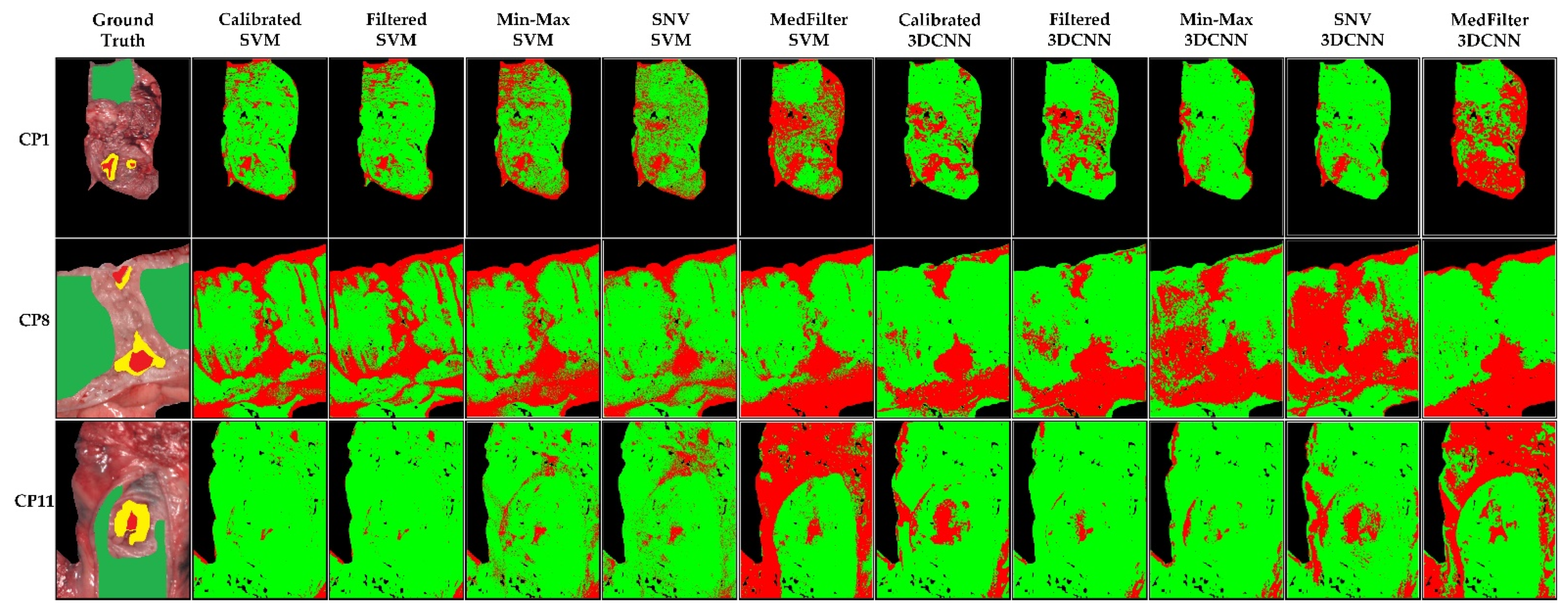
3.1. Colon Results
3.2. Esophagogastric Results
3.3. Brain Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- GLOBOCAN the Global Cancer Observatory—All Cancers; International Agency for Research on Cancer—WHO: Geneva, Switzerland, 2020; Volume 419, pp. 199–200.
- Gowen, A.; Odonnell, C.; Cullen, P.; Downey, G.; Frias, J. Hyperspectral Imaging–an Emerging Process Analytical Tool for Food Quality and Safety Control. Trends Food Sci. Technol. 2007, 18, 590–598. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Zhang, K.; Madani, K.; Sabourin, C. Morphological Band Selection for Hyperspectral Imagery. IEEE Geosci. Remote Sens. Lett. 2018, 15, 1259–1263. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Sun, D.-W. Introduction to Hyperspectral Imaging Technology. In Computer Vision Technology for Food Quality Evaluation; Elsevier: Amsterdam, The Netherlands, 2016; pp. 111–139. ISBN 9780128022320. [Google Scholar]
- Shippert, P. Introduction to Hyperspectral Image Analysis. Online J. Space Commun. 2003, 2, 8. [Google Scholar]
- Khan, U.; Paheding, S.; Elkin, C.P.; Devabhaktuni, V.K. Trends in Deep Learning for Medical Hyperspectral Image Analysis. IEEE Access 2021, 9, 79534–79548. [Google Scholar] [CrossRef]
- Zhu, S.; Su, K.; Liu, Y.; Yin, H.; Li, Z.; Huang, F.; Chen, Z.; Chen, W.; Zhang, G.; Chen, Y. Identification of Cancerous Gastric Cells Based on Common Features Extracted from Hyperspectral Microscopic Images. Biomed. Opt. Express 2015, 6, 1135. [Google Scholar] [CrossRef] [PubMed]
- Ortega, S.; Fabelo, H.; Camacho, R.; de la Luz Plaza, M.; Callicó, G.M.; Sarmiento, R. Detecting Brain Tumor in Pathological Slides Using Hyperspectral Imaging. Biomed. Opt. Express 2018, 9, 818. [Google Scholar] [CrossRef] [PubMed]
- Saiko, G.; Lombardi, P.; Au, Y.; Queen, D.; Armstrong, D.; Harding, K. Hyperspectral Imaging in Wound Care: A Systematic Review. Int. Wound J. 2020, 17, 1840–1856. [Google Scholar] [CrossRef] [PubMed]
- Reshef, E.R.; Miller, J.B.; Vavvas, D.G. Hyperspectral Imaging of the Retina: A Review. Int. Ophthalmol. Clin. 2020, 60, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, M.; Marx, S.; Bruckner, T.; Nickel, F.; Müller-Stich, B.P.; Hackert, T.; Weigand, M.A.; Uhle, F.; Brenner, T.; Schmidt, K. Bedside Hyperspectral Imaging for the Evaluation of Microcirculatory Alterations in Perioperative Intensive Care Medicine: A Study Protocol for an Observational Clinical Pilot Study (HySpI-ICU). BMJ Open 2020, 10, e035742. [Google Scholar] [CrossRef]
- Martinez-Vega, B.; Leon, R.; Fabelo, H.; Ortega, S.; Callico, G.M.; Suarez-Vega, D.; Clavo, B. Oxygen Saturation Measurement Using Hyperspectral Imaging Targeting Real-Time Monitoring. In Proceedings of the 2021 24th Euromicro Conference on Digital System Design (DSD), Palermo, Italy, 1–3 September 2021; pp. 480–487. [Google Scholar]
- Halicek, M.; Fabelo, H.; Ortega, S.; Callico, G.M.; Fei, B.; Halicek, M.; Fabelo, H.; Ortega, S.; Callico, G.M.; Fei, B. In-Vivo and Ex-Vivo Tissue Analysis through Hyperspectral Imaging Techniques: Revealing the Invisible Features of Cancer. Cancers 2019, 11, 756. [Google Scholar] [CrossRef]
- Jansen-winkeln, B.; Barberio, M.; Chalopin, C.; Schierle, K.; Diana, M.; Köhler, H.; Gockel, I.; Maktabi, M. Feedforward Artificial Neural Network-based Colorectal Cancer Detection Using Hyperspectral Imaging: A Step towards Automatic Optical Biopsy. Cancers 2021, 13, 967. [Google Scholar] [CrossRef] [PubMed]
- Fabelo, H.; Halicek, M.; Ortega, S.; Szolna, A.; Morera, J.; Sarmiento, R.; Callicó, G.M.; Fei, B. Surgical Aid Visualization System for Glioblastoma Tumor Identification Based on Deep Learning and In-Vivo Hyperspectral Images of Human Patients. In Proceedings of the Medical Imaging 2019: Image-Guided Procedures, Robotic Interventions, and Modeling, San Diego, CA, USA, 16–21 February 2019; Volume 10951, p. 35. [Google Scholar]
- Barberio, M.; Benedicenti, S.; Pizzicannella, M.; Felli, E.; Collins, T.; Jansen-Winkeln, B.; Marescaux, J.; Viola, M.G.; Diana, M. Intraoperative Guidance Using Hyperspectral Imaging: A Review for Surgeons. Diagnostics 2021, 11, 2066. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, X.; He, L.; Meng, C.; Du, S.; Bao, J.; Zheng, Y. Applications of Hyperspectral Imaging in the Detection and Diagnosis of Solid Tumors. Transl. Cancer Res. 2020, 9, 1265–1277. [Google Scholar] [CrossRef] [PubMed]
- Johansen, T.H.; Møllersen, K.; Ortega, S.; Fabelo, H.; Garcia, A.; Callico, G.M.; Godtliebsen, F. Recent Advances in Hyperspectral Imaging for Melanoma Detection. WIREs Comput. Stat. 2020, 12, e1465. [Google Scholar] [CrossRef]
- Eggert, D.; Bengs, M.; Westermann, S.; Gessert, N.; Gerstner, A.O.H.; Mueller, N.A.; Bewarder, J.; Schlaefer, A.; Betz, C.; Laffers, W. In Vivo Detection of Head and Neck Tumors by Hyperspectral Imaging Combined with Deep Learning Methods. J. Biophotonics 2022, 15, e202100167. [Google Scholar] [CrossRef] [PubMed]
- Halicek, M.; Fabelo, H.; Ortega, S.; Little, J.V.; Wang, X.; Chen, A.Y.; Callico, G.M.; Myers, L.; Sumer, B.D.; Fei, B. Hyperspectral Imaging for Head and Neck Cancer Detection: Specular Glare and Variance of the Tumor Margin in Surgical Specimens. J. Med. Imaging 2019, 6, 035004. [Google Scholar] [CrossRef] [PubMed]
- Panasyuk, S.V.; Yang, S.; Faller, D.V.; Ngo, D.; Lew, R.A.; Freeman, J.E.; Rogers, A.E. Medical Hyperspectral Imaging to Facilitate Residual Tumor Identification during Surgery. Cancer Biol. Ther. 2007, 6, 439–446. [Google Scholar] [CrossRef]
- Ortega, S.; Halicek, M.; Fabelo, H.; Guerra, R.; Lopez, C.; Lejeune, M.; Godtliebsen, F.; Callico, G.M.; Fei, B. Hyperspectral Imaging and Deep Learning for the Detection of Breast Cancer Cells in Digitized Histological Images. In Medical Imaging 2020: Digital Pathology; Tomaszewski, J.E., Ward, A.D., Eds.; SPIE: Bellingham, WA, USA, 2020; Volume 11320, p. 30. [Google Scholar]
- Martinez, B.; Leon, R.; Fabelo, H.; Ortega, S.; Piñeiro, J.F.; Szolna, A.; Hernandez, M.; Espino, C.; J. O’Shanahan, A.; Carrera, D.; et al. Most Relevant Spectral Bands Identification for Brain Cancer Detection Using Hyperspectral Imaging. Sensors 2019, 19, 5481. [Google Scholar] [CrossRef]
- Fabelo, H.; Ortega, S.; Lazcano, R.; Madroñal, D.; Callicó, G.M.; Juárez, E.; Salvador, R.; Bulters, D.; Bulstrode, H.; Szolna, A.; et al. An Intraoperative Visualization System Using Hyperspectral Imaging to Aid in Brain Tumor Delineation. Sensors 2018, 18, 430. [Google Scholar] [CrossRef]
- Fabelo, H.; Halicek, M.; Ortega, S.; Shahedi, M.; Szolna, A.; Piñeiro, J.F.; Sosa, C.; O’Shanahan, A.J.; Bisshopp, S.; Espino, C.; et al. Deep Learning-Based Framework for In Vivo Identification of Glioblastoma Tumor Using Hyperspectral Images of Human Brain. Sensors 2019, 19, 920. [Google Scholar] [CrossRef]
- Giannoni, L.; Lange, F.; Tachtsidis, I. Hyperspectral Imaging Solutions for Brain Tissue Metabolic and Hemodynamic Monitoring: Past, Current and Future Developments. J. Opt. 2018, 20, 044009. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Panchariya, P.C.; Patel, S.S.; Kiranmayee, A.H.; Ranjan, R. Application of Various Pre-Processing Techniques on Infrared (IR) Spectroscopy Data for Classification of Different Ghee Samples. In Proceedings of the 2018 Fourth International Conference on Computing Communication Control and Automation (ICCUBEA), Pune, India, 16–18 August 2018; pp. 1–6. [Google Scholar]
- Lu, G.; Wang, D.; Qin, X.; Muller, S.; Little, J.V.; Wang, X.; Chen, A.Y.; Chen, G.; Fei, B. Histopathology Feature Mining and Association with Hyperspectral Imaging for the Detection of Squamous Neoplasia. Sci. Rep. 2019, 9, 17863. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Joseph, J.; Waterhouse, D.J.; Luthman, A.S.; Gordon, G.S.D.; di Pietro, M.; Januszewicz, W.; Fitzgerald, R.C.; Bohndiek, S.E. A Clinically Translatable Hyperspectral Endoscopy (HySE) System for Imaging the Gastrointestinal Tract. Nat. Commun. 2019, 10, 1902. [Google Scholar] [CrossRef] [PubMed]
- De Landro, M.; Felli, E.; Collins, T.; Nkusi, R.; Baiocchini, A.; Barberio, M.; Orrico, A.; Pizzicannella, M.; Hostettler, A.; Diana, M.; et al. Prediction of in Vivo Laser-Induced Thermal Damage with Hyperspectral Imaging Using Deep Learning. Sensors 2021, 21, 6934. [Google Scholar] [CrossRef] [PubMed]
- Markgraf, W.; Lilienthal, J.; Feistel, P.; Thiele, C.; Malberg, H. Algorithm for Mapping Kidney Tissue Water Content during Normothermic Machine Perfusion Using Hyperspectral Imaging. Algorithms 2020, 13, 289. [Google Scholar] [CrossRef]
- Sato, D.; Takamatsu, T.; Umezawa, M.; Kitagawa, Y.; Maeda, K.; Hosokawa, N.; Okubo, K.; Kamimura, M.; Kadota, T.; Akimoto, T.; et al. Distinction of Surgically Resected Gastrointestinal Stromal Tumor by Near-Infrared Hyperspectral Imaging. Sci. Rep. 2020, 10, 21852. [Google Scholar] [CrossRef]
- Collins, T.; Maktabi, M.; Barberio, M.; Bencteux, V.; Jansen-Winkeln, B.; Chalopin, C.; Marescaux, J.; Hostettler, A.; Diana, M.; Gockel, I. Automatic Recognition of Colon and Esophagogastric Cancer with Machine Learning and Hyperspectral Imaging. Diagnostics 2021, 11, 1810. [Google Scholar] [CrossRef]
- Fabelo, H.; Ortega, S.; Szolna, A.; Bulters, D.; Pineiro, J.F.; Kabwama, S.; J.-O’Shanahan, A.; Bulstrode, H.; Bisshopp, S.; Kiran, B.R.; et al. In-Vivo Hyperspectral Human Brain Image Database for Brain Cancer Detection. IEEE Access 2019, 7, 39098–39116. [Google Scholar] [CrossRef]
- Zeaiter, M.; Rutledge, D. Preprocessing Methods. In Comprehensive Chemometrics; Elsevier: Amsterdam, The Netherlands, 2009; Volume 3, pp. 121–231. [Google Scholar]
- Tang, Y.; Zhang, Y.-Q.; Chawla, N.V.; Krasser, S. Correspondence SVMs Modeling for Highly Imbalanced Classification. Cybernetics 2009, 39, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Vapnik, V.N.; Kotz, S. Estimation of Dependences Based on Empirical Data; Information Science and Statistics; Springer: New York, NY, USA, 2006; Volume 40, ISBN 978-0-387-30865-4. [Google Scholar]
- Hu, W.; Huang, Y.; Wei, L.; Zhang, F.; Li, H. Deep Convolutional Neural Networks for Hyperspectral Image Classification. J. Sens. 2015, 2015, 258619. [Google Scholar] [CrossRef]
- Ben Hamida, A.; Benoit, A.; Lambert, P.; Ben Amar, C. 3-D Deep Learning Approach for Remote Sensing Image Classification. IEEE Trans. Geosci. Remote Sens. 2018, 56, 4420–4434. [Google Scholar] [CrossRef]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-Level Accuracy with 50x Fewer Parameters and <0.5MB Model Size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Rashmi, S.; Addamani, S.; Venkat, S.R. Spectral Angle Mapper Algorithm for Remote Sensing Image Classification. IJISET-Int. J. Innov. Sci. Eng. Technol. 2014, 1, 201–205. [Google Scholar] [CrossRef]
- Cruz-Guerrero, I.A.; Leon, R.; Campos-Delgado, D.U.; Ortega, S.; Fabelo, H.; Callico, G.M. Classification of Hyperspectral In Vivo Brain Tissue Based on Linear Unmixing. Appl. Sci. 2020, 10, 5686. [Google Scholar] [CrossRef]
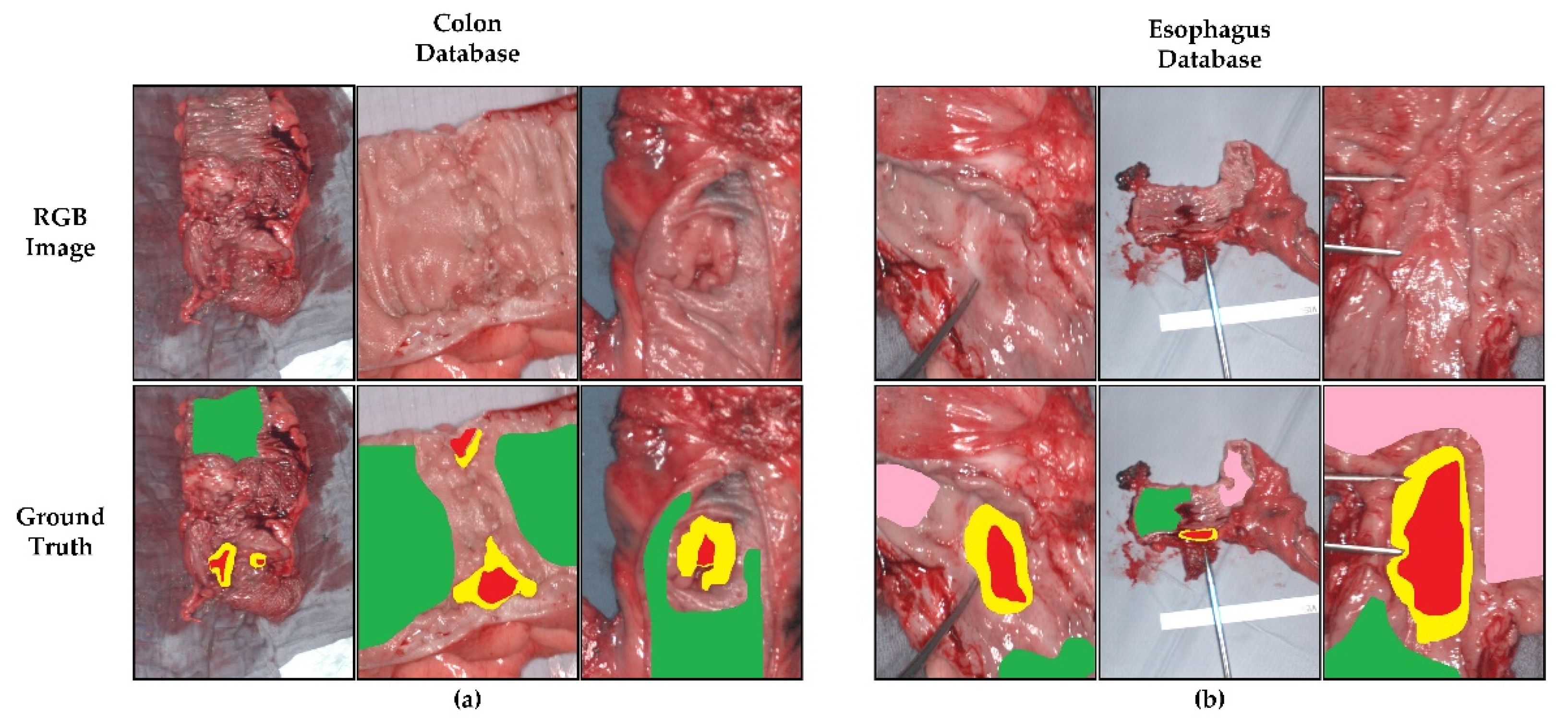







| Database | Patients | HS Cubes | #Labelled Pixels | |||
|---|---|---|---|---|---|---|
| TT | CT/ET/BT | ST/BV | BG | |||
| Colon | 12 | 12 | 75,588 | 668,927 | - | - |
| Esophagogastric | 10 | 10 | 61,414 | 369,256 | 286,554 | - |
| Brain | 16 | 26 | 11,054 | 101,706 | 38,784 | 118,132 |
| Name | Preprocessing Steps | Brief Comment |
|---|---|---|
| Calibrated | Extreme band removal + Glare removal | Calibration required for standardization of spectral signatures concerning equipment and illumination |
| Filtered | Calibrated + Gaussian smoothing filter | Application of a Gaussian filter for noise reduction in HS data |
| Min-Max | Filtered + Min-Max scaling | Reduces the range of values to [0, 1] to improve classification |
| SNV | Filtered + SNV normalization | SNV seeks to ensure that all spectra are comparable in terms of intensity. All spectra must have a mean of 0 and a standard deviation of 1. |
| MedFilter | Calibrated + Median filter spatial smoothing + SNV normalization | Homogenizes pixels with different intensities. |
| Models | Colon Threshold | Esophagogastric Threshold | Brain Threshold | |
|---|---|---|---|---|
| 3DCNN | Calibrated | 0.001 | 0.5 | 0.5 |
| Filtered | 0.0037 | 0.5 | 0.5 | |
| Min-Max | 0.0189 | 0.5 | 0.5 | |
| SNV | 0.0028 | 0.5 | 0.5 | |
| MedFilter | 0.0081 | 0.5 | 0.5 | |
| Models | F1-Score | AUC | MCC | ||
|---|---|---|---|---|---|
| TT | CT | ||||
| SVM | Calibrated | 0.36 ± 0.26 | 0.94 ± 0.12 | 0.69 ± 0.26 ** | 0.21 ± 0.27 |
| Filtered | 0.38 ± 0.28 | 0.87 ± 0.25 | 0.68 ± 0.27 ** | 0.22 ± 0.27 | |
| Min-Max | 0.31 ± 0.26 | 0.97 ± 0.03 | 0.71 ± 0.23 * | 0.19 ± 0.21 | |
| SNV | 0.32 ± 0.21 | 0.97 ± 0.03 | 0.74 ± 0.18 * | 0.19 ± 0.21 | |
| MedFilter | 0.43 ± 0.28 | 0.98 ± 0.03 | 0.84 ± 0.15* | 0.36 ± 0.27 | |
| 3DCNN | Calibrated | 0.52 ± 0.21 | 0.90 ± 0.11 | 0.94 ± 0.06 ** | 0.50 ± 0.19 |
| Filtered | 0.46 ± 0.26 | 0.88 ± 0.11 | 0.96 ± 0.03** | 0.48 ± 0.22 | |
| Min-Max | 0.38 ± 0.26 | 0.85 ± 0.12 | 0.93 ± 0.08 * | 0.37 ± 0.22 | |
| SNV 3 | 0.45 ± 0.22 | 0.87 ± 0.13 | 0.90 ± 0.08 * | 0.44 ± 0.20 | |
| MedFilter | 0.52 ± 0.23 | 0.91 ± 0.08 | 0.94 ± 0.08 * | 0.52 ± 0.20 | |
| Models | F1-Score | AUC | MCC | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TT | ET | ST | TT | ET | ST | TT | ET | ST | ||
| SVM | Calibrated | 0.56 ± 0.32 | 0.91 ± 0.09 | 0.90 ± 0.13 | 0.91 ± 0.06 | 0.92 ± 0.05 | 0.95 ± 0.11 | 0.42 ± 0.21 | 0.60 ± 0.13 | 0.71 ± 0.30 |
| Filtered | 0.53 ± 0.31 | 0.89 ± 0.10 | 0.89 ± 0.13 | 0.90 ± 0.06 ** | 0.89 ± 0.05 | 0.93 ± 0.11 | 0.40 ± 0.20 | 0.53 ± 0.11 | 0.62 ± 0.26 | |
| Min-Max | 0.58 ± 0.30 | 0.96 ± 0.05 | 0.84 ± 0.12 | 0.93 ± 0.05 | 0.94 ± 0.05 | 0.95 ± 0.10 | 0.41 ± 0.23 | 0.62 ± 0.12 | 0.64 ± 0.27 | |
| SNV | 0.56 ± 0.32 | 0.95 ± 0.06 | 0.86 ± 0.12 | 0.92 ± 0.05 | 0.93 ± 0.05 | 0.94 ± 0.11 | 0.44 ± 0.23 | 0.64 ± 0.15 | 0.68 ± 0.28 | |
| MedFilter | 0.58 ± 0.35 | 0.90 ± 0.11 | 0.88 ± 0.18 | 0.90 ± 0.08 | 0.93 ± 0.05 | 0.93 ± 0.13 | 0.41 ± 0.22 | 0.62 ± 0.19 | 0.72 ± 0.32 | |
| 3DCNN | Calibrated | 0.53 ± 0.30 | 0.89 ± 0.05 | 0.82 ± 0.34 | 0.90 ± 0.08 | 0.86 ± 0.22 | 0.95 ± 0.11 | 0.51 ± 0.29 | 0.77 ± 0.11 | 0.79 ± 0.35 |
| Filtered | 0.42 ± 0.29 | 0.84 ± 0.07 | 0.79 ± 0.33 | 0.92 ± 0.06 ** | 0.90 ± 0.09 | 0.95 ± 0.09 | 0.42 ± 0.26 | 0.70 ± 0.12 | 0.75 ± 0.31 | |
| Min-Max | 0.44 ± 0.31 | 0.78 ± 0.12 | 0.77 ± 0.33 | 0.92 ± 0.06 | 0.82 ± 0.16 | 0.95 ± 0.10 | 0.44 ± 0.27 | 0.63 ± 0.15 | 0.73 ± 0.32 | |
| SNV | 0.45 ± 0.30 | 0.84 ± 0.07 | 0.80 ± 0.33 | 0.89 ± 0.09 | 0.83 ± 0.24 | 0.94 ± 0.11 | 0.42 ± 0.28 | 0.69 ± 0.13 | 0.76 ± 0.32 | |
| MedFilter | 0.33 ± 0.15 | 0.77 ± 0.13 | 0.79 ± 0.26 | 0.86 ± 0.12 | 0.92 ± 0.08 | 0.92 ± 0.15 | 0.34 ± 0.16 | 0.60 ± 0.22 | 0.72 ± 0.32 | |
| Models | F1-Score | AUC | MCC | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BT | TT | BV | BG | BT | TT | BV | BG | BT | TT | BV | BG | ||
| SVM | Calibrated | 0.92 ± 0.09 | 0.68 ± 0.27 | 0.98 ± 0.02 | 0.79 ± 0.20 | 0.90 ± 0.11 | 0.92 ± 0.07 | 0.96 ± 0.07 | 0.98 ± 0.02 | 0.58 ± 0.28 | 0.55 ± 0.26 | 0.81 ± 0.20 | 0.72 ± 0.20 |
| Filtered | 0.92 ± 0.16 | 0.82 ± 0.29 | 0.97 ± 0.07 | 0.86 ± 0.23 | 0.95 ± 0.08 | 0.94 ± 0.05 | 0.97 ± 0.05 | 1.00 ± 0.00 | 0.75 ± 0.24 | 0.61 ± 0.29 | 0.84 ± 0.17 | 0.82 ± 0.23 | |
| Min-Max | 0.84 ± 0.14 | 0.80 ± 0.25 | 0.92 ± 0.12 | 0.88 ± 0.13 | 0.95 ± 0.05 | 0.92 ± 0.10 | 0.97 ± 0.05 | 0.98 ± 0.04 | 0.71 ± 0.17 | 0.55 ± 0.26 | 0.81 ± 0.21 | 0.79 ± 0.18 | |
| SNV | 0.87 ± 0.10 | 0.67 ± 0.32 | 0.90 ± 0.16 | 0.97 ± 0.03 | 0.97 ± 0.03 | 0.86 ± 0.20 | 0.96 ± 0.07 | 0.99 ± 0.02 | 0.79 ± 0.11 | 0.48 ± 0.39 | 0.78 ± 0.20 | 0.88 ± 0.14 | |
| MedFilter | 0.85 ± 0.17 | 0.78 ± 0.23 | 0.96 ± 0.06 | 0.83 ± 0.17 | 0.93 ± 0.07 | 0.90 ± 0.07 | 0.98 ± 0.03 | 0.98 ± 0.03 | 0.67 ± 0.22 | 0.52 ± 0.23 | 0.81 ± 0.07 | 0.80 ± 0.12 | |
| 3DCNN | Calibrated | 0.84 ± 0.19 | 0.52 ± 0.40 | 0.88 ± 0.12 | 0.97 ± 0.04 | 0.93 ± 0.13 | 0.86 ± 0.17 | 0.98 ± 0.04 | 0.99 ± 0.02 | 0.81 ± 0.21 | 0.51 ± 0.38 | 0.84 ± 0.15 | 0.96 ± 0.05 |
| Filtered | 0.82 ± 0.19 | 0.37 ± 0.40 | 0.89 ± 0.12 | 0.96 ± 0.04 | 0.93 ± 0.13 | 0.79 ± 0.17 | 0.99 ± 0.04 | 0.98 ± 0.02 | 0.79 ± 0.21 | 0.37 ± 0.32 | 0.86 ± 0.15 | 0.94 ± 0.05 | |
| Min-Max | 0.86 ± 0.20 | 0.44 ± 0.44 | 0.87 ± 0.1 | 0.98 ± 0.01 | 0.97 ± 0.04 | 0.87 ± 0.14 | 0.97 ± 0.03 | 0.98 ± 0.03 | 0.83 ± 0.23 | 0.54 ± 0.40 | 0.83 ± 0.23 | 0.96 ± 0.02 | |
| SNV | 0.84 ± 0.19 | 0.44 ± 0.42 | 0.87 ± 0.14 | 0.97 ± 0.02 | 0.97 ± 0.05 | 0.90 ± 0.10 | 0.95 ± 0.09 | 0.96 ± 0.01 | 0.80 ± 0.22 | 0.45 ± 0.41 | 0.82 ± 0.18 | 0.96 ± 0.03 | |
| MedFilter | 0.75 ± 0.30 | 0.41 ± 0.33 | 0.86 ± 0.10 | 0.92 ± 0.09 | 0.78 ± 0.17 | 0.83 ± 0.34 | 0.98 ± 0.03 | 0.99 ± 0.01 | 0.68 ± 0.39 | 0.40 ± 0.36 | 0.82 ± 0.12 | 0.91 ± 0.09 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinez-Vega, B.; Tkachenko, M.; Matkabi, M.; Ortega, S.; Fabelo, H.; Balea-Fernandez, F.; La Salvia, M.; Torti, E.; Leporati, F.; Callico, G.M.; et al. Evaluation of Preprocessing Methods on Independent Medical Hyperspectral Databases to Improve Analysis. Sensors 2022, 22, 8917. https://doi.org/10.3390/s22228917
Martinez-Vega B, Tkachenko M, Matkabi M, Ortega S, Fabelo H, Balea-Fernandez F, La Salvia M, Torti E, Leporati F, Callico GM, et al. Evaluation of Preprocessing Methods on Independent Medical Hyperspectral Databases to Improve Analysis. Sensors. 2022; 22(22):8917. https://doi.org/10.3390/s22228917
Chicago/Turabian StyleMartinez-Vega, Beatriz, Mariia Tkachenko, Marianne Matkabi, Samuel Ortega, Himar Fabelo, Francisco Balea-Fernandez, Marco La Salvia, Emanuele Torti, Francesco Leporati, Gustavo M. Callico, and et al. 2022. "Evaluation of Preprocessing Methods on Independent Medical Hyperspectral Databases to Improve Analysis" Sensors 22, no. 22: 8917. https://doi.org/10.3390/s22228917
APA StyleMartinez-Vega, B., Tkachenko, M., Matkabi, M., Ortega, S., Fabelo, H., Balea-Fernandez, F., La Salvia, M., Torti, E., Leporati, F., Callico, G. M., & Chalopin, C. (2022). Evaluation of Preprocessing Methods on Independent Medical Hyperspectral Databases to Improve Analysis. Sensors, 22(22), 8917. https://doi.org/10.3390/s22228917












